

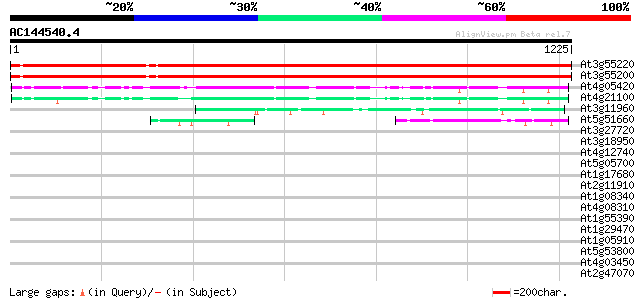
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144540.4 - phase: 0
(1225 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g55220 spliceosomal - like protein 2082 0.0
At3g55200 spliceosomal - like protein 2082 0.0
At4g05420 UV-damaged DNA binding factor - like protein 223 4e-58
At4g21100 UV-damaged DNA-binding protein- like 210 4e-54
At3g11960 hypothetical protein 117 5e-26
At5g51660 cleavage and polyadenylation specificity factor subunit 67 6e-11
At3g27720 hypothetical protein 36 0.15
At3g18950 unknown protein 35 0.34
At4g12740 adenine DNA glycosylase like protein 33 1.0
At5g05700 arginine-tRNA-protein transferase 1 homolog 32 1.7
At1g17680 unknown protein 32 1.7
At2g11910 unknown protein 31 3.8
At1g08340 unknown protein 31 3.8
At4g08310 unknown protein (At4g08310) 31 5.0
At1g55390 hypothetical protein 31 5.0
At1g29470 unknown protein 30 6.5
At1g05910 unknown protein 30 6.5
At5g53800 unknown protein 30 8.5
At4g03450 hypothetical protein 30 8.5
At2g47070 squamosa promoter binding protein-like 1 30 8.5
>At3g55220 spliceosomal - like protein
Length = 1214
Score = 2082 bits (5394), Expect = 0.0
Identities = 1022/1225 (83%), Positives = 1126/1225 (91%), Gaps = 11/1225 (0%)
Query: 1 MYLYNLTLQRPTGIVCAINGNFSGSDDGITQEIVVARGKVLELLRPDKFGRIQSILSVQV 60
MYLY+LTLQ+ TGIVCAINGNFSG G TQEI VARGK+L+LLRPD+ G+IQ+I SV+V
Sbjct: 1 MYLYSLTLQQATGIVCAINGNFSG---GKTQEIAVARGKILDLLRPDENGKIQTIHSVEV 57
Query: 61 FGTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQKNVFDKIHQETFGKSGCRRIVPG 120
FG IRSL+QFRLTGAQKD+IVVGSDSGRIVIL+YNK+KNVFDK+HQETFGKSGCRRIVPG
Sbjct: 58 FGAIRSLAQFRLTGAQKDYIVVGSDSGRIVILEYNKEKNVFDKVHQETFGKSGCRRIVPG 117
Query: 121 QYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSHTIVFSICAVDCGFE 180
QY+A+DPKGRAVMI ACEK+KLVYVLNRD+ ARLTISSPLEA+KSHTI +S+C VDCGF+
Sbjct: 118 QYVAVDPKGRAVMIGACEKQKLVYVLNRDTTARLTISSPLEAHKSHTICYSLCGVDCGFD 177
Query: 181 NPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDLGLNHVSRKWSDQVDNGANMLVTV 240
NPIFAAIELD S+ADQD TG AAS+AQKHL FYELDLGLNHVSRKWS+ VDNGANMLVTV
Sbjct: 178 NPIFAAIELDYSEADQDPTGQAASEAQKHLTFYELDLGLNHVSRKWSNPVDNGANMLVTV 237
Query: 241 PGGADGPSGVLVCAENFVIYKNQGHPDVRAVIPRRADLPAERGVLIVSAAMHKTKNLKPE 300
PGGADGPSGVLVCAENFVIY NQGHPDVRAVIPRR DLPAERGVL+VSAA+HK K +
Sbjct: 238 PGGADGPSGVLVCAENFVIYMNQGHPDVRAVIPRRTDLPAERGVLVVSAAVHKQKTM--- 294
Query: 301 EFKLFFLLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAASEFG 360
FFL+QTEYGD+FKVTL D GD VSEL +KYFDTI VA SICVLK GFLF+ASEFG
Sbjct: 295 ---FFFLIQTEYGDVFKVTL-DHNGDHVSELKVKYFDTIPVASSICVLKLGFLFSASEFG 350
Query: 361 NHALYQFKGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLVRIDQVESLMPVMDMK 420
NH LYQF+ IG++ DV +SS++LMETEEGFQPVFFQPRRLKNLVRIDQVESLMP+MDMK
Sbjct: 351 NHGLYQFQAIGEEP-DVESSSSNLMETEEGFQPVFFQPRRLKNLVRIDQVESLMPLMDMK 409
Query: 421 VSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGIPSAVWTVKKNVMDEFD 480
V N+FEEETPQIF+LCGRGPRSSLRI+R GLA++EMAVS+LPG PSAVWTVKKNV DEFD
Sbjct: 410 VLNIFEEETPQIFSLCGRGPRSSLRILRPGLAITEMAVSQLPGQPSAVWTVKKNVSDEFD 469
Query: 481 AYIVVSFTNATLVLSIGETADEVSDSGFLDTAPSLAVSLIGDDSLMQVHPNGIRHIREDG 540
AYIVVSFTNATLVLSIGE +EV+DSGFLDT PSLAVSLIGDDSLMQVHPNGIRHIREDG
Sbjct: 470 AYIVVSFTNATLVLSIGEQVEEVNDSGFLDTTPSLAVSLIGDDSLMQVHPNGIRHIREDG 529
Query: 541 RTNEWQTSGKRTIAKVGSNRLQVVIALNGGELIYFEVDVTGQLMEVERHEMSGDVACLDI 600
R NEW+T GKR+I KVG NRLQVVIAL+GGELIYFE D+TGQLMEVE+HEMSGDVACLDI
Sbjct: 530 RINEWRTPGKRSIVKVGYNRLQVVIALSGGELIYFEADMTGQLMEVEKHEMSGDVACLDI 589
Query: 601 APVPKGRLRSRFLAVGSYDKTIRILSLDPDDCMQTLGIQSLSSAPESLLFLEVQASVGGE 660
APVP+GR RSRFLAVGSYD T+RILSLDPDDC+Q L +QS+SSAPESLLFLEVQAS+GG+
Sbjct: 590 APVPEGRKRSRFLAVGSYDNTVRILSLDPDDCLQILSVQSVSSAPESLLFLEVQASIGGD 649
Query: 661 DGADHPASLFLNAGLQNGVLSRTVVDMVTGLLSDTRSRFLGLKAPKLFPIIVRGKRAMLC 720
DGADHPA+LFLN+GLQNGVL RTVVDMVTG LSD+RSRFLGLK PKLF I VRG+ AMLC
Sbjct: 650 DGADHPANLFLNSGLQNGVLFRTVVDMVTGQLSDSRSRFLGLKPPKLFSISVRGRSAMLC 709
Query: 721 LSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSDQCFEGVVSVASEALRIFTVERLGETF 780
