

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144517.6 + phase: 0
(366 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
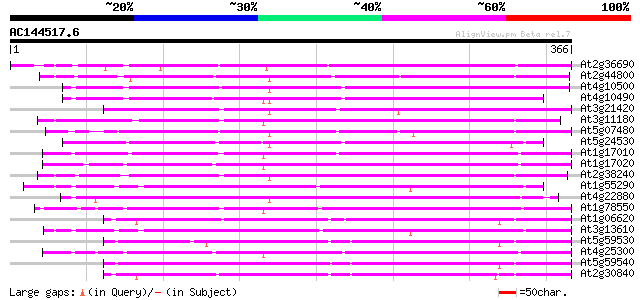
Score E
Sequences producing significant alignments: (bits) Value
At2g36690 putative giberellin beta-hydroxylase 207 6e-54
At2g44800 putative flavonol synthase 203 1e-52
At4g10500 putative Fe(II)/ascorbate oxidase 202 2e-52
At4g10490 putative flavanone 3-beta-hydroxylase 201 4e-52
At3g21420 unknown protein 197 1e-50
At3g11180 putative leucoanthocyanidin dioxygenase 193 1e-49
At5g07480 putative protein 192 2e-49
At5g24530 flavanone 3-hydroxylase-like protein 183 1e-46
At1g17010 SRG1-like protein 179 3e-45
At1g17020 SRG1-like protein 172 2e-43
At2g38240 putative anthocyanidin synthase 170 1e-42
At1g55290 leucoanthocyanidin dioxygenase 2, putative 169 2e-42
At4g22880 putative leucoanthocyanidin dioxygenase (LDOX) 168 5e-42
At1g78550 unknown protein 168 5e-42
At1g06620 oxidoreductase-like protein 166 2e-41
At3g13610 unknown protein 164 7e-41
At5g59530 1-aminocyclopropane-1-carboxylate oxidase - like protein 163 2e-40
At4g25300 SRG1-like protein 162 2e-40
At5g59540 1-aminocyclopropane-1-carboxylate oxidase - like protein 162 4e-40
At2g30840 putative dioxygenase 161 5e-40
>At2g36690 putative giberellin beta-hydroxylase
Length = 392
Score = 207 bits (528), Expect = 6e-54
Identities = 127/401 (31%), Positives = 212/401 (52%), Gaps = 51/401 (12%)
Query: 1 MASTLPPQVNQKSNNGITPFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVND 60
M + + + N+K G VK L E+ +P+ Y + P + I+ D++
Sbjct: 5 MIAPVMEEENKKYQKG------VKHLCEN-GLTKVPTKYIW---PEPDRPILTKSDKLIK 54
Query: 61 P-----IPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFIL------------------ 97
P +P+ID++ L+ N R + I +AC+ +GFF +
Sbjct: 55 PNKNLKLPLIDFAELLGPN---RPHVLRTIAEACKTYGFFQVGSLNPKAHAVISRFLAKQ 111
Query: 98 --------TNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSGD 149
NH + + + M+D FF L E++ Y +++ ++YGTSFN D
Sbjct: 112 VSYGNGQVVNHGMEGDVSKNMIDVCKRFFELPYEERSKYMSSDMSAP-VRYGTSFNQIKD 170
Query: 150 KNLFWRDFIKIIVHPKF----HSPDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEV 205
WRDF+K+ HP H P PS FR ++A Y+++T ++ ++K I ESL ++
Sbjct: 171 NVFCWRDFLKLYAHPLPDYLPHWPSSPSDFRSSAATYAKETKEMFEMMVKAILESLEIDG 230
Query: 206 NYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGK 265
+ + L+ G Q++ N YPPCP+P+L +GMPPHSD+G L LL+Q+ V GLQ+L+ +
Sbjct: 231 SD-EAAKELEEGSQVVVVNCYPPCPEPELTLGMPPHSDYGFLTLLLQDEVEGLQILYRDE 289
Query: 266 WINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPA 325
W+ V F+V V DHLEI SNG+YKSV+HR V++ R+S+A++ + L +VV+P+
Sbjct: 290 WVTVDPIPGSFVVNVGDHLEIFSNGRYKSVLHRVLVNSTKPRISVASLHSFPLTSVVKPS 349
Query: 326 SELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVKI 366
+L+D + NP+ Y+ +++ + + K+ L KI
Sbjct: 350 PKLVD-KHNPSQYMDTDFTTFLQYITSREPKWKNFLESRKI 389
>At2g44800 putative flavonol synthase
Length = 352
Score = 203 bits (516), Expect = 1e-52
Identities = 122/353 (34%), Positives = 190/353 (53%), Gaps = 19/353 (5%)
Query: 20 FTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQ-- 77
FTS TL+ S +P Y P + + +PVID SLL H
Sbjct: 8 FTSAMTLTNS-GVPQVPDRYVLP--PSQRPALGSSLGTSETTLPVIDLSLL----HQPFL 60
Query: 78 RTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDS 137
R+ IH+I AC+E+GFF + NH + S++ +D FF+L E+K + V +
Sbjct: 61 RSLAIHEISMACKEFGFFQVINHGIPSSVVNDALDAATQFFDLPVEEKMLLVSANV-HEP 119
Query: 138 IKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS----PDKPSGFRETSAEYSRKTWKLGREL 193
++YGTS N S D+ +WRDFIK HP P P +++ +Y+ T L ++L
