

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144517.16 - phase: 0
(445 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
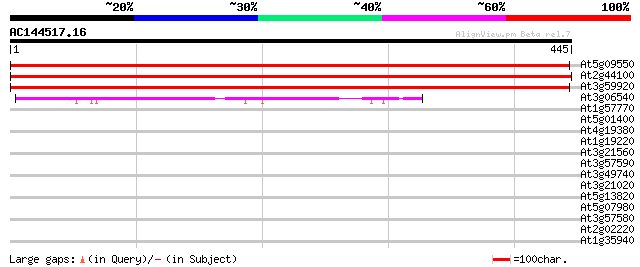
Score E
Sequences producing significant alignments: (bits) Value
At5g09550 GDP dissociation inhibitor 776 0.0
At2g44100 GDP dissociation inhibitor 725 0.0
At3g59920 Rab GDP dissociation inhibitor 721 0.0
At3g06540 Rab escort protein, putative 110 1e-24
At1g57770 unknown protein 35 0.064
At5g01400 putative protein 33 0.24
At4g19380 unknown protein 31 1.2
At1g19220 auxin response factor (ARF) like protein 30 2.7
At3g21560 UDP-glucose:indole-3-acetate beta-D-glucosyltransferas... 30 3.5
At3g57590 putative protein 29 4.6
At3g49740 putative protein 29 6.0
At3g21020 unknown protein 29 6.0
At5g13820 telomeric DNA-binding protein 1 (TBP1) 28 7.8
At5g07980 putative protein 28 7.8
At3g57580 unknown protein 28 7.8
At2g02220 putative receptor-like protein kinase 28 7.8
At1g35940 hypothetical protein 28 7.8
>At5g09550 GDP dissociation inhibitor
Length = 445
Score = 776 bits (2003), Expect = 0.0
Identities = 377/444 (84%), Positives = 405/444 (90%)
Query: 1 MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDQNDYYGGASTSLNLTQLFKRYRGDDK 60
MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMD+NDYYGG S+SLNLTQL+KR+RG D
Sbjct: 1 MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGESSSLNLTQLWKRFRGSDT 60
Query: 61 PPEELGSSREYNVDMIPKFMMANGALVRVLIHTDVTKYLNFKAVDGSFVYNKGKIYKVPA 120
P E LG+SREYNVDMIPKF+MANG LV+ LIHTDVTKYLNFKAVDGSFVY KGKIYKVPA
Sbjct: 61 PEENLGASREYNVDMIPKFIMANGLLVQTLIHTDVTKYLNFKAVDGSFVYKKGKIYKVPA 120
Query: 121 TDVEALKSPLMGLFEKRRARKFFIYVQDYEANDPKSHEGLDLNQVTARQLISKYGLEDDT 180
TDVEALKSPLMGLFEKRRARKFFIYVQDY+ DPKSHEGLDL++VTAR++ISKYGLEDDT
Sbjct: 121 TDVEALKSPLMGLFEKRRARKFFIYVQDYDEKDPKSHEGLDLSKVTAREIISKYGLEDDT 180
Query: 181 VDFIGHALALHLDDSYLDKPAKDFVDRVKTYAESLARFQGGSPYIYPLYGLGELPQAFAR 240
+DFIGHALALH DD YLD+PA DFV R+K YAESLARFQGGSPYIYPLYGLGELPQAFAR
Sbjct: 181 IDFIGHALALHNDDDYLDQPAIDFVKRIKLYAESLARFQGGSPYIYPLYGLGELPQAFAR 240
Query: 241 LSAVYGGTYMLNKPECKVEFDGDGKAIGVTSDGETAKCKKVVCDPSYLPDKVQNVGKVAR 300
LSAVYGGTYMLNKPECKVEFDG GKAIGVTS GETAKCKKVVCDPSYL +KV+ VGKV R
Sbjct: 241 LSAVYGGTYMLNKPECKVEFDGSGKAIGVTSAGETAKCKKVVCDPSYLSEKVKKVGKVTR 300
Query: 301 AICIMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCCSYAHNVAPKGKYIAFVTTEAET 360