LSSRPWLGYIH+GHF LTPLSYETLE+AA FSSDQC EGVVSVA +ALRIF ++RLGETF
Sbjct: 710 LSSRPWLGYIHRGHFHLTPLSYETLEFAAPFSSDQCAEGVVSVAGDALRIFMIDRLGETF 769
Query: 781 NQNVIPLRYTPRKFVLQPKRKLLVVIESDQGALTAEEREAARKECFEAAHAGENKTGSED 840
N+ V+PLRYTPRKFVL PKRKLLV+IESDQGA TAEEREAARKECFEA GEN G+ D
Sbjct: 770 NETVVPLRYTPRKFVLHPKRKLLVIIESDQGAFTAEEREAARKECFEAGGVGENGNGNAD 829
Query: 841 QMENGGEDEDNDDSLSDEHYGYPKSESDKWVSCIRVLDPRTGNTTCLLELQENEAAFSIC 900
QMENG +DED +D LSDE YGYPK+ES+KWVSCIRVLDP+T TTCLLELQ+NEAA+S+C
Sbjct: 830 QMENGADDEDKEDPLSDEQYGYPKAESEKWVSCIRVLDPKTATTTCLLELQDNEAAYSVC 889
Query: 901 TVNFHDKEYGTLLAVGTAKGLQFTPKRSLTAGFIHIYRFLDDGRSLELLHKTQVEGVPLA 960
TVNFHDKEYGTLLAVGT KG+QF PK++L AGFIHIYRF++DG+SLELLHKTQVEGVPLA
Sbjct: 890 TVNFHDKEYGTLLAVGTVKGMQFWPKKNLVAGFIHIYRFVEDGKSLELLHKTQVEGVPLA 949
Query: 961 LCQFQGRLLAGIGPVLRLYDLGKRRLLRKCENKSFPSSIVSIHAYRDRIYVGGIQESFHY 1020
LCQFQGRLLAGIGPVLRLYDLGK+RLLRKCENK FP++I+SI YRDRIYVG IQESFHY
Sbjct: 950 LCQFQGRLLAGIGPVLRLYDLGKKRLLRKCENKLFPNTIISIQTYRDRIYVGDIQESFHY 1009
Query: 1021 CKYRRDENQLYIFADDSVPRWLTSSYHIDFDTMAGADKFGNIFFARLPQDVSDEIEEDPT 1080
CKYRRDENQLYIFADD VPRWLT+S+H+DFDTMAGADKFGN++F RLPQD+S+EIEEDPT
Sbjct: 1010 CKYRRDENQLYIFADDCVPRWLTASHHVDFDTMAGADKFGNVYFVRLPQDLSEEIEEDPT 1069
Query: 1081 GGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLVPGGGECIVYGTVMGSVGALHA 1140
GGKIKWEQGKLNGAPNKV+EIVQFHVGDV+T LQKAS++PGG E I+YGTVMGS+GALHA
Sbjct: 1070 GGKIKWEQGKLNGAPNKVDEIVQFHVGDVVTCLQKASMIPGGSESIMYGTVMGSIGALHA 1129
Query: 1141 FTSRDDVDFFSHLEMHMRQDNPPLCGRDHMAYRSAYFPVKDVIDGDLCEQFPTLPMDLQR 1200
FTSRDDVDFFSHLEMHMRQ+ PPLCGRDHMAYRSAYFPVKDVIDGDLCEQFPTLPMDLQR
Sbjct: 1130 FTSRDDVDFFSHLEMHMRQEYPPLCGRDHMAYRSAYFPVKDVIDGDLCEQFPTLPMDLQR 1189
Query: 1201 KIADELDRTPGEILKKLEEVRNKII 1225
KIADELDRTP EILKKLE+ RNKII
Sbjct: 1190 KIADELDRTPAEILKKLEDARNKII 1214
>At3g55200 spliceosomal - like protein
Length = 1214
Score = 2082 bits (5394), Expect = 0.0
Identities = 1022/1225 (83%), Positives = 1126/1225 (91%), Gaps = 11/1225 (0%)
Query: 1 MYLYNLTLQRPTGIVCAINGNFSGSDDGITQEIVVARGKVLELLRPDKFGRIQSILSVQV 60
MYLY+LTLQ+ TGIVCAINGNFSG G TQEI VARGK+L+LLRPD+ G+IQ+I SV+V
Sbjct: 1 MYLYSLTLQQATGIVCAINGNFSG---GKTQEIAVARGKILDLLRPDENGKIQTIHSVEV 57
Query: 61 FGTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQKNVFDKIHQETFGKSGCRRIVPG 120
FG IRSL+QFRLTGAQKD+IVVGSDSGRIVIL+YNK+KNVFDK+HQETFGKSGCRRIVPG
Sbjct: 58 FGAIRSLAQFRLTGAQKDYIVVGSDSGRIVILEYNKEKNVFDKVHQETFGKSGCRRIVPG 117
Query: 121 QYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSHTIVFSICAVDCGFE 180
QY+A+DPKGRAVMI ACEK+KLVYVLNRD+ ARLTISSPLEA+KSHTI +S+C VDCGF+
Sbjct: 118 QYVAVDPKGRAVMIGACEKQKLVYVLNRDTTARLTISSPLEAHKSHTICYSLCGVDCGFD 177
Query: 181 NPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDLGLNHVSRKWSDQVDNGANMLVTV 240
NPIFAAIELD S+ADQD TG AAS+AQKHL FYELDLGLNHVSRKWS+ VDNGANMLVTV
Sbjct: 178 NPIFAAIELDYSEADQDPTGQAASEAQKHLTFYELDLGLNHVSRKWSNPVDNGANMLVTV 237
Query: 241 PGGADGPSGVLVCAENFVIYKNQGHPDVRAVIPRRADLPAERGVLIVSAAMHKTKNLKPE 300
PGGADGPSGVLVCAENFVIY NQGHPDVRAVIPRR DLPAERGVL+VSAA+HK K +
Sbjct: 238 PGGADGPSGVLVCAENFVIYMNQGHPDVRAVIPRRTDLPAERGVLVVSAAVHKQKTM--- 294
Query: 301 EFKLFFLLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAASEFG 360
FFL+QTEYGD+FKVTL D GD VSEL +KYFDTI VA SICVLK GFLF+ASEFG
Sbjct: 295 ---FFFLIQTEYGDVFKVTL-DHNGDHVSELKVKYFDTIPVASSICVLKLGFLFSASEFG 350
Query: 361 NHALYQFKGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLVRIDQVESLMPVMDMK 420
NH LYQF+ IG++ DV +SS++LMETEEGFQPVFFQPRRLKNLVRIDQVESLMP+MDMK
Sbjct: 351 NHGLYQFQAIGEEP-DVESSSSNLMETEEGFQPVFFQPRRLKNLVRIDQVESLMPLMDMK 409
Query: 421 VSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGIPSAVWTVKKNVMDEFD 480
V N+FEEETPQIF+LCGRGPRSSLRI+R GLA++EMAVS+LPG PSAVWTVKKNV DEFD
Sbjct: 410 VLNIFEEETPQIFSLCGRGPRSSLRILRPGLAITEMAVSQLPGQPSAVWTVKKNVSDEFD 469
Query: 481 AYIVVSFTNATLVLSIGETADEVSDSGFLDTAPSLAVSLIGDDSLMQVHPNGIRHIREDG 540
AYIVVSFTNATLVLSIGE +EV+DSGFLDT PSLAVSLIGDDSLMQVHPNGIRHIREDG
Sbjct: 470 AYIVVSFTNATLVLSIGEQVEEVNDSGFLDTTPSLAVSLIGDDSLMQVHPNGIRHIREDG 529