Sbjct: 120 VRYGTSLNHSTDRVHYWRDFIKHYSHPLSKWIDMWPSNPPCYKDKVGKYAEATHLLHKQL 179
Query: 194 LKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQN 253
++ ISESLGLE NY+ + ++ G Q++A N YP CP+P++A+GMPPHSD L +L+Q+
Sbjct: 180 IEAISESLGLEKNYLQE--EIEEGSQVMAVNCYPACPEPEMALGMPPHSDFSSLTILLQS 237
Query: 254 GVSGLQVLH-NGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLAT 312
GLQ++ N W+ V +V + D +E+MSNG YKSV+HR V+ R+S A+
Sbjct: 238 S-KGLQIMDCNKNWVCVPYIEGALIVQLGDQVEVMSNGIYKSVIHRVTVNKEVKRLSFAS 296
Query: 313 VIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVK 365
+ + L + PA +L+ N +N AY D++ +N + ++ +K
Sbjct: 297 LHSLPLHKKISPAPKLV-NPNNAPAYGEFSFNDFLNYISSNDFIQERFIDTIK 348
>At4g10500 putative Fe(II)/ascorbate oxidase
Length = 349
Score = 202 bits (515), Expect = 2e-52
Identities = 120/335 (35%), Positives = 179/335 (52%), Gaps = 13/335 (3%)
Query: 35 IPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGF 94
IPS+Y P + +++ + D IP+ID L N R + + AC +GF
Sbjct: 20 IPSNYV---RPISDRPNLSEVESSGDSIPLIDLRDLHGPN---RAVIVQQLASACSTYGF 73
Query: 95 FILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFW 154
F + NH V + + KM FF+ E ++ + + T + + TSFNV DK L W
Sbjct: 74 FQIKNHGVPDTTVNKMQTVAREFFHQPESERVKHYSADPTKTT-RLSTSFNVGADKVLNW 132
Query: 155 RDFIKIIVHPKFHS----PDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDK 210
RDF+++ P P P FRE +AEY+ L LL+ ISESLGLE ++I
Sbjct: 133 RDFLRLHCFPIEDFIEEWPSSPISFREVTAEYATSVRALVLRLLEAISESLGLESDHISN 192
Query: 211 TMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVS 270
+ + Q +A N YPPCP+P+L G+P H D ++ +L+Q+ VSGLQV + KW+ VS
Sbjct: 193 ILGKHA--QHMAFNYYPPCPEPELTYGLPGHKDPTVITVLLQDQVSGLQVFKDDKWVAVS 250
Query: 271 STSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLD 330
N F+V + D ++++SN KYKSV+HRA V+ R+S+ T PS D V+ PA EL++
Sbjct: 251 PIPNTFIVNIGDQMQVISNDKYKSVLHRAVVNTENERLSIPTFYFPSTDAVIGPAHELVN 310
Query: 331 NESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVK 365
+ + A Y ++Y N L S L+ K
Sbjct: 311 EQDSLAIYRTYPFVEYWDKFWNRSLATASCLDAFK 345
>At4g10490 putative flavanone 3-beta-hydroxylase
Length = 348
Score = 201 bits (512), Expect = 4e-52
Identities = 118/319 (36%), Positives = 177/319 (54%), Gaps = 14/319 (4%)
Query: 35 IPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGF 94
+PS+Y P + +++ D IP+ID L N R I+ AC GF
Sbjct: 18 VPSNYV---RPVSDRPKMSEVQTSGDSIPLIDLHDLHGPN---RADIINQFAHACSSCGF 71
Query: 95 FILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFW 154
F + NH V + ++KM++ FF E ++ + + T + + TSFNVS +K W
Sbjct: 72 FQIKNHGVPEETIKKMMNAAREFFRQSESERVKHYSAD-TKKTTRLSTSFNVSKEKVSNW 130
Query: 155 RDFIKIIVHP--KFHS--PDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDK 210
RDF+++ +P F + P P FRE +AEY+ L LL+ ISESLGL + +
Sbjct: 131 RDFLRLHCYPIEDFINEWPSTPISFREVTAEYATSVRALVLTLLEAISESLGLAKDRVSN 190
Query: 211 TMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVS 270
T+ Q +A N YP CPQP+L G+P H D L+ +L+Q+ VSGLQV +GKWI V+
Sbjct: 191 TIGKHG--QHMAINYYPRCPQPELTYGLPGHKDANLITVLLQDEVSGLQVFKDGKWIAVN 248
Query: 271 STSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELL- 329
N F+V + D ++++SN KYKSV+HRA V++ R+S+ T PS D V+ PA EL+
Sbjct: 249 PVPNTFIVNLGDQMQVISNEKYKSVLHRAVVNSDMERISIPTFYCPSEDAVISPAQELIN 308
Query: 330 DNESNPAAYVGMKHIDYMK 348
+ E +PA Y + +Y +
Sbjct: 309 EEEDSPAIYRNFTYAEYFE 327
>At3g21420 unknown protein
Length = 364
Score = 197 bits (500), Expect = 1e-50
Identities = 113/312 (36%), Positives = 170/312 (54%), Gaps = 11/312 (3%)
Query: 62 IPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLK 121
IPVID S L ++D I + +ACE+WGFF + NH + ++E + + FF++
Sbjct: 55 IPVIDLSKLSKPDNDDFFFEILKLSQACEDWGFFQVINHGIEVEVVEDIEEVASEFFDMP 114
