

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.11 - phase: 0
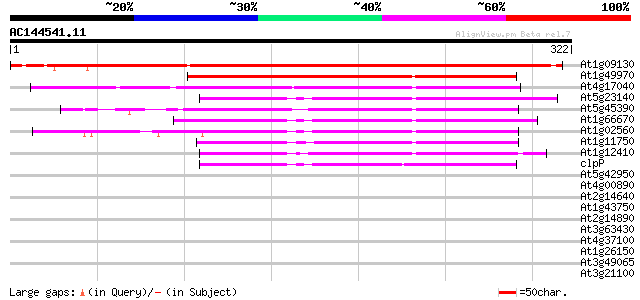
(322 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g09130 ClpP protease complex subunit ClpR3 409 e-115
At1g49970 ClpP protease complex subunit ClpR1 166 1e-41
At4g17040 ClpP protease complex subunit ClpR4 163 1e-40
At5g23140 ATP-dependent protease proteolytic subunit ClpP-like p... 122 2e-28
At5g45390 ATP-dependent Clp protease-like protein 114 5e-26
At1g66670 nClpP3 protein 110 1e-24
At1g02560 ATP-dependent clp protease proteolytic subunit (nClpP1) 109 2e-24
At1g11750 putative ATP-dependent Clp protease proteolytic subunit 102 2e-22
At1g12410 ClpP protease complex subunit ClpR2 88 5e-18
clpP -chloroplast genome- ATP-dependent protease subunit 77 1e-14
At5g42950 putative protein 32 0.61
At4g00890 30 1.3
At2g14640 putative retroelement pol polyprotein 30 1.3
At1g43750 hypothetical protein 30 1.3
At2g14890 arabinogalactan-protein AGP9 30 2.3
At3g63430 unknown protein 28 5.1
At4g37100 putative protein 28 6.7
At1g26150 Pto kinase interactor, putative 28 6.7
At3g49065 unknown protein 28 8.7
At3g21100 unknown protein 28 8.7
>At1g09130 ClpP protease complex subunit ClpR3
Length = 330
Score = 409 bits (1052), Expect = e-115
Identities = 221/333 (66%), Positives = 264/333 (78%), Gaps = 22/333 (6%)
Query: 1 MATCFRVPISMPTSMPASSTRFTP-----------TRLRTLR-TVNAISKSKIPF---NP 45
MA+C + SM + +P SS+ F+P L+T R A + +KIP NP
Sbjct: 1 MASCLQA--SMNSLLPRSSS-FSPHPPLSSNSSGRRNLKTFRYAFRAKASAKIPMPPINP 57
Query: 46 NDPFLSKLASVAESSPETLFNPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPK 105
DPFLS LAS+A +SPE L N + D P+LDIFDSPQLM++PAQ+ERS +Y++ R P+
Sbjct: 58 KDPFLSTLASIAANSPEKLLNRPVNADVPPYLDIFDSPQLMSSPAQVERSVAYNEHR-PR 116
Query: 106 RPPPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADG 165
PPPDLPS+LL+GRIVYIGMPLVPAVTELV+AELM+LQW+ PKEPIYIYINSTGTTR DG
Sbjct: 117 TPPPDLPSMLLDGRIVYIGMPLVPAVTELVVAELMYLQWLDPKEPIYIYINSTGTTRDDG 176
Query: 166 ETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQP 225
ETV MESEGFAIYD++MQ+K E++TV +GAA+GQACLLLSAGTKG+R+M PHAKAMIQQP
Sbjct: 177 ETVGMESEGFAIYDSLMQLKNEVHTVCVGAAIGQACLLLSAGTKGKRFMMPHAKAMIQQP 236
Query: 226 RVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEF 285
RVPSSGL PASDVLI AKEV+ NRD LV+LL+KHT NS ETV+NVM+R YYMDA AKEF
Sbjct: 237 RVPSSGLMPASDVLIRAKEVITNRDILVELLSKHTGNSVETVANVMRRPYYMDAPKAKEF 296
Query: 286 GVIDKILWRGQEKIMAD-VPSRDDRENGTAGAK 317
GVID+ILWRGQEKI+AD VPS + +N AG K
Sbjct: 297 GVIDRILWRGQEKIIADVVPSEEFDKN--AGIK 327
>At1g49970 ClpP protease complex subunit ClpR1
Length = 387
Score = 166 bits (421), Expect = 1e-41
Identities = 87/189 (46%), Positives = 123/189 (65%), Gaps = 1/189 (0%)
Query: 103 RPKRPPPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTR 162
RP+ PPDLPSLLL+ RI Y+GMP+VPAVTEL++A+ M+L + P +PIY+YINS GT