Query: 541 RTNEWQTSGKRTIAKVGSNRLQVVIALNGGELIYFEVDVTGQLMEVERHEMSGDVACLDI 600
R NEW+T GKR+I KVG NRLQVVIAL+GGELIYFE D+TGQLMEVE+HEMSGDVACLDI
Sbjct: 530 RINEWRTPGKRSIVKVGYNRLQVVIALSGGELIYFEADMTGQLMEVEKHEMSGDVACLDI 589
Query: 601 APVPKGRLRSRFLAVGSYDKTIRILSLDPDDCMQTLGIQSLSSAPESLLFLEVQASVGGE 660
APVP+GR RSRFLAVGSYD T+RILSLDPDDC+Q L +QS+SSAPESLLFLEVQAS+GG+
Sbjct: 590 APVPEGRKRSRFLAVGSYDNTVRILSLDPDDCLQILSVQSVSSAPESLLFLEVQASIGGD 649
Query: 661 DGADHPASLFLNAGLQNGVLSRTVVDMVTGLLSDTRSRFLGLKAPKLFPIIVRGKRAMLC 720
DGADHPA+LFLN+GLQNGVL RTVVDMVTG LSD+RSRFLGLK PKLF I VRG+ AMLC
Sbjct: 650 DGADHPANLFLNSGLQNGVLFRTVVDMVTGQLSDSRSRFLGLKPPKLFSISVRGRSAMLC 709
Query: 721 LSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSDQCFEGVVSVASEALRIFTVERLGETF 780
LSSRPWLGYIH+GHF LTPLSYETLE+AA FSSDQC EGVVSVA +ALRIF ++RLGETF
Sbjct: 710 LSSRPWLGYIHRGHFHLTPLSYETLEFAAPFSSDQCAEGVVSVAGDALRIFMIDRLGETF 769
Query: 781 NQNVIPLRYTPRKFVLQPKRKLLVVIESDQGALTAEEREAARKECFEAAHAGENKTGSED 840
N+ V+PLRYTPRKFVL PKRKLLV+IESDQGA TAEEREAARKECFEA GEN G+ D
Sbjct: 770 NETVVPLRYTPRKFVLHPKRKLLVIIESDQGAFTAEEREAARKECFEAGGVGENGNGNAD 829
Query: 841 QMENGGEDEDNDDSLSDEHYGYPKSESDKWVSCIRVLDPRTGNTTCLLELQENEAAFSIC 900
QMENG +DED +D LSDE YGYPK+ES+KWVSCIRVLDP+T TTCLLELQ+NEAA+S+C
Sbjct: 830 QMENGADDEDKEDPLSDEQYGYPKAESEKWVSCIRVLDPKTATTTCLLELQDNEAAYSVC 889
Query: 901 TVNFHDKEYGTLLAVGTAKGLQFTPKRSLTAGFIHIYRFLDDGRSLELLHKTQVEGVPLA 960
TVNFHDKEYGTLLAVGT KG+QF PK++L AGFIHIYRF++DG+SLELLHKTQVEGVPLA
Sbjct: 890 TVNFHDKEYGTLLAVGTVKGMQFWPKKNLVAGFIHIYRFVEDGKSLELLHKTQVEGVPLA 949
Query: 961 LCQFQGRLLAGIGPVLRLYDLGKRRLLRKCENKSFPSSIVSIHAYRDRIYVGGIQESFHY 1020
LCQFQGRLLAGIGPVLRLYDLGK+RLLRKCENK FP++I+SI YRDRIYVG IQESFHY
Sbjct: 950 LCQFQGRLLAGIGPVLRLYDLGKKRLLRKCENKLFPNTIISIQTYRDRIYVGDIQESFHY 1009
Query: 1021 CKYRRDENQLYIFADDSVPRWLTSSYHIDFDTMAGADKFGNIFFARLPQDVSDEIEEDPT 1080
CKYRRDENQLYIFADD VPRWLT+S+H+DFDTMAGADKFGN++F RLPQD+S+EIEEDPT
Sbjct: 1010 CKYRRDENQLYIFADDCVPRWLTASHHVDFDTMAGADKFGNVYFVRLPQDLSEEIEEDPT 1069
Query: 1081 GGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLVPGGGECIVYGTVMGSVGALHA 1140
GGKIKWEQGKLNGAPNKV+EIVQFHVGDV+T LQKAS++PGG E I+YGTVMGS+GALHA
Sbjct: 1070 GGKIKWEQGKLNGAPNKVDEIVQFHVGDVVTCLQKASMIPGGSESIMYGTVMGSIGALHA 1129
Query: 1141 FTSRDDVDFFSHLEMHMRQDNPPLCGRDHMAYRSAYFPVKDVIDGDLCEQFPTLPMDLQR 1200
FTSRDDVDFFSHLEMHMRQ+ PPLCGRDHMAYRSAYFPVKDVIDGDLCEQFPTLPMDLQR
Sbjct: 1130 FTSRDDVDFFSHLEMHMRQEYPPLCGRDHMAYRSAYFPVKDVIDGDLCEQFPTLPMDLQR 1189
Query: 1201 KIADELDRTPGEILKKLEEVRNKII 1225
KIADELDRTP EILKKLE+ RNKII
Sbjct: 1190 KIADELDRTPAEILKKLEDARNKII 1214
>At4g05420 UV-damaged DNA binding factor - like protein
Length = 1088
Score = 223 bits (569), Expect = 4e-58
Identities = 285/1245 (22%), Positives = 513/1245 (40%), Gaps = 194/1245 (15%)
Query: 4 YNLTLQRPTGIVCAINGNFSGSDDGITQEIVVARGKVLE--LLRPDKFGRIQSILSVQVF 61
Y +T +PT + + GNF+ + ++VA+ +E LL P +Q +L V ++
Sbjct: 6 YVVTAHKPTSVTHSCVGNFTSPQE---LNLIVAKCTRIEIHLLTPQG---LQPMLDVPIY 59
Query: 62 GTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQKN-VFDKIHQETFGKSGCRRIVPG 120
G I +L FR G +DF+ + ++ + +L ++ + + + + + + G R G
Sbjct: 60 GRIATLELFRPHGEAQDFLFIATERYKFCVLQWDPESSELITRAMGDVSDRIG-RPTDNG 118
Query: 121 QYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSHTIVFSICAVDCGFE 180
Q IDP R + + + V + + + LE + I F G
Sbjct: 119 QIGIIDPDCRLIGLHLYDGLFKVIPFDNKGQLKEAFNIRLEELQVLDIKFLF-----GCA 173
Query: 181 NPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDL-GLNHVSRKWS-DQVDNGANMLV 238
P A + D DA +H+ YE+ L + V WS + +DNGA++L+
Sbjct: 174 KPTIAVLYQDNKDA-------------RHVKTYEVSLKDKDFVEGPWSQNSLDNGADLLI 220
Query: 239 TVPGGADGPSGVLVCAENFVIYKNQGHPDVRAVIPRRADLPAERGVLIVSAAMHKTKNLK 298
VP GVL+ E ++Y + IP R + G + V +
Sbjct: 221 PVPPPL---CGVLIIGEETIVYCS---ASAFKAIPIRPSITKAYGRVDVDGSR------- 267
Query: 299 PEEFKLFFLLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAASE 358
+LL G I + +T ++V+ L I+ ++A +I L + +F S
Sbjct: 268 -------YLLGDHAGMIHLLVITH-EKEKVTGLKIELLGETSIASTISYLDNAVVFVGSS 319