Query: 122 EEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFR 176
E+K+ Y + T YG +F S D+ L W + + VHP P KP+ F
Sbjct: 115 LEEKKKYPMEPGTVQG--YGQAFIFSEDQKLDWCNMFALGVHPPQIRNPKLWPSKPARFS 172
Query: 177 ETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAM 236
E+ YS++ +L + LLK I+ SLGL+ ++ +Q + N YPPC PDL +
Sbjct: 173 ESLEGYSKEIRELCKRLLKYIAISLGLKEERFEEMFG--EAVQAVRMNYYPPCSSPDLVL 230
Query: 237 GMPPHSDHGLLNLLIQ--NGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKS 294
G+ PHSD L +L Q N GLQ+L + W+ V N ++ + D +E++SNGKYKS
Sbjct: 231 GLSPHSDGSALTVLQQSKNSCVGLQILKDNTWVPVKPLPNALVINIGDTIEVLSNGKYKS 290
Query: 295 VVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQ 354
V HRA + R+++ T AP+ + +EP SEL+D+E+NP Y H DY +N+
Sbjct: 291 VEHRAVTNREKERLTIVTFYAPNYEVEIEPMSELVDDETNPCKYRSYNHGDYSYHYVSNK 350
Query: 355 LYGKSVLNKVKI 366
L GK L+ KI
Sbjct: 351 LQGKKSLDFAKI 362
>At3g11180 putative leucoanthocyanidin dioxygenase
Length = 400
Score = 193 bits (490), Expect = 1e-49
Identities = 122/348 (35%), Positives = 178/348 (51%), Gaps = 15/348 (4%)
Query: 19 PFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVND-PIPVIDYSLLINGNHDQ 77
P V++L+ES + S+P Y + + I+ Q EV D IP+ID L +GN D
Sbjct: 52 PIVRVQSLAES-NLTSLPDRYIKPPSQRPQTTIIDHQPEVADINIPIIDLDSLFSGNEDD 110
Query: 78 RTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDS 137
+ + I +AC EWGFF + NH V LM+ + +FFNL E K+VY++ T +
Sbjct: 111 KKR----ISEACREWGFFQVINHGVKPELMDAARETWKSFFNLPVEAKEVYSNSPRTYEG 166
Query: 138 IKYGTSFNVSGDKNLFWRDFIKIIVHP----KFHS-PDKPSGFRETSAEYSRKTWKLGRE 192
YG+ V L W D+ + P F+ P PS RE + EY ++ KLG
Sbjct: 167 --YGSRLGVEKGAILDWNDYYYLHFLPLALKDFNKWPSLPSNIREMNDEYGKELVKLGGR 224
Query: 193 LLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQ 252
L+ +S +LGL + + + L N YP CPQP+LA+G+ PHSD G + +L+
Sbjct: 225 LMTILSSNLGLRAEQLQEAFGGEDVGACLRVNYYPKCPQPELALGLSPHSDPGGMTILLP 284
Query: 253 NG-VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLA 311
+ V GLQV H WI V+ + F+V + D ++I+SN KYKSV HR V++ R+SLA
Sbjct: 285 DDQVVGLQVRHGDTWITVNPLRHAFIVNIGDQIQILSNSKYKSVEHRVIVNSEKERVSLA 344
Query: 312 TVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKS 359
P D ++P +L+ + P Y M Y R GKS
Sbjct: 345 FFYNPKSDIPIQPMQQLV-TSTMPPLYPPMTFDQYRLFIRTQGPRGKS 391
>At5g07480 putative protein
Length = 356
Score = 192 bits (488), Expect = 2e-49
Identities = 111/354 (31%), Positives = 192/354 (53%), Gaps = 29/354 (8%)
Query: 24 KTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIH 83
++L PD +P S + P D N + +P ID S L G D+R I
Sbjct: 19 RSLPYVPDCYVVPPS----SKPCDSNSGI---------VPTIDVSRL-KGGDDERRGVIQ 64
Query: 84 DIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTS 143
++ AC+ GFF + NH +++++++ ++ +FF L ++K+ + +V ++Y TS
Sbjct: 65 ELSLACQHLGFFQIVNHGINQNILDDALEVANSFFELPAKEKKQFMSNDVYAP-VRYSTS 123
Query: 144 FNVSGDKNLFWRDFIKIIVHPKFHS----PDKPSGFRETSAEYSRKTWKLGRELLKGISE 199
D FWR F+K HP P+ P G+RE ++ + KL EL+ I+E
Sbjct: 124 LKDGLDTIQFWRIFLKHYAHPLHRWIHLWPENPPGYREKMGKFCEEVRKLSIELMGAITE 183
Query: 200 SLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQ 259
SLGL +Y+ M+ ++G+Q++ N YPPCP P+ A+G+PPHSD+ + LL+QN + GL+
Sbjct: 184 SLGLGRDYLSSRMD-ENGMQVMTVNCYPPCPDPETALGLPPHSDYSCITLLLQN-LDGLK 241
Query: 260 VLH------NGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATV 313
+ +G+W+ V + V + DH+E++SNG YKS+VH+ ++ TR+SLA++
Sbjct: 242 IFDPMAHGGSGRWVGVPQVTGVLKVHIGDHVEVLSNGLYKSIVHKVTLNEEKTRISLASL 301
Query: 314 IAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMK-LQRNNQLYGKSVLNKVKI 366
+ +D + EL+ N+ NP Y D++ L +N+ G ++ ++I