Sbjct: 164 RPRTAPPDLPSLLLDARICYLGMPIVPAVTELLVAQFMWLDYDNPTKPIYLYINSPGTQN 223
Query: 163 ADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMI 222
ETV E+E +AI D + K+++ T+ G A GQA +LLS G KG R + PH+ +
Sbjct: 224 EKMETVGSETEAYAIADTISYCKSDVYTINCGMAFGQAAMLLSLGKKGYRAVQPHSSTKL 283
Query: 223 QQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLA 282
P+V S A D+ I AKE+ N + ++LLAK T S+E ++ +KR Y+ A A
Sbjct: 284 YLPKVNRSS-GAAIDMWIKAKELDANTEYYIELLAKGTGKSKEQINEDIKRPKYLQAQAA 342
Query: 283 KEFGVIDKI 291
++G+ DKI
Sbjct: 343 IDYGIADKI 351
>At4g17040 ClpP protease complex subunit ClpR4
Length = 305
Score = 163 bits (413), Expect = 1e-40
Identities = 106/282 (37%), Positives = 158/282 (55%), Gaps = 9/282 (3%)
Query: 13 TSMPASSTRFTPTRLRTLRTVNAISKSK-IPFNPNDPFLSKLASVAESSPETLFNPSSSP 71
T++ A ++ P+ R LR+ S S + + ++ FLS + SS L
Sbjct: 12 TTLRARTSAIIPSSTRNLRSKPRFSSSSSLRASLSNGFLSPYTGGSISSD--LCGAKLRA 69
Query: 72 DNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLPSLLLNGRIVYIGMPLVPAV 131
++L L+ S + +S+ ++PPPDL S L RIVY+GM LVP+V
Sbjct: 70 ESLNPLNFSSSKPKRGVVTMV---IPFSKGSAHEQPPPDLASYLFKNRIVYLGMSLVPSV 126
Query: 132 TELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTV 191
TEL++AE ++LQ+ ++PIY+YINSTGTT+ +GE + ++E FAIYD M +K I T+
Sbjct: 127 TELILAEFLYLQYEDEEKPIYLYINSTGTTK-NGEKLGYDTEAFAIYDVMGYVKPPIFTL 185
Query: 192 ALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDT 251
+G A G+A LLL+AG KG R P + MI+QP G A+DV I KE+ +
Sbjct: 186 CVGNAWGEAALLLTAGAKGNRSALPSSTIMIKQPIARFQG--QATDVEIARKEIKHIKTE 243
Query: 252 LVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKILW 293
+VKL +KH S E + MKR Y A E+G+IDK+++
Sbjct: 244 MVKLYSKHIGKSPEQIEADMKRPKYFSPTEAVEYGIIDKVVY 285
>At5g23140 ATP-dependent protease proteolytic subunit ClpP-like
protein
Length = 241
Score = 122 bits (307), Expect = 2e-28
Identities = 72/205 (35%), Positives = 115/205 (55%), Gaps = 11/205 (5%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RI+ I P+ + +V+A+L++L+ P +PI++Y+NS G G A
Sbjct: 48 DIFSRLLKERIICINGPINDDTSHVVVAQLLYLESENPSKPIHMYLNSPG-----GHVTA 102
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G AIYD M +++ I+T+ LG A A LLL+AG KG+R P+A MI QP
Sbjct: 103 ----GLAIYDTMQYIRSPISTICLGQAASMASLLLAAGAKGQRRSLPNATVMIHQPSGGY 158
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
SG A D+ IH K+++ D L +L KHT + V+N M R ++M AK FG+ID
Sbjct: 159 SG--QAKDITIHTKQIVRVWDALNELYVKHTGQPLDVVANNMDRDHFMTPEEAKAFGIID 216
Query: 290 KILWRGQEKIMADVPSRDDRENGTA 314
+++ +++ D + ++ ++
Sbjct: 217 EVIDERPLELVKDAVGNESKDKSSS 241
>At5g45390 ATP-dependent Clp protease-like protein
Length = 292
Score = 114 bits (286), Expect = 5e-26
Identities = 84/266 (31%), Positives = 136/266 (50%), Gaps = 37/266 (13%)
Query: 30 LRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNP---SSSPDNLPFLDIFDSPQLM 86
L + ++ S S P PN+ +L P L +P ++SP L F +