Query: 359 FGNHALYQFKGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLVRIDQVESLMPVMD 418
+G+ L + D + S +E +++ +L P++D
Sbjct: 320 YGDSQLVKLNLHPD-------AKGSYVEV-------------------LERYINLGPIVD 353
Query: 419 MKVSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGIPSAVWTVKKNVMDE 478
V +L + Q+ T G SLR++R G+ ++E A +L GI +W++K ++ +
Sbjct: 354 FCVVDLERQGQGQVVTCSGAFKDGSLRVVRNGIGINEQASVELQGI-KGMWSLKSSIDEA 412
Query: 479 FDAYIVVSFTNAT--LVLSIGETADEVSDSGFLDTAPSLAVSLIGDDSLMQVHPNGIRHI 536
FD ++VVSF + T L +++ + +E GFL +L + L+QV N +R +
Sbjct: 413 FDTFLVVSFISETRILAMNLEDELEETEIEGFLSQVQTLFCHDAVYNQLVQVTSNSVRLV 472
Query: 537 REDGR--TNEWQTSGKRTIAKVGSNRLQVVIALNGGELIYFEVDVTGQLMEVERHEMSGD 594
R +EW T+ +N QV++A GG L+Y E+ G+L EV+ + +
Sbjct: 473 SSTTRELRDEWHAPAGFTVNVATANASQVLLATGGGHLVYLEIG-DGKLTEVQHALLEYE 531
Query: 595 VACLDIAPVPKGRLRSRFLAVGSY-DKTIRILSLDPDDCMQTLGIQSLSSAPESLLFLEV 653
V+CLDI P+ S+ AVG + D ++RI SL P+ + T P S+L
Sbjct: 532 VSCLDINPIGDNPNYSQLAAVGMWTDISVRIFSL-PELTLITKEQLGGEIIPRSVLLCAF 590
Query: 654 QASVGGEDGADHPASLFLNAGLQNGVLSRTVVDMVTGLLSDTRSRFLGLKAPKLFPIIVR 713
+ +L L +G L +D TG L D + LG + L +
Sbjct: 591 E------------GISYLLCALGDGHLLNFQMDTTTGQLKDRKKVSLGTQPITLRTFSSK 638
Query: 714 GKRAMLCLSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSDQCFEGVVSVASEA-LRIFT 772
+ S RP + Y L + ++ + + + F+S F +++A E L I T
Sbjct: 639 SATHVFAASDRPTVIYSSNKKLLYSNVNLKEVSHMCPFNS-AAFPDSLAIAREGELTIGT 697
Query: 773 VERLGETFNQNVIPLRYTPRKFVLQPKRKLLVVIESDQGALTAEEREAARKECFEAAHAG 832
++ + + + IPL E AR+ C H
Sbjct: 698 IDDI-QKLHIRTIPL------------------------------GEHARRIC----HQE 722
Query: 833 ENKTGSEDQMENGGEDEDNDDSLSDEHYGYPKSESDKWVSCIRVLDPRTGNTTCLLELQE 892
+ +T + N E+ S+ H+ +R+LD +T L
Sbjct: 723 QTRTFGICSLGNQSNSEE-----SEMHF-------------VRLLDDQTFEFMSTYPLDS 764
Query: 893 NEAAFSICTVNFHDKEYGTLLAVGTAKGLQFTPKRSLTAGFIHIYRFLDDGRSLELLHKT 952
E SI + +F ++ VGTA L + T G I ++ ++DGR L+L+ +
Sbjct: 765 FEYGCSILSCSF-TEDKNVYYCVGTAYVL--PEENEPTKGRILVF-IVEDGR-LQLIAEK 819
Query: 953 QVEGVPLALCQFQGRLLAGIGPVLRLY-----DLGKRRLLRKCENKSFPSSIVSIHAYRD 1007
+ +G +L F G+LLA I ++LY D G R L +C + ++ + D
Sbjct: 820 ETKGAVYSLNAFNGKLLAAINQKIQLYKWMLRDDGTRELQSECGHHGHILALY-VQTRGD 878
Query: 1008 RIYVGGIQESFHYCKYRRDENQLYIFADDSVPRWLTSSYHIDFDTMAGADKFGNIFFARL 1067
I VG + +S Y+ +E + A D W+++ +D D GA+ N+ +
Sbjct: 879 FIVVGDLMKSISLLLYKHEEGAIEERARDYNANWMSAVEILDDDIYLGAENNFNLLTVK- 937
Query: 1068 PQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLVP------- 1120
K +G + ++E + ++H+G+ + + SLV
Sbjct: 938 -----------------KNSEGATDEERGRLEVVGEYHLGEFVNRFRHGSLVMRLPDSEI 980
Query: 1121 GGGECIVYGTVMGSVGALHAFTSRDDVDFFSHLEMHMRQDNPPLCGRDHMAYRS-----A 1175
G +++GTV G +G + A ++ F L+ +R+ + G H +RS
Sbjct: 981 GQIPTVIFGTVNGVIGVI-ASLPQEQYTFLEKLQSSLRKVIKGVGGLSHEQWRSFNNEKR 1039
Query: 1176 YFPVKDVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKKLEEV 1220
++ +DGDL E F L + I+ ++ E+ K++EE+
Sbjct: 1040 TAEARNFLDGDLIESFLDLSRNKMEDISKSMNVQVEELCKRVEEL 1084
>At4g21100 UV-damaged DNA-binding protein- like
Length = 1102
Score = 210 bits (535), Expect = 4e-54
Identities = 292/1259 (23%), Positives = 506/1259 (39%), Gaps = 208/1259 (16%)
Query: 4 YNLTLQRPTGIVCAINGNFSGSDDGITQEIVVARGKVLE--LLRPDKFGRIQSILSVQVF 61
Y +T Q+PT + + GNF+ + ++VA+ +E LL P +Q+IL V ++
Sbjct: 6 YAVTAQKPTCVTHSCVGNFTSPQE---LNLIVAKSTRIEIHLLSPQG---LQTILDVPLY 59
Query: 62 GTIRSLSQFR----LTGAQKDFIVVGSDSGRIVILDYNKQKNVFDK-------IHQETFG 110
G I ++ FR + F+ + +L +VF + Q G
Sbjct: 60 GRIATMELFRPHVSIVFLLNTFLCLRVKHKTFCLLQLKDINSVFFNGIMSPLSLLQGAMG 119
Query: 111 KSGCRRIVP---GQYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSHT 167
R P GQ IDP R + + + V + + + LE +
Sbjct: 120 DVSDRIGRPTDNGQIGIIDPDCRVIGLHLYDGLFKVIPFDNKGQLKEAFNIRLEELQVLD 179
Query: 168 IVFSICAVDCGFENPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDL-GLNHVSRKW 226
I F G P A + D DA +H+ YE+ L N V W
Sbjct: 180 IKFLY-----GCTKPTIAVLYQDNKDA-------------RHVKTYEVSLKDKNFVEGPW 221
Query: 227 S-DQVDNGANMLVTVPGGADGPSGVLVCAENFVIYKNQGHPDVRAVIPRRADLPAERGVL 285