Sbjct: 302 HSLGMDDKMSVPRELV-NDENPVRYKESSFNDFLDFLVKNDISQGDRFIDTLRI 354
>At5g24530 flavanone 3-hydroxylase-like protein
Length = 341
Score = 183 bits (464), Expect = 1e-46
Identities = 108/324 (33%), Positives = 187/324 (57%), Gaps = 17/324 (5%)
Query: 35 IPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGF 94
I + + +TT P + ++D+ +++ + D+ L+ + D R+ I I +AC +GF
Sbjct: 6 ISTGFRHTTLPENYVRPISDRPRLSEVSQLEDFPLIDLSSTD-RSFLIQQIHQACARFGF 64
Query: 95 FILTNHSVSKSLMEKMVDQVFAFFNLKEEDK-QVYADKEVTDDSIKYGTSFNVSGDKNLF 153
F + NH V+K ++++MV FF++ E+K ++Y+D + + TSFNV ++
Sbjct: 65 FQVINHGVNKQIIDEMVSVAREFFSMSMEEKMKLYSDDPTK--TTRLSTSFNVKKEEVNN 122
Query: 154 WRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYI 208
WRD++++ +P H P P F+E ++YSR+ ++G ++ + ISESLGLE +Y+
Sbjct: 123 WRDYLRLHCYP-IHKYVNEWPSNPPSFKEIVSKYSREVREVGFKIEELISESLGLEKDYM 181
Query: 209 DKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNG-VSGLQVLHNGKWI 267
K + Q +A N YPPCP+P+L G+P H+D L +L+Q+ V GLQ+L +G+W
Sbjct: 182 KKVLGEQG--QHMAVNYYPPCPEPELTYGLPAHTDPNALTILLQDTTVCGLQILIDGQWF 239
Query: 268 NVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPAS- 326
V+ + F++ + D L+ +SNG YKSV HRA + R+S+A+ + P+ V+ PA
Sbjct: 240 AVNPHPDAFVINIGDQLQALSNGVYKSVWHRAVTNTENPRLSVASFLCPADCAVMSPAKP 299
Query: 327 --ELLDNESNPAAYVGMKHIDYMK 348
E D+E+ P Y + +Y K
Sbjct: 300 LWEAEDDETKP-VYKDFTYAEYYK 322
>At1g17010 SRG1-like protein
Length = 361
Score = 179 bits (453), Expect = 3e-45
Identities = 115/352 (32%), Positives = 186/352 (52%), Gaps = 16/352 (4%)
Query: 22 SVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKT 81
SV+ + + ++P Y D+ E+V + IP+ID + L +
Sbjct: 16 SVQEMVKDKMITTVPPRYV--RYDQDKTEVVVHDSGLISEIPIIDMNRLCSSTAVD--SE 71
Query: 82 IHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLK-EEDKQVYADKEVTDDSIKY 140
+ + AC+E+GFF L NH + S ++K+ ++ FFNL EE K+++ V + +
Sbjct: 72 VEKLDFACKEYGFFQLVNHGIDPSFLDKIKSEIQDFFNLPMEEKKKLWQTPAVMEG---F 128
Query: 141 GTSFNVSGDKNLFWRDFIKIIVHP----KFHS-PDKPSGFRETSAEYSRKTWKLGRELLK 195
G +F VS D+ L W D +I+ P K H P P FR+T YS + + + LL
Sbjct: 129 GQAFVVSEDQKLDWADLFFLIMQPVQLRKRHLFPKLPLPFRDTLDMYSTRVKSIAKILLA 188
Query: 196 GISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQ-NG 254
++++L ++ +++ D +Q + N YPPCPQP+L G+ PHSD L +L+Q N
Sbjct: 189 KMAKALQIKPEEVEEIFG-DDMMQSMRMNYYPPCPQPNLVTGLIPHSDAVGLTILLQVNE 247
Query: 255 VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVI 314
V GLQ+ NGKW V N F+V V D LEI++NG Y+S+ HRA V+ R+S+AT
Sbjct: 248 VDGLQIKKNGKWFFVKPLQNAFIVNVGDVLEIITNGTYRSIEHRAMVNLEKERLSIATFH 307
Query: 315 APSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVKI 366
+D + PA L+ + A + +K DY+ + +L GK+ L+ ++I
Sbjct: 308 NTGMDKEIGPARSLVQRQ-EAAKFRSLKTKDYLNGLFSRELKGKAYLDAMRI 358
>At1g17020 SRG1-like protein
Length = 358
Score = 172 bits (437), Expect = 2e-43
Identities = 110/351 (31%), Positives = 180/351 (50%), Gaps = 14/351 (3%)
Query: 22 SVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKT 81
SV+ + + ++P Y + E V D +V IP+ID L +
Sbjct: 16 SVQEMVKEKTITTVPPRYVRSDQDKTE---VDDDFDVKIEIPIIDMKRLCSST--TMDSE 70
Query: 82 IHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYG 141
+ + AC+EWGFF L NH + S ++K+ ++ FFNL E+K+ + + D+ +G
Sbjct: 71 VEKLDFACKEWGFFQLVNHGIDSSFLDKVKSEIQDFFNLPMEEKKKFWQRP--DEIEGFG 128
Query: 142 TSFNVSGDKNLFWRDFIKIIVHP----KFHS-PDKPSGFRETSAEYSRKTWKLGRELLKG 196
+F VS D+ L W D V P K H P P FR+T YS + + + L+
Sbjct: 129 QAFVVSEDQKLDWADLFFHTVQPVELRKPHLFPKLPLPFRDTLEMYSSEVQSVAKILIAK 188
Query: 197 ISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQ-NGV 255
++ +L ++ ++K + +Q + N YPPCPQPD +G+ PHSD L +L+Q N V