Sbjct: 19 LNSSSSASSSSFP-KPNNLYLK---------PTKLISPPLRTTSPSPLRFAN-------- 60
Query: 87 ATPAQLERSASYSQQRRPKRPPPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMA 146
A +E S + Q+ + D+ LLL RIV++G + V + ++++L+ L
Sbjct: 61 ---ASIEMSQT--QESAIRGAESDVMGLLLRERIVFLGSSIDDFVADAIMSQLLLLDAKD 115
Query: 147 PKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSA 206
PK+ I ++INS G + S AIYD + ++ +++T+ALG A A ++L A
Sbjct: 116 PKKDIKLFINSPGGSL---------SATMAIYDVVQLVRADVSTIALGIAASTASIILGA 166
Query: 207 GTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEET 266
GTKG+R+ P+ + MI QP +SG A DV I AKEVM N++ + ++A T S E
Sbjct: 167 GTKGKRFAMPNTRIMIHQPLGGASG--QAIDVEIQAKEVMHNKNNVTSIIAGCTSRSFEQ 224
Query: 267 VSNVMKRSYYMDALLAKEFGVIDKIL 292
V + R YM + A E+G+ID ++
Sbjct: 225 VLKDIDRDRYMSPIEAVEYGLIDGVI 250
>At1g66670 nClpP3 protein
Length = 309
Score = 110 bits (274), Expect = 1e-24
Identities = 67/209 (32%), Positives = 113/209 (54%), Gaps = 11/209 (5%)
Query: 95 SASYSQQRRPKRPPPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIY 154
S + S R P D ++LL RIV++G + +LVI++L+ L + I ++
Sbjct: 70 SVAQSPSRLPSFEELDTTNMLLRQRIVFLGSQVDDMTADLVISQLLLLDAEDSERDITLF 129
Query: 155 INSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYM 214
INS G G A G IYDAM Q K +++TV LG A LL++G+KG+RY
Sbjct: 130 INSPG-----GSITA----GMGIYDAMKQCKADVSTVCLGLAASMGAFLLASGSKGKRYC 180
Query: 215 TPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRS 274
P++K MI QP + G A+++ I +E+M ++ L K+ ++ T E + + R
Sbjct: 181 MPNSKVMIHQPLGTAGG--KATEMSIRIREMMYHKIKLNKIFSRITGKPESEIESDTDRD 238
Query: 275 YYMDALLAKEFGVIDKILWRGQEKIMADV 303
+++ AKE+G+ID ++ G+ ++A +
Sbjct: 239 NFLNPWEAKEYGLIDAVIDDGKPGLIAPI 267
>At1g02560 ATP-dependent clp protease proteolytic subunit (nClpP1)
Length = 298
Score = 109 bits (273), Expect = 2e-24
Identities = 84/296 (28%), Positives = 143/296 (47%), Gaps = 34/296 (11%)
Query: 14 SMPASSTRFTPTRLRTLRTVNAISKSKI--PFNP-----NDPFLSKLASVAESSPETLFN 66
S ASS RFT + ++ K+ PF P + +S + S+P+ +++
Sbjct: 7 STSASSLRFTAGFVSASPNGSSFDSPKLSLPFEPLRSRKTNKLVSDRKNWKNSTPKAVYS 66
Query: 67 PSSSPDNLPFLDIFDSPQ---LMATPAQLERSASYSQQRRPKRPPP-------DLPSLLL 116
+ +P SPQ + Q+ S + + + PPP + S L
Sbjct: 67 GNLWTPEIP------SPQGVWSIRDDLQVPSSPYFPAYAQGQGPPPMVQERFQSIISQLF 120
Query: 117 NGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVAMESEGFA 176
RI+ G + + +++A+L++L + P + I +Y+NS G G A G A
Sbjct: 121 QYRIIRCGGAVDDDMANIIVAQLLYLDAVDPTKDIVMYVNSPG-----GSVTA----GMA 171
Query: 177 IYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPSSGLRPAS 236
I+D M ++ +++TV +G A LLSAGTKG+RY P+++ MI QP + G +
Sbjct: 172 IFDTMRHIRPDVSTVCVGLAASMGAFLLSAGTKGKRYSLPNSRIMIHQPLGGAQG--GQT 229
Query: 237 DVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKIL 292
D+ I A E++ ++ L LA HT S E ++ R ++M A AKE+G+ID ++
Sbjct: 230 DIDIQANEMLHHKANLNGYLAYHTGQSLEKINQDTDRDFFMSAKEAKEYGLIDGVI 285
>At1g11750 putative ATP-dependent Clp protease proteolytic subunit