A+CIMSHPIPDTND+HS Q+ILPQKQLGRKSDMYLFCCSYAHNVAPKGKYIAFV+ EAET
Sbjct: 301 AVCIMSHPIPDTNDAHSVQIILPQKQLGRKSDMYLFCCSYAHNVAPKGKYIAFVSAEAET 360
Query: 361 DQPQVELKPGIDLLGPVDEIFYDIYDRFEPTNDHATDGCFISTSYDPTTHFETTVKDVVQ 420
D P+ ELKPGI+LLGP DEIFY YD + PTN D CFIS +YD TTHFE+TV DV+
Sbjct: 361 DNPEEELKPGIELLGPTDEIFYHSYDTYVPTNIQEEDNCFISATYDATTHFESTVVDVLD 420
Query: 421 MYSKITGKVLDLSVDLSAASAAAE 444
MY+KITGK LDLSVDLSAASAAAE
Sbjct: 421 MYTKITGKTLDLSVDLSAASAAAE 444
>At2g44100 GDP dissociation inhibitor
Length = 445
Score = 725 bits (1872), Expect = 0.0
Identities = 347/445 (77%), Positives = 401/445 (89%)
Query: 1 MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDQNDYYGGASTSLNLTQLFKRYRGDDK 60
MDEEY+VIVLGTGLKECILSGLLSVDGLKVLHMD+NDYYGG STSLNL QL+K++RG++K
Sbjct: 1 MDEEYEVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGESTSLNLNQLWKKFRGEEK 60
Query: 61 PPEELGSSREYNVDMIPKFMMANGALVRVLIHTDVTKYLNFKAVDGSFVYNKGKIYKVPA 120
P LGSSR+YNVDM+PKFMMANG LVRVLIHTDVTKYL+FKAVDGS+V+ +GK+ KVPA
Sbjct: 61 APAHLGSSRDYNVDMMPKFMMANGKLVRVLIHTDVTKYLSFKAVDGSYVFVQGKVQKVPA 120
Query: 121 TDVEALKSPLMGLFEKRRARKFFIYVQDYEANDPKSHEGLDLNQVTARQLISKYGLEDDT 180
T +EALKSPLMG+FEKRRA KFF YVQ+Y+ DPK+H+G+DL +VT + LI+K+GL++DT
Sbjct: 121 TPMEALKSPLMGIFEKRRAGKFFSYVQEYDEKDPKTHDGMDLRRVTTKDLIAKFGLKEDT 180
Query: 181 VDFIGHALALHLDDSYLDKPAKDFVDRVKTYAESLARFQGGSPYIYPLYGLGELPQAFAR 240
+DFIGHA+ALH +D++L +PA D V R+K YAESLARFQGGSPYIYPLYGLGELPQAFAR
Sbjct: 181 IDFIGHAVALHCNDNHLHQPAYDTVMRMKLYAESLARFQGGSPYIYPLYGLGELPQAFAR 240
Query: 241 LSAVYGGTYMLNKPECKVEFDGDGKAIGVTSDGETAKCKKVVCDPSYLPDKVQNVGKVAR 300
LSAVYGGTYMLNKPECKVEFD +GK GVTS+GETAKCKKVVCDPSYL +KV+ +G+VAR
Sbjct: 241 LSAVYGGTYMLNKPECKVEFDEEGKVSGVTSEGETAKCKKVVCDPSYLTNKVRKIGRVAR 300
Query: 301 AICIMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCCSYAHNVAPKGKYIAFVTTEAET 360
AI IMSHPIP+TNDS S QVILPQKQLGRKSDMY+FCCSY+HNVAPKGK+IAFV+T+AET
Sbjct: 301 AIAIMSHPIPNTNDSQSVQVILPQKQLGRKSDMYVFCCSYSHNVAPKGKFIAFVSTDAET 360
Query: 361 DQPQVELKPGIDLLGPVDEIFYDIYDRFEPTNDHATDGCFISTSYDPTTHFETTVKDVVQ 420
D PQ EL+PGIDLLGPVDE+F+DIYDR+EP N+ D CFISTSYD TTHF+TTV DV+
Sbjct: 361 DNPQTELQPGIDLLGPVDELFFDIYDRYEPVNEPTLDNCFISTSYDATTHFDTTVVDVLN 420
Query: 421 MYSKITGKVLDLSVDLSAASAAAEE 445
MY ITGK LDLSVDL+AASAA EE
Sbjct: 421 MYKLITGKELDLSVDLNAASAAEEE 445
>At3g59920 Rab GDP dissociation inhibitor
Length = 444
Score = 721 bits (1862), Expect = 0.0