Sbjct: 189 MARALEIKPEELEKLFDDVDSVQSMRMNYYPPCPQPDQVIGLTPHSDSVGLTVLMQVNDV 248
Query: 256 SGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIA 315
GLQ+ +GKW+ V N F+V + D LEI++NG Y+S+ HR V++ R+S+AT
Sbjct: 249 EGLQIKKDGKWVPVKPLPNAFIVNIGDVLEIITNGTYRSIEHRGVVNSEKERLSIATFHN 308
Query: 316 PSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVKI 366
+ V PA L++ + A + + +Y + L GK+ L+ ++I
Sbjct: 309 VGMYKEVGPAKSLVERQ-KVARFKRLTMKEYNDGLFSRTLDGKAYLDALRI 358
>At2g38240 putative anthocyanidin synthase
Length = 353
Score = 170 bits (430), Expect = 1e-42
Identities = 115/353 (32%), Positives = 176/353 (49%), Gaps = 18/353 (5%)
Query: 19 PFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQR 78
P SV++LS++ ++P+ Y H Q + IPV+D + + +
Sbjct: 8 PIVSVQSLSQT-GVPTVPNRYVKPA--HQRPVFNTTQSDAGIEIPVLDMNDVWG-----K 59
Query: 79 TKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSI 138
+ + + ACEEWGFF + NH V+ SLME++ FF L E+K+ YA+ T +
Sbjct: 60 PEGLRLVRSACEEWGFFQMVNHGVTHSLMERVRGAWREFFELPLEEKRKYANSPDTYEG- 118
Query: 139 KYGTSFNVSGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGREL 193
YG+ V D L W D+ + P P +P RE +Y + KL L
Sbjct: 119 -YGSRLGVVKDAKLDWSDYFFLNYLPSSIRNPSKWPSQPPKIRELIEKYGEEVRKLCERL 177
Query: 194 LKGISESLGLEVNYIDKTMNL-DSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQ 252
+ +SESLGL+ N + + + D L N YP CPQP L +G+ HSD G + +L+
Sbjct: 178 TETLSESLGLKPNKLMQALGGGDKVGASLRTNFYPKCPQPQLTLGLSSHSDPGGITILLP 237
Query: 253 NG-VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLA 311
+ V+GLQV W+ + S N +V + D L+I+SNG YKSV H+ V++G R+SLA
Sbjct: 238 DEKVAGLQVRRGDGWVTIKSVPNALIVNIGDQLQILSNGIYKSVEHQVIVNSGMERVSLA 297
Query: 312 TVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKV 364
P D V P EL+ + PA Y ++ +Y L R GK+ ++ +
Sbjct: 298 FFYNPRSDIPVGPIEELV-TANRPALYKPIRFDEYRSLIRQKGPCGKNQVDSL 349
>At1g55290 leucoanthocyanidin dioxygenase 2, putative
Length = 361
Score = 169 bits (429), Expect = 2e-42
Identities = 112/344 (32%), Positives = 181/344 (52%), Gaps = 18/344 (5%)
Query: 10 NQKSNNGITPFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDP-IPVIDYS 68
+Q +N + VK LSE+ +P Y P +E I E +D IPVID S
Sbjct: 13 DQVTNFVVHEGNGVKGLSET-GIKVLPDQYI---QPFEERLINFHVKEDSDESIPVIDIS 68
Query: 69 LLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVY 128
N + +K + D A EEWGFF + NH VS ++E M FF L E+K+ +
Sbjct: 69 ---NLDEKSVSKAVCD---AAEEWGFFQVINHGVSMEVLENMKTATHRFFGLPVEEKRKF 122
Query: 129 ADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKI-IVHPKFHSPDKPSGFRETSAEYSRKTW 187
+ ++ ++++GTSF+ +K L W+D++ + V S P R + EY +T
Sbjct: 123 SREKSLSTNVRFGTSFSPHAEKALEWKDYLSLFFVSEAEASQLWPDSCRSETLEYMNETK 182
Query: 188 KLGRELLKGISESLGLEVNYIDKTM-NLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGL 246
L ++LL+ + E+ L V +DKT + G + N YP CP P+L +G+ HSD
Sbjct: 183 PLVKKLLRFLGEN--LNVKELDKTKESFFMGSTRINLNYYPICPNPELTVGVGRHSDVSS 240
Query: 247 LNLLIQNGVSGLQV--LHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNG 304
L +L+Q+ + GL V L G+W++V S ++ + D ++IMSNG+YKSV HR +
Sbjct: 241 LTILLQDEIGGLHVRSLTTGRWVHVPPISGSLVINIGDAMQIMSNGRYKSVEHRVLANGS 300
Query: 305 ATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMK 348
R+S+ ++P ++V+ P E+++N P Y + + DY+K
Sbjct: 301 YNRISVPIFVSPKPESVIGPLLEVIENGEKP-VYKDILYTDYVK 343
>At4g22880 putative leucoanthocyanidin dioxygenase (LDOX)
Length = 356
Score = 168 bits (425), Expect = 5e-42
Identities = 104/337 (30%), Positives = 171/337 (49%), Gaps = 20/337 (5%)
Query: 34 SIPSSYTYTTNPHDENEIVAD-----QDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKA 88
SIP Y P +E E + D + E +P ID + + + R I ++ KA