S + +DNGA++L+ VP GVL+ E ++Y + + IP R + G +
Sbjct: 222 SQNNLDNGADLLIPVPSPL---CGVLIIGEETIVYCS---ANAFKAIPIRPSITKAYGRV 275
Query: 286 IVSAAMHKTKNLKPEEFKLFFLLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSI 345
+ + +LL G I + +T ++V+ L I+ ++A SI
Sbjct: 276 DLDGSR--------------YLLGDHAGLIHLLVITH-EKEKVTGLKIELLGETSIASSI 320
Query: 346 CVLKSGFLFAASEFGNHALYQFKGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLV 405
L + +F S +G+ L + QP + V
Sbjct: 321 SYLDNAVVFVGSSYGDSQLIKLN---------------------------LQPDAKGSYV 353
Query: 406 RI-DQVESLMPVMDMKVSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGI 464
I ++ +L P++D V +L + Q+ T G SLRI+R G+ ++E A +L GI
Sbjct: 354 EILEKYVNLGPIVDFCVVDLERQGQGQVVTCSGAYKDGSLRIVRNGIGINEQASVELQGI 413
Query: 465 PSAVWTVKKNVMDEFDAYIVVSFTNAT--LVLSIGETADEVSDSGFLDTAPSLAVSLIGD 522
+W++K ++ + FD ++VVSF + T L ++I + +E GFL +L
Sbjct: 414 -KGMWSLKSSIDEAFDTFLVVSFISETRILAMNIEDELEETEIEGFLSEVQTLFCHDAVY 472
Query: 523 DSLMQVHPNGIRHIREDGR--TNEWQTSGKRTIAKVGSNRLQVVIALNGGELIYFEVDVT 580
+ L+QV N +R + R N+W ++ +N QV++A GG L+Y E+
Sbjct: 473 NQLVQVTSNSVRLVSSTTRELRNKWDAPAGFSVNVATANASQVLLATGGGHLVYLEIG-D 531
Query: 581 GQLMEVERHEMSGDVACLDIAPVPKGRLRSRFLAVGSY-DKTIRILSLDPDDCMQTLGIQ 639
G L EV+ + +V+CLDI P+ S+ AVG + D ++RI L PD + T
Sbjct: 532 GTLTEVKHVLLEYEVSCLDINPIGDNPNYSQLAAVGMWTDISVRIFVL-PDLTLITKEEL 590
Query: 640 SLSSAPESLLFLEVQASVGGEDGADHPASLFLNAGLQNGVLSRTVVDMVTGLLSDTRSRF 699
P S+L + +L L +G L +D G L D +
Sbjct: 591 GGEIIPRSVLLCAFE------------GISYLLCALGDGHLLNFQLDTSCGKLRDRKKVS 638
Query: 700 LGLKAPKLFPIIVRGKRAMLCLSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSDQCFEG 759
LG + L + + S RP + Y + L + ++ + + + F+S F
Sbjct: 639 LGTRPITLRTFSSKSATHVFAASDRPAVIYSNNKKLLYSNVNLKEVSHMCPFNS-AAFPD 697
Query: 760 VVSVASEA-LRIFTVERLGETFNQNVIPLRYTPRKFVLQPKRKLLVVIESDQGALTAEER 818
+++A E L I T++ + + + IP+
Sbjct: 698 SLAIAREGELTIGTIDDI-QKLHIRTIPI------------------------------G 726
Query: 819 EAARKECFEAAHAGENKTGSEDQMENGGEDEDNDDSLSDEHYGYPKSESDKWVSCIRVLD 878
E AR+ C H + +T + + N E+ S+ H+ +R+LD
Sbjct: 727 EHARRIC----HQEQTRTFAISCLRNEPSAEE-----SESHF-------------VRLLD 764
Query: 879 PRTGNTTCLLELQENEAAFSICTVNFHDKEYGTLLAVGTAKGLQFTPKRSLTAGFIHIYR 938
++ L E SI + +F D + VGTA L + T G I ++
Sbjct: 765 AQSFEFLSSYPLDAFECGCSILSCSFTD-DKNVYYCVGTAYVL--PEENEPTKGRILVF- 820
Query: 939 FLDDGRSLELLHKTQVEGVPLALCQFQGRLLAGIGPVLRLY-----DLGKRRLLRKCENK 993
+++GR L+L+ + + +G +L F G+LLA I ++LY D G R L +C +
Sbjct: 821 IVEEGR-LQLITEKETKGAVYSLNAFNGKLLASINQKIQLYKWMLRDDGTRELQSECGHH 879
Query: 994 SFPSSIVSIHAYRDRIYVGGIQESFHYCKYRRDENQLYIFADDSVPRWLTSSYHIDFDTM 1053
++ + D I VG + +S Y+ +E + A D W+T+ ++ D
Sbjct: 880 GHILALY-VQTRGDFIAVGDLMKSISLLIYKHEEGAIEERARDYNANWMTAVEILNDDIY 938
Query: 1054 AGADKFGNIFFARLPQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSL 1113
G D NIF + K +G + ++E + ++H+G+ +
Sbjct: 939 LGTDNCFNIFTVK------------------KNNEGATDEERARMEVVGEYHIGEFVNRF 980
Query: 1114 QKASLVP-------GGGECIVYGTVMGSVGALHAFTSRDDVDFFSHLEMHMRQDNPPLCG 1166
+ SLV G +++GTV G +G + A ++ F L+ +R+ + G
Sbjct: 981 RHGSLVMKLPDSDIGQIPTVIFGTVSGMIGVI-ASLPQEQYAFLEKLQTSLRKVIKGVGG 1039
Query: 1167 RDHMAYRS-----AYFPVKDVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKKLEEV 1220
H +RS K +DGDL E F L +I+ +D E+ K++EE+
Sbjct: 1040 LSHEQWRSFNNEKRTAEAKGYLDGDLIESFLDLSRGKMEEISKGMDVQVEELCKRVEEL 1098
>At3g11960 hypothetical protein
Length = 1331
Score = 117 bits (292), Expect = 5e-26
Identities = 195/910 (21%), Positives = 351/910 (38%), Gaps = 151/910 (16%)
Query: 407 IDQVESLMPVMDMKVSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEM--AVSKLPGI 464
+ ++++ P++D V + E+ QIF CG P SLRI+R+G+ V ++ GI
Sbjct: 435 MSSIQNIAPILDFSVMDDQNEKRDQIFACCGVTPEGSLRIIRSGINVEKLLKTAPVYQGI 494
Query: 465 PSAVWTVKKNVMDEFDAYIVVSFTNATLVLSIGETADEVSDS-GFLDTAPSLAVSLIGDD 523
+ WTVK + D + +++V+SF T VLS+G + +V+DS GF + A L+ D
Sbjct: 495 -TGTWTVKMKLTDVYHSFLVLSFVEETRVLSVGLSFKDVTDSVGFQSDVCTFACGLVADG 553
Query: 524 SLMQVHPNGIR------HIREDG-------RTNEWQTSGKRTIAKVGSNRLQVVIALNGG 570