Length = 271
Score = 102 bits (255), Expect = 2e-22
Identities = 65/185 (35%), Positives = 94/185 (50%), Gaps = 11/185 (5%)
Query: 108 PPDLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGET 167
P DL S+L RI++IG P+ V + VI++L+ L + K I +Y+N G G T
Sbjct: 94 PLDLSSVLFRNRIIFIGQPINAQVAQRVISQLVTLASIDDKSDILMYLNCPG-----GST 148
Query: 168 VAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRV 227
++ AIYD M +K ++ TVA G A Q LLL+ G KG RY P+ + MI QP+
Sbjct: 149 YSV----LAIYDCMSWIKPKVGTVAFGVAASQGALLLAGGEKGMRYAMPNTRVMIHQPQT 204
Query: 228 PSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGV 287
G DV E + R + ++ A T E V +R ++ A A EFG+
Sbjct: 205 GCGG--HVEDVRRQVNEAIEARQKIDRMYAAFTGQPLEKVQQYTERDRFLSASEALEFGL 262
Query: 288 IDKIL 292
ID +L
Sbjct: 263 IDGLL 267
>At1g12410 ClpP protease complex subunit ClpR2
Length = 279
Score = 88.2 bits (217), Expect = 5e-18
Identities = 61/199 (30%), Positives = 99/199 (49%), Gaps = 13/199 (6%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ + L R+++IG + + ++A +++L + IY+Y+N G G+
Sbjct: 89 DIWNALYRERVIFIGQNIDEEFSNQILATMLYLDTLDDSRRIYMYLNGPG-----GDL-- 141
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
+ AIYD M +K+ + T +G A A LL+AG KG R+ P ++ +Q P +
Sbjct: 142 --TPSLAIYDTMKSLKSPVGTHCVGLAYNLAGFLLAAGEKGHRFAMPLSRIALQSPAGAA 199
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G A D+ AKE+ RD L LAK+T E V + R +A A E+G+ID
Sbjct: 200 RG--QADDIQNEAKELSRIRDYLFNELAKNTGQPAERVFKDLSRVKRFNAEEAIEYGLID 257
Query: 290 KILWRGQEKIMADVPSRDD 308
KI+ +I D P +D+
Sbjct: 258 KIV--RPPRIKEDAPRQDE 274
>clpP -chloroplast genome- ATP-dependent protease subunit
Length = 196
Score = 77.4 bits (189), Expect = 1e-14
Identities = 46/182 (25%), Positives = 92/182 (50%), Gaps = 10/182 (5%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ + L R+ ++G + ++ +I+ +++L + +Y++INS G G ++
Sbjct: 22 DIYNRLYRERLFFLGQEVDTEISNQLISLMIYLSIEKDTKDLYLFINSPG-----GWVIS 76
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G AIYD M ++ ++ T+ +G A A +L G +R PHA+ MI QP S
Sbjct: 77 ----GMAIYDTMQFVRPDVQTICMGLAASIASFILVGGAITKRIAFPHARVMIHQP-ASS 131
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
+ ++ A+E++ R+T+ ++ + T +S M+R +M A A+ G++D
Sbjct: 132 FYEAQTGEFILEAEELLKLRETITRVYVQRTGKPIWVISEDMERDVFMSATEAQAHGIVD 191
Query: 290 KI 291
+
Sbjct: 192 LV 193
>At5g42950 putative protein
Length = 1714
Score = 31.6 bits (70), Expect = 0.61
Identities = 18/57 (31%), Positives = 28/57 (48%), Gaps = 2/57 (3%)
Query: 256 LAKHTENSEETVSNVMKRSYYMDALLAKEFGVIDKILWRGQEKIMADVPSRDDRENG 312
+ + T N EET+ N+ K+ + +LL E G D+ WR +E+ D NG
Sbjct: 68 VVRTTGNGEETLDNLKKKDVFRPSLLDAESGRRDR--WRDEERDTLSSVRNDRWRNG 122
>At4g00890
Length = 431
Score = 30.4 bits (67), Expect = 1.3
Identities = 32/136 (23%), Positives = 48/136 (34%), Gaps = 22/136 (16%)
Query: 9 ISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSP------E 62