Sbjct: 17 SIPKEYI---RPKEELESINDVFLEEKKEDGPQVPTIDLKNIESDDEKIRENCIEELKKA 73
Query: 89 CEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSG 148
+WG L NH + LME++ FF+L E+K+ YA+ + T YG+ +
Sbjct: 74 SLDWGVMHLINHGIPADLMERVKKAGEEFFSLSVEEKEKYANDQATGKIQGYGSKLANNA 133
Query: 149 DKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGRELLKGISESLGL 203
L W D+ + +P+ P PS + E ++EY++ L ++ K +S LGL
Sbjct: 134 SGQLEWEDYFFHLAYPEEKRDLSIWPKTPSDYIEATSEYAKCLRLLATKVFKALSVGLGL 193
Query: 204 EVNYIDKTMN-LDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLH 262
E + ++K + L+ L + N YP CPQP+LA+G+ H+D L ++ N V GLQ+ +
Sbjct: 194 EPDRLEKEVGGLEELLLQMKINYYPKCPQPELALGVEAHTDVSALTFILHNMVPGLQLFY 253
Query: 263 NGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVV 322
GKW+ + ++ + D LEI+SNGKYKS++HR V+ R+S A P D +V
Sbjct: 254 EGKWVTAKCVPDSIVMHIGDTLEILSNGKYKSILHRGLVNKEKVRISWAVFCEPPKDKIV 313
Query: 323 -EPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGK 358
+P E++ ES PA + +++ ++L+GK
Sbjct: 314 LKPLPEMVSVES-PAKFPPRTFAQHIE----HKLFGK 345
>At1g78550 unknown protein
Length = 356
Score = 168 bits (425), Expect = 5e-42
Identities = 114/356 (32%), Positives = 181/356 (50%), Gaps = 18/356 (5%)
Query: 17 ITPFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHD 76
I PF V + + +F +IP Y ++ EI+ D ++ IPVID + L + +
Sbjct: 13 IVPF--VLEIVKEKNFTTIPPRYVRVDQ--EKTEILNDSS-LSSEIPVIDMTRLCSVS-- 65
Query: 77 QRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDD 136
+ + AC++WGFF L NH + S +EK+ +V FFNL ++KQ + +
Sbjct: 66 AMDSELKKLDFACQDWGFFQLVNHGIDSSFLEKLETEVQEFFNLPMKEKQKLWQRSGEFE 125
Query: 137 SIKYGTSFNVSGDKNLFWRDFIKIIVHP----KFHSPDK-PSGFRETSAEYSRKTWKLGR 191
+G VS ++ L W D + P K H K P FRET YS + + +
Sbjct: 126 G--FGQVNIVSENQKLDWGDMFILTTEPIRSRKSHLFSKLPPPFRETLETYSSEVKSIAK 183
Query: 192 ELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLI 251
L ++ L E+ + + D Q + N YPPCPQPD MG+ HSD L +L+
Sbjct: 184 ILFAKMASVL--EIKHEEMEDLFDDVWQSIKINYYPPCPQPDQVMGLTQHSDAAGLTILL 241
Query: 252 Q-NGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSL 310
Q N V GLQ+ +GKW+ V + +V V + LEI++NG+Y+S+ HRA V++ R+S+
Sbjct: 242 QVNQVEGLQIKKDGKWVVVKPLRDALVVNVGEILEIITNGRYRSIEHRAVVNSEKERLSV 301
Query: 311 ATVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVKI 366
A +P +T++ PA L+D + + M +Y +L GKS L+ ++I
Sbjct: 302 AMFHSPGKETIIRPAKSLVDRQKQ-CLFKSMSTQEYFDAFFTQKLNGKSHLDLMRI 356
>At1g06620 oxidoreductase-like protein
Length = 365
Score = 166 bits (420), Expect = 2e-41
Identities = 106/311 (34%), Positives = 163/311 (52%), Gaps = 13/311 (4%)
Query: 62 IPVIDYSLLINGNHDQRTKT--IHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFN 119
IP ID L G D T+ + IG A E+WGFF + NH + ++EKM+D + F
Sbjct: 62 IPTID---LKGGGTDSITRRSLVEKIGDAAEKWGFFQVINHGIPMDVLEKMIDGIREFHE 118
Query: 120 LKEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHSP-DKPSGFRET 178
E K+ + ++ + Y ++F++ WRD + P P D P+ E
Sbjct: 119 QDTEVKKGFYSRDPASKMV-YSSNFDLFSSPAANWRDTLGCYTAPDPPRPEDLPATCGEM 177
Query: 179 SAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGM 238
EYS++ KLG+ L + +SE+LGL N++ K M+ + L +L + YPPCPQPDL +G+
Sbjct: 178 MIEYSKEVMKLGKLLFELLSEALGLNTNHL-KDMDCTNSL-LLLGHYYPPCPQPDLTLGL 235
Query: 239 PPHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHR 298
HSD+ L +L+Q+ + GLQVLH+ W++V +V V D L++++N K+ SV HR
Sbjct: 236 TKHSDNSFLTILLQDHIGGLQVLHDQYWVDVPPVPGALVVNVGDLLQLITNDKFISVEHR 295
Query: 299 AAVSNGATRMSLATVIAPSL---DTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQL 355
+ R+S+A + L V P E+L +E NP Y +Y K R+
Sbjct: 296 VLANVAGPRISVACFFSSYLMANPRVYGPIKEIL-SEENPPNYRDTTITEYAKFYRSKGF 354
Query: 356 YGKSVLNKVKI 366