Identities = 344/444 (77%), Positives = 398/444 (89%)
Query: 1 MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDQNDYYGGASTSLNLTQLFKRYRGDDK 60
MDEEY+VIVLGTGLKECILSGLLSVDG+KVLHMD+NDYYGG STSLNL QL+K++RG++K
Sbjct: 1 MDEEYEVIVLGTGLKECILSGLLSVDGVKVLHMDRNDYYGGESTSLNLNQLWKKFRGEEK 60
Query: 61 PPEELGSSREYNVDMIPKFMMANGALVRVLIHTDVTKYLNFKAVDGSFVYNKGKIYKVPA 120
PE LG+SR+YNVDM+PKFMM NG LVR LIHTDVTKYL+FKAVDGS+V+ KGK+ KVPA
Sbjct: 61 APEHLGASRDYNVDMMPKFMMGNGKLVRTLIHTDVTKYLSFKAVDGSYVFVKGKVQKVPA 120
Query: 121 TDVEALKSPLMGLFEKRRARKFFIYVQDYEANDPKSHEGLDLNQVTARQLISKYGLEDDT 180
T +EALKS LMG+FEKRRA KFF +VQ+Y+ DPK+H+G+DL +VT ++LI+KYGL+ +T
Sbjct: 121 TPMEALKSSLMGIFEKRRAGKFFSFVQEYDEKDPKTHDGMDLTRVTTKELIAKYGLDGNT 180
Query: 181 VDFIGHALALHLDDSYLDKPAKDFVDRVKTYAESLARFQGGSPYIYPLYGLGELPQAFAR 240
+DFIGHA+ALH +D +LD+PA D V R+K YAESLARFQG SPYIYPLYGLGELPQAFAR
Sbjct: 181 IDFIGHAVALHTNDQHLDQPAFDTVMRMKLYAESLARFQGTSPYIYPLYGLGELPQAFAR 240
Query: 241 LSAVYGGTYMLNKPECKVEFDGDGKAIGVTSDGETAKCKKVVCDPSYLPDKVQNVGKVAR 300
LSAVYGGTYMLNKPECKVEFD GK IGVTS+GETAKCKK+VCDPSYLP+KV+ +G+VAR
Sbjct: 241 LSAVYGGTYMLNKPECKVEFDEGGKVIGVTSEGETAKCKKIVCDPSYLPNKVRKIGRVAR 300
Query: 301 AICIMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCCSYAHNVAPKGKYIAFVTTEAET 360
AI IMSHPIP+TNDSHS QVI+PQKQL RKSDMY+FCCSY+HNVAPKGK+IAFV+T+AET
Sbjct: 301 AIAIMSHPIPNTNDSHSVQVIIPQKQLARKSDMYVFCCSYSHNVAPKGKFIAFVSTDAET 360
Query: 361 DQPQVELKPGIDLLGPVDEIFYDIYDRFEPTNDHATDGCFISTSYDPTTHFETTVKDVVQ 420
D PQ ELKPG DLLGPVDEIF+D+YDR+EP N+ D CFISTSYD TTHFETTV DV+
Sbjct: 361 DNPQTELKPGTDLLGPVDEIFFDMYDRYEPVNEPELDNCFISTSYDATTHFETTVADVLN 420
Query: 421 MYSKITGKVLDLSVDLSAASAAAE 444
MY+ ITGK LDLSVDLSAASAA E
Sbjct: 421 MYTLITGKQLDLSVDLSAASAAEE 444
>At3g06540 Rab escort protein, putative
Length = 536
Score = 110 bits (276), Expect = 1e-24
Identities = 105/395 (26%), Positives = 162/395 (40%), Gaps = 99/395 (25%)
Query: 5 YDVIVLGTGLKECILSGLLSVDGLKVLHMDQNDYYGGASTSLNLTQL--FKRYRGDDKPP 62
YD+IV+GTG+ E +L+ S G VLH+D N +YG SL+L L F PP
Sbjct: 15 YDLIVVGTGVSESVLAAAASSSGSSVLHLDPNPFYGSHFASLSLPDLTSFLHSNSVSPPP 74
Query: 63 E----------------------------ELGS---------SREYNVDMI-PKFMMANG 84
E+ S SR +NVD+ P+ +
Sbjct: 75 SPSSPPLPPSNNHDFISVDLVNRSLYSSVEISSFESEILEEHSRRFNVDLAGPRVVFCAD 134
Query: 85 ALVRVLIHTDVTKYLNFKAVDGSFVYNK-GKIYKVPATDVEALKSPLMGLFEKRRARKFF 143
+ +++ + Y+ FK++D SFV + G++ VP + K + L EK + KFF