I P P ST L ++ ++ K P +P+ + S ++ SP E
Sbjct: 94 IQSPPKSPPESTESQQQFLASVPLRPLTTEPKTPLSPSSTLKATEESQSQPSPPLESIVE 153
Query: 63 TLFNPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLPSLLLNGRIVY 122
T F PS + D QL P Q ++ PP PS++LN +
Sbjct: 154 TWFRPSPPTST----ETGDETQLPIPPPQEAKT------------PPSSPSMMLNATEEF 197
Query: 123 IGMPLVPAVTELVIAE 138
P P + I E
Sbjct: 198 ESQPKPPLLPSKSIDE 213
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 30.4 bits (67), Expect = 1.3
Identities = 34/120 (28%), Positives = 47/120 (38%), Gaps = 20/120 (16%)
Query: 23 TPTRLRTLRTVN---AISKSKIPFNPNDPFLSKLASVAESSPETLFNPSSSPDNLPFL-- 77
TPT+L + R + A+ K P N P+ E S + PSSSP P L
Sbjct: 493 TPTQLPSERPIVHRIALKKGTDPVNVR-PYRYAFYQKDEMSRAGIIRPSSSPFLSPVLLE 551
Query: 78 --------DIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLPSLLL---NGRIVYIGMP 126
D+ D +L+ +A Y Q R PD+P + NG Y+ MP
Sbjct: 552 RFPIPTVDDMLDELNGAVYFTKLDLTAGYQQVRMHS---PDIPKIAFQTHNGHYEYLVMP 608
>At1g43750 hypothetical protein
Length = 625
Score = 30.4 bits (67), Expect = 1.3
Identities = 21/59 (35%), Positives = 32/59 (53%), Gaps = 3/59 (5%)
Query: 60 SPETLFNPSSSPDNLPFLDI-FDSPQLMATPAQLERSASYSQQRRPKRPPPDLPSLLLN 117
+P F S+S P D FD P+ A+ +L RS+++S +R RPP + L+LN
Sbjct: 521 TPRPEFRRSNSTSVAPRRDARFDPPR--ASNYELGRSSTFSDRRFNSRPPGPVNQLVLN 577
>At2g14890 arabinogalactan-protein AGP9
Length = 191
Score = 29.6 bits (65), Expect = 2.3
Identities = 35/127 (27%), Positives = 50/127 (38%), Gaps = 19/127 (14%)
Query: 6 RVPISMPTSMPASSTRFTPTRLRTLRTVNA----------ISKSKIPFNPNDPFLS-KLA 54
+ P S PT+ PA T TP T V+A ++ + P NP P S A
Sbjct: 21 QAPTSPPTATPAPPTPTTPPPAATPPPVSAPPPVTTSPPPVTTAPPPANPPPPVSSPPPA 80
Query: 55 SVAESSPETLFNP-----SSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPP- 108
S ++P + +P S P P + L + PAQ+ A ++ P P
Sbjct: 81 SPPPATPPPVASPPPPVASPPPATPPPVATPPPAPLASPPAQVPAPAPTTKPDSPSPSPS 140
Query: 109 --PDLPS 113
P LPS
Sbjct: 141 SSPPLPS 147
>At3g63430 unknown protein
Length = 540
Score = 28.5 bits (62), Expect = 5.1
Identities = 21/64 (32%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Query: 59 SSPETLFNPSSSPDNLPFLDIFDSPQLMA-TPAQLERSASYSQQRRPKRPPPDLPSLLLN 117
SSP +S N ++ +P+L + PAQ S+S S RP R + P L L+
Sbjct: 36 SSPSAESESASGRTNQQ--SVYSTPELRSPAPAQQRSSSSSSSSSRPWRFSKEAPRLSLD 93
Query: 118 GRIV 121
R V
Sbjct: 94 SRAV 97
>At4g37100 putative protein
Length = 896
Score = 28.1 bits (61), Expect = 6.7
Identities = 36/135 (26%), Positives = 54/135 (39%), Gaps = 14/135 (10%)
Query: 13 TSMPASSTRFTPTRLR----TLRTVNAISKSKIPFNPNDPFL-SKLASVAESSPETLFNP 67
T MPA S +T ++R T + IS + F ++ SV E +F+
Sbjct: 439 TQMPAFSGAYTSAQVRDVFETELLEDNISSDRDGTTSTTIFEETESVSVGELMKSPVFSE 498
Query: 68 SSSPDNLPFLDIFDSP------QLMATPAQ---LERSASYSQQRRPKRPPPDLPSLLLNG 118
S DN ++D+ SP +A+P L Q++ PK P S L +G
Sbjct: 499 DESSDNSFWIDLGQSPLGSDQHNKIASPLPPIWLTNKRKQKQRQSPKPIPKSYSSPLYDG 558