G S L +KI
Sbjct: 355 DGTSGLLYLKI 365
>At3g13610 unknown protein
Length = 361
Score = 164 bits (415), Expect = 7e-41
Identities = 108/348 (31%), Positives = 182/348 (52%), Gaps = 17/348 (4%)
Query: 23 VKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTI 82
VK LSE+ ++P Y P +E I +E ++ IPVID S N + D+ + +
Sbjct: 27 VKGLSET-GIKALPEQYI---QPLEERLINKFVNETDEAIPVIDMS---NPDEDRVAEAV 79
Query: 83 HDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGT 142
D A E+WGFF + NH V +++ + FFNL E+K+ + + ++++GT
Sbjct: 80 CD---AAEKWGFFQVINHGVPLEVLDDVKAATHKFFNLPVEEKRKFTKENSLSTTVRFGT 136
Query: 143 SFNVSGDKNLFWRDFIKIIVHPKFHSPDK-PSGFRETSAEYSRKTWKLGRELLKGISESL 201
SF+ ++ L W+D++ + + + P R + EY K+ K+ R LL+ + ++L
Sbjct: 137 SFSPLAEQALEWKDYLSLFFVSEAEAEQFWPDICRNETLEYINKSKKMVRRLLEYLGKNL 196
Query: 202 GLEVNYIDKTM-NLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQV 260
V +D+T +L G + N YP CP PDL +G+ HSD L +L+Q+ + GL V
Sbjct: 197 N--VKELDETKESLFMGSIRVNLNYYPICPNPDLTVGVGRHSDVSSLTILLQDQIGGLHV 254
Query: 261 --LHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSL 318
L +G W++V + F++ + D ++IMSNG YKSV HR + R+S+ + P
Sbjct: 255 RSLASGNWVHVPPVAGSFVINIGDAMQIMSNGLYKSVEHRVLANGYNNRISVPIFVNPKP 314
Query: 319 DTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVKI 366
++V+ P E++ N P Y + + DY+K GK ++ KI
Sbjct: 315 ESVIGPLPEVIANGEEP-IYRDVLYSDYVKYFFRKAHDGKKTVDYAKI 361
>At5g59530 1-aminocyclopropane-1-carboxylate oxidase - like protein
Length = 364
Score = 163 bits (412), Expect = 2e-40
Identities = 105/313 (33%), Positives = 169/313 (53%), Gaps = 14/313 (4%)
Query: 62 IPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLK 121
IP ID++ +N + R + + A E WGFF + NH V +++E++ D V F +
Sbjct: 58 IPTIDFAS-VNVDTPSREAIVEKVKYAVENWGFFQVINHGVPLNVLEEIKDGVRRFH--E 114
Query: 122 EEDKQV---YADKEVTDDSIKYGTSFNV-SGDKNLFWRDFIKIIVHPKFHSPDK-PSGFR 176
EED +V Y + T + Y ++F++ S +L WRD I + P +P++ P R
Sbjct: 115 EEDPEVKKSYYSLDFTKNKFAYSSNFDLYSSSPSLTWRDSISCYMAPDPPTPEELPETCR 174
Query: 177 ETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAM 236
+ EYS+ LG L + +SE+LGL+ + K+M+ L M+ + YPPCPQPDL +
Sbjct: 175 DAMIEYSKHVLSLGDLLFELLSEALGLKSEIL-KSMDCLKSLLMIC-HYYPPCPQPDLTL 232
Query: 237 GMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVV 296
G+ HSD+ L +L+Q+ + GLQ+LH W++VS +V V D L++++N K+ SV
Sbjct: 233 GISKHSDNSFLTVLLQDNIGGLQILHQDSWVDVSPLPGALVVNVGDFLQLITNDKFISVE 292
Query: 297 HRAAVSNGATRMSLATVIAPSL---DTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNN 353
HR + R+S+A+ + S+ TV P EL+ +E NP Y +Y +
Sbjct: 293 HRVLANTRGPRISVASFFSSSIRENSTVYGPMKELV-SEENPPKYRDTTLREYSEGYFKK 351
Query: 354 QLYGKSVLNKVKI 366
L G S L+ +I
Sbjct: 352 GLDGTSHLSNFRI 364
>At4g25300 SRG1-like protein
Length = 356
Score = 162 bits (411), Expect = 2e-40
Identities = 111/351 (31%), Positives = 178/351 (50%), Gaps = 15/351 (4%)
Query: 22 SVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKT 81
SV+ + + ++P Y + D EI D + + IP+ID SLL +
Sbjct: 15 SVQEMVKEKMITTVPPRYV--RSDQDVAEIAVDSG-LRNQIPIIDMSLLCSST--SMDSE 69
Query: 82 IHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYG 141
I + AC+EWGFF L NH + S + K+ +V FFNL E+K+ + D+ +G
Sbjct: 70 IDKLDSACKEWGFFQLVNHGMESSFLNKVKSEVQDFFNLPMEEKKNLWQQP--DEIEGFG 127
Query: 142 TSFNVSGDKNLFWRDFIKIIVHP----KFHS-PDKPSGFRETSAEYSRKTWKLGRELLKG 196
F VS ++ L W D + + P K H P P FR+T YS + + + LL
Sbjct: 128 QVFVVSEEQKLDWADMFFLTMQPVRLRKPHLFPKLPLPFRDTLDMYSAEVKSIAKILLGK 187
Query: 197 ISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQ-NGV 255
I+ +L ++ +DK + + G Q + N YP CP+PD +G+ PHSD L +L+Q N V