Sbjct: 135 ESINLMLKSGANNYVEFKSIDASFVGDSSGELRNVPDSRAAIFKDKSLTLLEKNQLMKFF 194
Query: 144 IYVQDYEANDPKSHEGLDLNQVTARQLISKYGLEDDTVDFIG-------------HALA- 189
VQ + A+ + + + IS+ +E VDF+ +A+A
Sbjct: 195 KLVQSHLASSTEKDDSTTVK-------ISEEDMESPFVDFLSKMRLPPKIKSIILYAIAM 247
Query: 190 LHLDDSYLDK-----PAKDFVDRVKTYAESLARFQGG-SPYIYPLYGLGELPQAFARLSA 243
L D + K+ +DR+ Y S+ RF IYP+YG GELPQAF R +A
Sbjct: 248 LDYDQDNTETCRHLLKTKEGIDRLALYITSMGRFSNALGALIYPIYGQGELPQAFCRRAA 307
Query: 244 VYGGTYMLNKPECKVEFDGDGKAIGVTSDGETAKCKKVVCDP---------SYLPDKVQN 294
V G Y+L P + D K++ DP S L D+
Sbjct: 308 VKGCIYVLRMPITALLLD------------------KLILDPCVTVGLESLSSLTDQQNE 349
Query: 295 V--GKVARAICIMSHPIPDTNDSHSAQVILPQKQL 327
K+AR +C++ + + +A V+ P K L
Sbjct: 350 TLSEKIARGVCVIRGSV--KANVSNALVVYPPKSL 382
>At1g57770 unknown protein
Length = 574
Score = 35.4 bits (80), Expect = 0.064
Identities = 69/326 (21%), Positives = 127/326 (38%), Gaps = 52/326 (15%)
Query: 2 DEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDQNDYYGGASTSLNLTQLFKRYRGDDKP 61
+ E DV+V+G+G+ LL+ V+ ++ +D+ GGA+ S + K Y+ D P
Sbjct: 50 EPEADVVVIGSGIGGLCCGALLARYDQDVIVLESHDHPGGAAHSFEI----KGYKFDSGP 105
Query: 62 P-------------------EELGSS---REYNVDMIPKFMMANGALVRVLIHTDVTKYL 99
+ LG S ++Y+ M+ + G + + TD K L
Sbjct: 106 SLFSGLQSRGPQANPLAQVLDALGESFPCKKYDSWMV---YLPEGDFLSRIGPTDFFKDL 162
Query: 100 NFKAVDGSFVYNKGKIYK--VPATDVEALKSPL-----MGLFEKRRARKFFIYVQDYEAN 152
K S V K+ +P + PL +G+ AR ++ +
Sbjct: 163 E-KYAGPSAVQEWEKLLGAILPLSSAAMALPPLSIRGDLGVLSTAAARYAPSLLKSFIKM 221
Query: 153 DPKSHEGLDLNQVTARQLISKYGLED----DTVDFIGHALALHLDDSYLDKPAKDFVDRV 208
PK G +++ L+D + +D + LA D L + +
Sbjct: 222 GPKGALGATKLLRPFSEIVDSLELKDPFIRNWIDLLAFLLAGVKSDGILS------AEMI 275
Query: 209 KTYAESLARFQGGSPYIYPLYGLGELPQAFARLSAVYGGTYMLNKPECKVEFDGDGKAIG 268
+AE ++ G YP+ G G + +A R +GG L + + +GKA+G
Sbjct: 276 YMFAE---WYKPGCTLEYPIDGTGAVVEALVRGLEKFGGRLSLKSHVENIVIE-NGKAVG 331
Query: 269 V-TSDGETAKCKKVVCDPSYLPDKVQ 293
V +G+ + +K V + + D ++
Sbjct: 332 VKLRNGQFVRARKAVVSNASMWDTLK 357
>At5g01400 putative protein
Length = 1467
Score = 33.5 bits (75), Expect = 0.24
Identities = 22/80 (27%), Positives = 39/80 (48%), Gaps = 3/80 (3%)
Query: 146 VQDYEANDPKSHEGLDLNQVTARQLISKYGLEDDTVDFIGHALALHLDDSYLDKPAKDFV 205
+Q++ +D +HEG +++T R L YG + DF A +S+L A+
Sbjct: 709 LQEHVLSDYLNHEG---HELTVRVLYRLYGEAEAEQDFFSSTTAASAYESFLLTVAEALR 765