L+Q+H + IR DG ++ + + ++ VG N L VV N
Sbjct: 554 LLVQIHQDAIRLCMPTMDAHSDGIPVSSPFFSSWFPENVSISLGAVGQN-LIVVSTSNPC 612
Query: 571 ELIYFEVDVTG----QLMEVERHEMSGDVACLDIAPVPKGRLRSR--------------- 611
L V ++ E++R + +V+C+ + G+ RSR
Sbjct: 613 FLSILGVKSVSSQCCEIYEIQRVTLQYEVSCISVPQKHIGKKRSRDSSPDNFCKAAIPSA 672
Query: 612 -----FLAVGSYDKTIRILSLDPDDCMQTLGIQSLSSAPESL---LFLEVQASVGGEDGA 663
+G++ ++ +LS D +G++ L+S SL + + + +
Sbjct: 673 MEQGYTFLIGTHKPSVEVLSFTEDG----VGVRVLASGLVSLTNTMGTVISGCIPQDVRL 728
Query: 664 DHPASLFLNAGLQNGVLSR--------------------------TVVDMVTGL---LSD 694
L++ +GL+NG+L R TVV L L
Sbjct: 729 VLVDQLYVLSGLRNGMLLRFEWAPFSNSSGLNCPDYFSHCKEEMDTVVGKKDNLPVNLLL 788
Query: 695 TRSRFLGLKAPKLFPIIVRGKRAMLCLSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSD 754
+R +G+ L P ++ LS RPWL + T +S++ +A
Sbjct: 789 IATRRIGITPVFLVPFSDSLDSDIIALSDRPWLLQTARQSLSYTSISFQPSTHATP---- 844
Query: 755 QCFEGVVSVASEALRIFTVERLGETFNQNVIPLRYTPRKFVLQPKRKLLVVIESDQGALT 814
V + R N L TPRK + + KLL+V+ +D
Sbjct: 845 ------VEMVHSKRR-----------NAQKFQLGGTPRKVIYHSESKLLIVMRTDLYDTC 887
Query: 815 AEE---REAARKECFEAAHAGENKTGSEDQMENGGEDE--DNDDSLSDEHYGYPKSESDK 869
+ + + +TG ++ G + SLS P E++
Sbjct: 888 TSDICCVDPLSGSVLSSYKLKPGETGKSMELVRVGNEHVLVVGTSLSSGPAILPSGEAES 947
Query: 870 WVSCIRVLDPRTGNTTCLLELQENEA-AFSIC---------TVNFHDKEYGTLLAVG-TA 918
+ +L CL Q +++ + +IC T FHD VG T
Sbjct: 948 TKGRVIIL--------CLEHTQNSDSGSMTICSKACSSSQRTSPFHD-------VVGYTT 992
Query: 919 KGLQFTPKRSLTAGFIHIYRFLDDGRS--LELLHKTQVEGVPLALCQFQGR-LLAGIGPV 975
+ L + S + + LD+ + L L T G+ LA+C + LA G
Sbjct: 993 ENLSSSSLCSSPDDYSYDGIKLDEAETWQLRLASSTTWPGMVLAICPYLDHYFLASAGNA 1052
Query: 976 LRLYDL---GKRRLLRKCENKSFPSSIVSIHAYRDRIYVGGIQESFHYCKYRRDENQLYI 1032
+ R+ R ++ I S+ Y RI VG ++ + Y + +L+
Sbjct: 1053 FYVCGFPNDSPERMKRFAVGRT-RFMITSLRTYFTRIVVGDCRDGVLFYSYHEESKKLHQ 1111
Query: 1033 FADDSVPRWLTSSYHIDFDTMAGADKFGNIFFARLPQDVSD--------EIEEDPTGGKI 1084
D R + + +D +++A +D+ G+I +D SD E
Sbjct: 1112 IYCDPAQRLVADCFLMDANSVAVSDRKGSIAILSC-KDHSDFGMKHLEYSSPESNLNLNC 1170
Query: 1085 KWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLVPGGGECIVYGTVMGSVGALHAFTSR 1144
+ G++ + K I + DV+ S + + + I+ GT++GS+ + A S
Sbjct: 1171 AYYMGEIAMSIKKGCNIYKLPADDVLRSYGLSKSIDTADDTIIAGTLLGSI-FVFAPISS 1229
Query: 1145 DDVDFFSHLE--MHMRQDNPPLCGRDHMAYRSAYFP--VKDVIDGDLCEQFPTLPMDLQR 1200
++ + ++ + + P+ G DH +R P + ++DGD+ QF L Q
Sbjct: 1230 EEYELLEGVQAKLGIHPLTAPVLGNDHNEFRGRENPSQARKILDGDMLAQFLELTNRQQE 1289
Query: 1201 KIADELDRTP 1210
+ +P
Sbjct: 1290 SVLSTPQPSP 1299
Score = 33.9 bits (76), Expect = 0.59
Identities = 26/96 (27%), Positives = 47/96 (48%), Gaps = 11/96 (11%)
Query: 10 RPTGIVCAINGNFSGSDDGITQEIVVARGKVLELLRPDKFGRIQSILSVQVFGTIRSL-- 67
RP+ ++ G F +++IV + +EL+ + G ++S+ VFGTI+ L
Sbjct: 43 RPSVVLQVAYGYFRSPS---SRDIVFGKETCIELVVIGEDGIVESVCEQYVFGTIKDLAV 99
Query: 68 ----SQFRLTGAQ--KDFIVVGSDSGRIVILDYNKQ 97
S+ Q KD + V SDSG++ L ++ +
Sbjct: 100 IPQSSKLYSNSLQMGKDLLAVLSDSGKLSFLSFSNE 135
>At5g51660 cleavage and polyadenylation specificity factor subunit
Length = 1448
Score = 67.0 bits (162), Expect = 6e-11
Identities = 95/410 (23%), Positives = 179/410 (43%), Gaps = 64/410 (15%)
Query: 843 ENGGEDEDNDDSLSDE-HYGYPKSESDKWVSCIRVLDP-RTGN---TTCLLELQENEAAF 897
+ G+ DN + SD+ Y E + I++L+P R+G T + +Q +E A
Sbjct: 1064 QEAGQQLDNHNMSSDDLQRTYTVEEFE-----IQILEPERSGGPWETKAKIPMQTSEHAL 1118
Query: 898 SICTVNFHDKEYG---TLLAVGTAKGLQFTPKRSLTA-GFIHIYRFLDDGRSLELL---- 949
++ V + G TLLAVGTA + + A G + ++ F +G + + +
Sbjct: 1119 TVRVVTLLNASTGENETLLAVGTA----YVQGEDVAARGRVLLFSFGKNGDNSQNVVTEV 1174
Query: 950 HKTQVEGVPLALCQFQGRLLAGIGPVLRLYDLGKRRLLRKCENKSFPSSIVSIHAYRDRI 1009
+ +++G A+ QG LL GP + L+ L + P +VS++ + I
Sbjct: 1175 YSRELKGAISAVASIQGHLLISSGPKIILHKWNGTELNGVAFFDAPPLYVVSMNVVKSFI 1234
Query: 1010 YVGGIQESFHYCKYRRDENQLYIFADD--SVPRWLTSSYHIDFDTM--AGADKFGNI-FF 1064
+G + +S ++ ++ +QL + A D S+ + T + ID T+ A +D+ NI F
Sbjct: 1235 LLGDVHKSIYFLSWKEQGSQLSLLAKDFESLDCFATE-FLIDGSTLSLAVSDEQKNIQVF 1293
Query: 1065 ARLPQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLVPGGGE 1124