Query: 119 RIVYIGMPLVPAVTE 133
V V +VTE
Sbjct: 559 NDVLSFDAAVMSVTE 573
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 28.1 bits (61), Expect = 6.7
Identities = 23/106 (21%), Positives = 38/106 (35%), Gaps = 1/106 (0%)
Query: 8 PISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAE-SSPETLFN 66
P ++P S P + P N +S +P P ++ +SP N
Sbjct: 87 PTTIPVSPPPEPSPPPPLPTEAPPPANPVSSPPPESSPPPPPPTEAPPTTPITSPSPPTN 146
Query: 67 PSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLP 112
P P++ P L D P P +L + + P P ++P
Sbjct: 147 PPPPPESPPSLPAPDPPSNPLPPPKLVPPSHSPPRHLPSPPASEIP 192
Score = 28.1 bits (61), Expect = 6.7
Identities = 30/108 (27%), Positives = 42/108 (38%), Gaps = 10/108 (9%)
Query: 6 RVPISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLF 65
R +S+ S+ + P + NA S ++ P N N P + + + PET
Sbjct: 9 REEVSLSPSLASPPLMALPPPQPSFPGDNATSPTREPTNGNPPETTNTPAQSSPPPET-- 66
Query: 66 NPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPPPDLPS 113
P SSP P SP L P + S P PPP LP+
Sbjct: 67 -PLSSPPPEPSP---PSPSLTGPP---PTTIPVSPPPEPS-PPPPLPT 106
Score = 27.7 bits (60), Expect = 8.7
Identities = 31/116 (26%), Positives = 40/116 (33%), Gaps = 11/116 (9%)
Query: 8 PISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPET---- 63
P + P S P + P I+ P NP P S + A P
Sbjct: 110 PPANPVSSPPPESSPPPPPPTEAPPTTPITSPSPPTNPPPPPESPPSLPAPDPPSNPLPP 169
Query: 64 --LFNPSSSPD----NLPFLDIFDSPQLMATPAQLER-SASYSQQRRPKRPPPDLP 112
L PS SP + P +I P+ + +P ER S S P PPP P
Sbjct: 170 PKLVPPSHSPPRHLPSPPASEIPPPPRHLPSPPASERPSTPPSDSEHPSPPPPGHP 225
>At3g49065 unknown protein
Length = 805
Score = 27.7 bits (60), Expect = 8.7
Identities = 19/57 (33%), Positives = 28/57 (48%), Gaps = 4/57 (7%)
Query: 52 KLASVAESSPETLFNPSSSPDNLPFLDIFDSPQLMATPAQLERSASYSQQRRPKRPP 108
KLA+VA + NP + PD L D + P+ ++SY+ Q P+RPP
Sbjct: 684 KLANVAIRCCKK--NPMNRPDLAVVLRFIDRMKAPEVPSS--ETSSYANQNVPRRPP 736
>At3g21100 unknown protein
Length = 528
Score = 27.7 bits (60), Expect = 8.7
Identities = 22/78 (28%), Positives = 37/78 (47%), Gaps = 15/78 (19%)
Query: 65 FNPSSSPDNLPFLDIFDS---PQLM----------ATPAQLERSASYSQQR--RPKRPPP 109
F+P SSP + D+FDS P++ A A L+++ + ++R + P
Sbjct: 415 FSPGSSPSGMDSRDLFDSHLAPRMFSNTQEMMRRKAEQADLQQAIEFQRRRFLSLQLPDM 474
Query: 110 DLPSLLLNGRIVYIGMPL 127
D S L + R + IG P+
Sbjct: 475 DSESFLHHQRSLSIGSPV 492
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,983,369
Number of Sequences: 26719
Number of extensions: 292256
Number of successful extensions: 964
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 930
Number of HSP's gapped (non-prelim): 26
length of query: 322
length of database: 11,318,596
effective HSP length: 99
effective length of query: 223
effective length of database: 8,673,415
effective search space: 1934171545
effective search space used: 1934171545
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144541.11