Query: 206 DRVKTYAESLARFQGGSPYI 225
D +SL++ G SP++
Sbjct: 766 DSFPPSDKSLSKLLGDSPHL 785
>At4g19380 unknown protein
Length = 678
Score = 31.2 bits (69), Expect = 1.2
Identities = 15/46 (32%), Positives = 27/46 (58%)
Query: 1 MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDQNDYYGGASTSL 46
M + D +V+G+G + +G+L+ G KVL ++ +YY + SL
Sbjct: 172 MKIQCDAVVVGSGSGGGVAAGVLAKAGYKVLVIESGNYYARSKLSL 217
>At1g19220 auxin response factor (ARF) like protein
Length = 1086
Score = 30.0 bits (66), Expect = 2.7
Identities = 28/108 (25%), Positives = 52/108 (47%), Gaps = 7/108 (6%)
Query: 63 EELGSSREYNVDMIPKFMMANGALVRVLIHTDVTKYLNFKAVDGSFVYNKGKIYKVPATD 122
E++ +S + D IP + L+ L+H+ VT + + + + VY + + V D
Sbjct: 51 EQVAASMQKQTDFIPNYPNLPSKLI-CLLHS-VTLHADTETDE---VYAQMTLQPVNKYD 105
Query: 123 VEALKSPLMGLFEKRRARKFFIYVQDYEANDPKSHEGLDLNQVTARQL 170
EAL + MGL R+ +FF + A+D +H G + + A ++
Sbjct: 106 REALLASDMGLKLNRQPTEFF--CKTLTASDTSTHGGFSVPRRAAEKI 151
>At3g21560 UDP-glucose:indole-3-acetate beta-D-glucosyltransferase,
putative
Length = 496
Score = 29.6 bits (65), Expect = 3.5
Identities = 23/85 (27%), Positives = 41/85 (48%), Gaps = 2/85 (2%)
Query: 344 VAPKGKYIAFVTTEAETDQPQVELKPGIDLLGPVDE--IFYDIYDRFEPTNDHATDGCFI 401
+A KG I FVTTE+ + ++ K +L PV + + YD +D P +D A+
Sbjct: 34 LASKGLLITFVTTESWGKKMRISNKIQDRVLKPVGKGYLRYDFFDDGLPEDDEASRTNLT 93
Query: 402 STSYDPTTHFETTVKDVVQMYSKIT 426
+ +K++V+ Y ++T
Sbjct: 94 ILRPHLELVGKREIKNLVKRYKEVT 118
>At3g57590 putative protein
Length = 404
Score = 29.3 bits (64), Expect = 4.6
Identities = 14/40 (35%), Positives = 19/40 (47%)
Query: 249 YMLNKPECKVEFDGDGKAIGVTSDGETAKCKKVVCDPSYL 288
Y+ PE +V D GVT+ G+ C K C P Y+
Sbjct: 289 YVYTLPENEVLDSCDFSVAGVTTRGDIVLCMKYTCKPFYV 328
>At3g49740 putative protein
Length = 737
Score = 28.9 bits (63), Expect = 6.0
Identities = 17/54 (31%), Positives = 26/54 (47%), Gaps = 2/54 (3%)
Query: 314 DSHSAQVILPQKQLGRKSDMY--LFCCSYAHNVAPKGKYIAFVTTEAETDQPQV 365
D + V + +K +G + D++ LF AH GK +A + E E D P V
Sbjct: 644 DEAESLVKISEKTIGSRVDVWWALFSACAAHGDLKLGKMVAKLLMEKEKDDPSV 697
>At3g21020 unknown protein
Length = 310
Score = 28.9 bits (63), Expect = 6.0
Identities = 23/92 (25%), Positives = 44/92 (47%), Gaps = 11/92 (11%)
Query: 286 SYLPDKVQNVGKVARAIC-------IMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCC 338
SYL DKV+ + + R + +++ + ++SH L ++Q+ K DM+L