P+ + W+ KL +FHVG ++ + +V G +
Sbjct: 1294 YYAPKMIES------------WKGLKLLSR-------AEFHVGAHVSKFLRLQMVSSGAD 1334
Query: 1125 -----CIVYGTVMGSVGALHAFTSRDDVDF--FSHLEMHMRQDNPPLCGRDHMAYRSAYF 1177
+++GT+ GS G + D+V F L+ + P + G + +A+R
Sbjct: 1335 KINRFALLFGTLDGSFGCIAPL---DEVTFRRLQSLQKKLVDAVPHVAGLNPLAFRQFRS 1391
Query: 1178 PVK-------DVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKKLEEV 1220
K ++D +L + LP++ Q ++A ++ T ILK L ++
Sbjct: 1392 SGKARRSGPDSIVDCELLCHYEMLPLEEQLELAHQIGTTRYSILKDLVDL 1441
Score = 45.1 bits (105), Expect = 3e-04
Identities = 60/288 (20%), Positives = 105/288 (35%), Gaps = 62/288 (21%)
Query: 307 LLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAASEFGNHALYQ 366
LL T+ G++ +TL G V L++ +A I + + F S G+ L Q
Sbjct: 381 LLSTKSGELLLLTLIYDGR-AVQRLDLSKSKASVLASDITSVGNSLFFLGSRLGDSLLVQ 439
Query: 367 FK----------GIGDDDNDV---GASSASLMETEEGFQPVF------------------ 395
F G+ D+D D+ G + L T + FQ
Sbjct: 440 FSCRSGPAASLPGLRDEDEDIEGEGHQAKRLRMTSDTFQDTIGNEELSLFGSTPNNSDSA 499
Query: 396 ---------FQPRRLKNLVRIDQVES----LMPVMDMKVSNLFEEETPQIFTLCGRGPRS 442
F +LV + V+ L D + + ++ ++ G G
Sbjct: 500 QVTSSVLKSFSFAVRDSLVNVGPVKDFAYGLRINADANATGVSKQSNYELVCCSGHGKNG 559
Query: 443 SLRIMRTGLAVSEMAVSKLPGIPSAVWTVKKNVM--------------DEFDAYIVVSFT 488
+L ++R + + +LPG +WTV DE+ AY+++S
Sbjct: 560 ALCVLRQSIRPEMITEVELPGC-KGIWTVYHKSSRGHNADSSKMAADEDEYHAYLIISLE 618
Query: 489 NATLVLSIGETADEVSDS--GFLDTAPSLAVSLIGDDSLMQVHPNGIR 534
T+VL + EV++S ++ A +L G ++QV +G R
Sbjct: 619 ARTMVLETADLLTEVTESVDYYVQGRTIAAGNLFGRRRVIQVFEHGAR 666
>At3g27720 hypothetical protein
Length = 493
Score = 35.8 bits (81), Expect = 0.15
Identities = 23/61 (37%), Positives = 31/61 (50%), Gaps = 5/61 (8%)
Query: 803 LVVIESDQGA----LTAEEREAARKECFEAAHAGENKTGSEDQ-MENGGEDEDNDDSLSD 857
L + +DQG AEE E KE E E + G ED+ E GGEDE+ ++ + D
Sbjct: 70 LFSVYTDQGKDVLFSRAEEEEGEDKEEEEGGEDEEEEEGGEDEEAEEGGEDEEAEEGVED 129
Query: 858 E 858
E
Sbjct: 130 E 130
>At3g18950 unknown protein
Length = 473
Score = 34.7 bits (78), Expect = 0.34
Identities = 34/154 (22%), Positives = 72/154 (46%), Gaps = 26/154 (16%)
Query: 506 SGFLDTAPSL-AVSLIGDDSLMQVHPNGIRHIREDGRTNEWQTSGKRT--IAKVGS-NRL 561
+GF T+ + A+ + GD+ + H +DG+ W+ S +RT +++GS L
Sbjct: 170 TGFKSTSGLVKAIVITGDNRIFTGH--------QDGKIRVWRGSKRRTGGYSRIGSLPTL 221
Query: 562 QVVIALNGGELIYFEVDVTGQLMEVERHEMSGDVACLDIAPVPKGRLRSRFLAVGSYDKT 621
+ + + Y EV ++++ ++ V+CL + L GS+DKT
Sbjct: 222 KEFLTKSVNPKNYVEVRRRKNVLKIRHYDA---VSCLSLNE------ELGLLYSGSWDKT 272
Query: 622 IRILSLDPDDCMQTL-----GIQSLSSAPESLLF 650
+++ L C++++ I ++++ + LLF
Sbjct: 273 LKVWRLSDSKCLESIQAHDDAINTVAAGFDDLLF 306
>At4g12740 adenine DNA glycosylase like protein
Length = 608
Score = 33.1 bits (74), Expect = 1.0
Identities = 22/83 (26%), Positives = 40/83 (47%), Gaps = 3/83 (3%)
Query: 779 TFNQNVIPLRYTPRKFVLQPKRKLLVVIESDQGALTAEEREAARKECFEAAHAGENKTGS 838
+F + + PR+ ++ RK E+++ A EREA +E E A A +K +
Sbjct: 37 SFKTKTMSQSFAPREKLM---RKCREKKEAEREAEREAEREAEEEEKAEEAEAEADKEEA 93
Query: 839 EDQMENGGEDEDNDDSLSDEHYG 861
E++ E E+E+ + +E G
Sbjct: 94 EEESEEEEEEEEEEAEAEEEALG 116
>At5g05700 arginine-tRNA-protein transferase 1 homolog
Length = 632
Score = 32.3 bits (72), Expect = 1.7
Identities = 17/54 (31%), Positives = 25/54 (45%)
Query: 814 TAEEREAARKECFEAAHAGENKTGSEDQMENGGEDEDNDDSLSDEHYGYPKSES 867
T E A+ E E +N G D+ E+ ED+D+DD +E Y +S
Sbjct: 513 TLVEPAASEHEDMEQGETNDNFMGCSDEDEDEDEDDDDDDDDDEEMYETESEDS 566
>At1g17680 unknown protein
Length = 896
Score = 32.3 bits (72), Expect = 1.7
Identities = 21/67 (31%), Positives = 32/67 (47%), Gaps = 3/67 (4%)
Query: 805 VIESDQGALTAEEREA-ARKECFEAAHAGENKTGSED--QMENGGEDEDNDDSLSDEHYG 861
V+E D+G L +E E + EC + G+ +D E G D+D+DDS D+
Sbjct: 8 VVEGDEGNLISELEEGPSNMECDKQVLGGDTNYDDKDLNSDEEGLVDDDDDDSDDDDEGD 67
Query: 862 YPKSESD 868
+ E D
Sbjct: 68 ESEEEDD 74
>At2g11910 unknown protein
Length = 168
Score = 31.2 bits (69), Expect = 3.8
Identities = 21/64 (32%), Positives = 35/64 (53%), Gaps = 5/64 (7%)