Sbjct: 188 IAVALKIKPEEMDKLFDDELG-QRIRLNYYPRCPEPDKVIGLTPHSDSTGLTILLQANEV 246
Query: 256 SGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIA 315
GLQ+ N KW++V N +V V D LEI++NG Y+S+ HR V++ R+S+A
Sbjct: 247 EGLQIKKNAKWVSVKPLPNALVVNVGDILEIITNGTYRSIEHRGVVNSEKERLSVAAFHN 306
Query: 316 PSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVKI 366
L + P L++ A + + +Y + +L GK+ L+ +++
Sbjct: 307 IGLGKEIGPMRSLVERH-KAAFFKSVTTEEYFNGLFSRELDGKAYLDVMRL 356
>At5g59540 1-aminocyclopropane-1-carboxylate oxidase - like protein
Length = 366
Score = 162 bits (409), Expect = 4e-40
Identities = 96/310 (30%), Positives = 168/310 (53%), Gaps = 9/310 (2%)
Query: 62 IPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLK 121
IP+ID++ ++ + R + + A E WGFF + NHS+ +++E++ D V F
Sbjct: 61 IPIIDFAS-VHADTASREAIVEKVKYAVENWGFFQVINHSIPLNVLEEIKDGVRRFHEED 119
Query: 122 EEDKQVYADKEVTDDSIKYGTSFNV-SGDKNLFWRDFIKIIVHPKFHSPDK-PSGFRETS 179
E K+ + ++ + Y ++F++ S ++ WRD + P +P++ P R+
Sbjct: 120 PEVKKSFFSRDAGNKKFVYNSNFDLYSSSPSVNWRDSFSCYIAPDPPAPEEIPETCRDAM 179
Query: 180 AEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMP 239
EYS+ G L + +SE+LGL+ ++ +M+ L M+ + YPPCPQPDL +G+
Sbjct: 180 FEYSKHVLSFGGLLFELLSEALGLKSQTLE-SMDCVKTLLMIC-HYYPPCPQPDLTLGIT 237
Query: 240 PHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRA 299
HSD+ L LL+Q+ + GLQ+LH W++VS +V + D L++++N K+ SV HR
Sbjct: 238 KHSDNSFLTLLLQDNIGGLQILHQDSWVDVSPIHGALVVNIGDFLQLITNDKFVSVEHRV 297
Query: 300 AVSNGATRMSLATVIAPSL---DTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLY 356
+ R+S+A+ + S+ V P EL+ +E NP Y + +Y K+ L
Sbjct: 298 LANRQGPRISVASFFSSSMRPNSRVYGPMKELV-SEENPPKYRDITIKEYSKIFFEKGLD 356
Query: 357 GKSVLNKVKI 366
G S L+ ++I
Sbjct: 357 GTSHLSNIRI 366
>At2g30840 putative dioxygenase
Length = 362
Score = 161 bits (408), Expect = 5e-40
Identities = 111/314 (35%), Positives = 164/314 (51%), Gaps = 17/314 (5%)
Query: 62 IPVIDYSLLINGNHDQRTKT----IHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAF 117
IP ID L G D+ T T I I A E +GFF + NH +S +MEKM D + F
Sbjct: 57 IPTID---LKGGVFDEYTVTRESVIAMIRDAVERFGFFQVINHGISNDVMEKMKDGIRGF 113
Query: 118 FNLKEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKF-HSPDKPSGFR 176
+ ++ + ++VT ++KY ++F++ + WRD + + P + D P
Sbjct: 114 HEQDSDVRKKFYTRDVTK-TVKYNSNFDLYSSPSANWRDTLSCFMAPDVPETEDLPDICG 172
Query: 177 ETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAM 236
E EY+++ KLG + + +SE+LGL N++ K M+ GL ML+ + YPPCP+P L
Sbjct: 173 EIMLEYAKRVMKLGELIFELLSEALGLNPNHL-KEMDCTKGLLMLS-HYYPPCPEPGLTF 230
Query: 237 GMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVV 296
G PHSD L +L+Q+ + GLQV NG W++V LV + D L++M+N ++ SV
Sbjct: 231 GTSPHSDRSFLTILLQDHIGGLQVRQNGYWVDVPPVPGALLVNLGDLLQLMTNDQFVSVE 290
Query: 297 HRAAVSNG-ATRMSLATVIA---PSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRN 352
HR + G R+S+A+ PSL V P ELL +E N Y +Y
Sbjct: 291 HRVLANKGEKPRISVASFFVHPLPSL-RVYGPIKELL-SEQNLPKYRDTTVTEYTSHYMA 348
Query: 353 NQLYGKSVLNKVKI 366
LYG SVL KI
Sbjct: 349 RGLYGNSVLLDFKI 362
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,648,747
Number of Sequences: 26719
Number of extensions: 383567
Number of successful extensions: 1179
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 95
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 862
Number of HSP's gapped (non-prelim): 114
length of query: 366
length of database: 11,318,596
effective HSP length: 101
effective length of query: 265
effective length of database: 8,619,977
effective search space: 2284293905
effective search space used: 2284293905
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144517.6