Sbjct: 132 SYL-DKVRKIAEQLRRLKNGKSDYEVVTKVLGSLSESHEDLATLLEEQVDLKKDMWLLSS 190
Query: 339 SYAHNVAPKGKYIAFVTTEAETDQPQVELKPG 370
++++P Y+ F TT + V+L G
Sbjct: 191 GTTNHMSP---YLKFFTTLDRRHRAPVKLING 219
>At5g13820 telomeric DNA-binding protein 1 (TBP1)
Length = 640
Score = 28.5 bits (62), Expect = 7.8
Identities = 14/36 (38%), Positives = 19/36 (51%), Gaps = 1/36 (2%)
Query: 172 SKYGLEDDTVDFIGHALALHLDDSYLDKPAK-DFVD 206
S Y E D +D +G + L+D Y KP K +F D
Sbjct: 65 STYASEADNLDHLGGLIKQELEDGYTTKPCKSEFFD 100
>At5g07980 putative protein
Length = 1501
Score = 28.5 bits (62), Expect = 7.8
Identities = 10/31 (32%), Positives = 17/31 (54%)
Query: 192 LDDSYLDKPAKDFVDRVKTYAESLARFQGGS 222
+ D Y+ K A+DF+ R + AR + G+
Sbjct: 1409 ISDQYISKAAEDFISRTRKLETDFARLENGT 1439
>At3g57580 unknown protein
Length = 408
Score = 28.5 bits (62), Expect = 7.8
Identities = 13/40 (32%), Positives = 19/40 (47%)
Query: 249 YMLNKPECKVEFDGDGKAIGVTSDGETAKCKKVVCDPSYL 288
Y+ PE +V + GVT+ G+ C K C P Y+
Sbjct: 293 YVYTLPENEVLASCEFSVAGVTATGDIVLCMKYTCKPYYV 332
>At2g02220 putative receptor-like protein kinase
Length = 1008
Score = 28.5 bits (62), Expect = 7.8
Identities = 13/38 (34%), Positives = 21/38 (55%)
Query: 14 LKECILSGLLSVDGLKVLHMDQNDYYGGASTSLNLTQL 51
+K+ I + ++ L+ L + ND GG TS+NL L
Sbjct: 112 IKDSIPLSIFNLKNLQTLDLSSNDLSGGIPTSINLPAL 149
>At1g35940 hypothetical protein
Length = 1678
Score = 28.5 bits (62), Expect = 7.8
Identities = 21/84 (25%), Positives = 37/84 (44%), Gaps = 3/84 (3%)
Query: 5 YDVIVLGTGLKECILSGLLSVDGLKVLHMDQNDYYGGASTSLNLTQLFKRYRGDDKPPEE 64
YD +V T + C G+L D + + + + + + L + K R + K P+
Sbjct: 1128 YDGVVYKTFKEACFARGILDDDQVFIDGLVEATHVRSQTWHLLAEDILKTKRDEFKNPDL 1187
Query: 65 LGSSRE---YNVDMIPKFMMANGA 85
+ E Y + I K M++NGA
Sbjct: 1188 TLTETEIKNYTLQEIEKIMLSNGA 1211
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.138 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,434,914
Number of Sequences: 26719
Number of extensions: 476240
Number of successful extensions: 1013
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 999
Number of HSP's gapped (non-prelim): 19
length of query: 445
length of database: 11,318,596
effective HSP length: 102
effective length of query: 343
effective length of database: 8,593,258
effective search space: 2947487494
effective search space used: 2947487494
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144517.16