Query: 802 LLVVIESDQGALTAEEREAARKECFE-AAHAGENKTGSE----DQMENGGEDEDNDDSLS 856
LL+V ES L A ++ + KE ++ + ENK S+ D E+ ED+D++D +
Sbjct: 36 LLLVAESLLIGLVAVDQRLSVKEGYKRGSRIEENKDASDSDDDDDDEDADEDDDDEDDAN 95
Query: 857 DEHY 860
DE +
Sbjct: 96 DEDF 99
>At1g08340 unknown protein
Length = 404
Score = 31.2 bits (69), Expect = 3.8
Identities = 16/49 (32%), Positives = 27/49 (54%), Gaps = 4/49 (8%)
Query: 814 TAEEREAAR----KECFEAAHAGENKTGSEDQMENGGEDEDNDDSLSDE 858
T EREA+ + C + A GE + +E++ E+ E+E+ +D DE
Sbjct: 297 TVREREASSSVVDRRCSKEAEDGEKEKDNEEEEEDEEEEEEEEDEDEDE 345
>At4g08310 unknown protein (At4g08310)
Length = 504
Score = 30.8 bits (68), Expect = 5.0
Identities = 24/79 (30%), Positives = 32/79 (40%), Gaps = 4/79 (5%)
Query: 788 RYTPRKFVLQPKRKLLVVIESDQGALTAEEREAARKECFEAAHAGENKTGSEDQMENGGE 847
R + FV PK ESD + E + + E E++ GSED GGE
Sbjct: 430 RRSSASFVPPPKPIKAEESESDDSEDSENEEDEDEEVVVEEEEEEEDEGGSED----GGE 485
Query: 848 DEDNDDSLSDEHYGYPKSE 866
N+ L E G +SE
Sbjct: 486 GSQNEGELKTEDGGEEESE 504
>At1g55390 hypothetical protein
Length = 684
Score = 30.8 bits (68), Expect = 5.0
Identities = 12/29 (41%), Positives = 19/29 (65%)
Query: 832 GENKTGSEDQMENGGEDEDNDDSLSDEHY 860
GE++ G D +GG+D+D+DD D +Y
Sbjct: 148 GEDRDGDSDGDGDGGDDDDDDDFQQDIYY 176
>At1g29470 unknown protein
Length = 768
Score = 30.4 bits (67), Expect = 6.5
Identities = 18/63 (28%), Positives = 33/63 (51%), Gaps = 7/63 (11%)
Query: 807 ESDQGALTAEEREAARKECFEAAHAGENKTGSEDQMENGGEDEDNDDSLSDEHYGYPKSE 866
ESD+ +++E + E + E+ G+E EN GE E+N + S+E+ G +
Sbjct: 139 ESDE----TKQKEKTQLEESSEENKSEDSNGTE---ENAGESEENTEKKSEENAGETEES 191
Query: 867 SDK 869
++K
Sbjct: 192 TEK 194
>At1g05910 unknown protein
Length = 1210
Score = 30.4 bits (67), Expect = 6.5
Identities = 29/118 (24%), Positives = 54/118 (45%), Gaps = 17/118 (14%)
Query: 758 EGVVSVASEALRIFTVERLGETFNQNVIPLRYTPRKFVLQPKRKLLVV---IESDQGAL- 813
E ++S A LR + + ++ + PR+ L+P+R + ++++ GA
Sbjct: 107 EDMMSPAYRTLRRRVHKNFSTSKSRKDMDAELAPRREGLRPRRSTTIANKRLKTESGADQ 166
Query: 814 -TAEER----EAARKECFEAAHAGENKTGSEDQM--------ENGGEDEDNDDSLSDE 858
T+EE+ E + A GEN+ +ED+ E+ GE++ +DD DE
Sbjct: 167 DTSEEKDGQDETENGNELDDADDGENEVEAEDEGNGEDEGDGEDEGEEDGDDDEEGDE 224
>At5g53800 unknown protein
Length = 339
Score = 30.0 bits (66), Expect = 8.5
Identities = 17/59 (28%), Positives = 29/59 (48%)
Query: 808 SDQGALTAEEREAARKECFEAAHAGENKTGSEDQMENGGEDEDNDDSLSDEHYGYPKSE 866
S G + EE++ +R+ + N +GSE +E+G E E + S + G KS+
Sbjct: 32 SGSGKDSGEEKDVSRRRESKRRTKDGNDSGSESGLESGSESEKEERRRSRKDRGKRKSD 90
>At4g03450 hypothetical protein
Length = 641
Score = 30.0 bits (66), Expect = 8.5
Identities = 26/120 (21%), Positives = 47/120 (38%), Gaps = 19/120 (15%)
Query: 414 MPVMDMKVSNL-----FEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGIPSAV 468
+ +D+ SNL F E + LC PR I +G+ + + K+ G
Sbjct: 400 LSALDIAESNLQSNYVFRERMTLMVLLCTCSPRGFKMIPTSGITLKSRS-EKVAG----- 453
Query: 469 WTVKKNVMDEFDAYIVVSFTNATLVLSIGETADEVSDSGFLDTAPSLAVSLIGDDSLMQV 528
D + ++V ATLV ++ A GF + P ++++ DD + +
Sbjct: 454 ----NKYKDSINVLLLV----ATLVATVAFAAGIAIPGGFSSSTPKRGIAILDDDDFLSI 505
>At2g47070 squamosa promoter binding protein-like 1
Length = 881
Score = 30.0 bits (66), Expect = 8.5
Identities = 22/70 (31%), Positives = 32/70 (45%), Gaps = 9/70 (12%)
Query: 1139 HAFTSRDDVDFFSHLEM----HMRQDNPPLCGRDHMAYR-----SAYFPVKDVIDGDLCE 1189
H T+R+D DF + E+ + D P L GR M S++FP V D D+C
Sbjct: 522 HDSTTREDDDFKDNSEIVECVNFSCDMPILSGRGFMEIEDQGLSSSFFPFLVVEDDDVCS 581
Query: 1190 QFPTLPMDLQ 1199
+ L L+
Sbjct: 582 EIRILETTLE 591
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,045,730
Number of Sequences: 26719
Number of extensions: 1264742
Number of successful extensions: 3674
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 3597
Number of HSP's gapped (non-prelim): 53
length of query: 1225
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1114
effective length of database: 8,352,787
effective search space: 9305004718
effective search space used: 9305004718
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144540.4


