

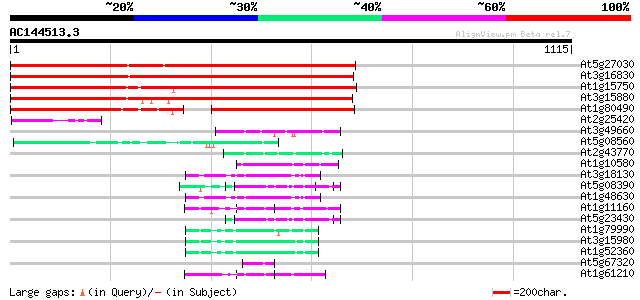
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144513.3 - phase: 0 /pseudo
(1115 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g27030 unknown protein 1099 0.0
At3g16830 unknown protein 1048 0.0
At1g15750 unknown protein (At1g15750) 925 0.0
At3g15880 putative WD-repeat protein 890 0.0
At1g80490 unknown protein 421 e-118
At2g25420 hypothetical protein 92 2e-18
At3g49660 putative WD-40 repeat - protein 59 2e-08
At5g08560 WD-repeat protein-like 55 2e-07
At2g43770 putative splicing factor 53 8e-07
At1g10580 putative pre-mRNA splicing factor 51 3e-06
At3g18130 protein kinase C-receptor/G-protein, putative 50 5e-06
At5g08390 katanin p80 subunit - like protein 49 1e-05
At1g48630 unknown protein 49 1e-05
At1g11160 hypothetical protein 49 2e-05
At5g23430 unknown protein 49 2e-05
At1g79990 coatomer protein like complex, subunit beta 2 (beta pr... 49 2e-05
At3g15980 putative coatomer complex subunit 48 3e-05
At1g52360 coatomer complex subunit, putative 48 4e-05
At5g67320 unknown protein 47 5e-05
At1g61210 47 6e-05
>At5g27030 unknown protein
Length = 1108
Score = 1099 bits (2842), Expect = 0.0
Identities = 540/688 (78%), Positives = 604/688 (87%), Gaps = 5/688 (0%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFLEEEKFKESVH+LEKESGFFFN KYF+EKV AGEW++VE YLSGF
Sbjct: 1 MSSLSRELVFLILQFLEEEKFKESVHRLEKESGFFFNTKYFDEKVLAGEWDDVETYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALDRQ+K KAVEILV DL+VFSTFNEELYKEITQLLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQEKAKAVEILVQDLRVFSTFNEELYKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
L NFRENEQLSKYGDTKTARGIML ELKKLIEANPLFRDKL+FPTL+SSRLRTLINQSLN
Sbjct: 121 LQNFRENEQLSKYGDTKTARGIMLGELKKLIEANPLFRDKLMFPTLRSSRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLPVAAVAKPAAYTSLGVGAHGPF 240
WQHQLCKNPRPNPDIKTLF DH+CT NGPLAP+ VN PV + KPAAY SLG H PF
Sbjct: 181 WQHQLCKNPRPNPDIKTLFTDHTCTLPNGPLAPSAVNQPVTTLTKPAAYPSLG--PHVPF 238
Query: 241 PPAAATANANALAGWMANASVSSSVQAAVVTASTIPVPHNQVSILKRPITPSTTPGMVEY 300
PP A ANA ALA WMA AS +S+VQAAVVT + +P P NQ+SILKRP TP TPG+V+Y
Sbjct: 239 PPGPAAANAGALASWMAAASGASAVQAAVVTPALMPQPQNQMSILKRPRTPPATPGIVDY 298
Query: 301 QSADHEQLMKRLRPAPSVEEVSYPSARQ-ASWSLDDLPRTVAMSLHQGSSVTSMDFHPSH 359
Q+ DHE LMKRLRPAPSVEEV+YP+ RQ A WSL+DLP A++LHQGS+VTSM+F+P
Sbjct: 299 QNPDHE-LMKRLRPAPSVEEVTYPAPRQQAPWSLEDLPTKAALALHQGSTVTSMEFYPMQ 357
Query: 360 QTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP-SVSRVTWSL 418
TLLLVGS GEI+LWEL RERLVS+PFKIWD+S CS FQA + K+TP SV+RV WS
Sbjct: 358 NTLLLVGSATGEITLWELAARERLVSRPFKIWDMSNCSHQFQALIAKETPISVTRVAWSP 417
Query: 419 DGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKL 478
DG+F+GVAFTKHLI +YA++G N+L Q EIDAH+G VNDLAFA+PN+QLCV+TCGDDKL
Sbjct: 418 DGNFIGVAFTKHLIQLYAFSGPNDLRQHTEIDAHVGAVNDLAFANPNRQLCVITCGDDKL 477
Query: 479 IKVWDLTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDA 538
IKVWD++GR+ F FEGH+APVYSICPH+KENIQFIFSTA+DGKIKAWLYDN+GSRVDYDA
Sbjct: 478 IKVWDVSGRKHFTFEGHDAPVYSICPHYKENIQFIFSTAIDGKIKAWLYDNLGSRVDYDA 537
Query: 539 PGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTT 598
PG WCT MLYSADGTRLFSCGTSKDGDSFLVEWNESEG+IKRTY F+KK AGVVQFDT+
Sbjct: 538 PGKWCTRMLYSADGTRLFSCGTSKDGDSFLVEWNESEGSIKRTYKEFQKKLAGVVQFDTS 597
Query: 599 QNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKIL 658
+N FLA GED QIKFWDM+N+N LTST+AEGGL LPHLRFNK+GNLLAVTTADNGFKIL
Sbjct: 598 KNHFLAVGEDGQIKFWDMNNINVLTSTDAEGGLPALPHLRFNKDGNLLAVTTADNGFKIL 657
Query: 659 ANAGGLRSLRTVETPAFEALRSPIESAA 686
AN G RSLR +ETPA E +R+P++ A
Sbjct: 658 ANPAGFRSLRAMETPASETMRTPVDFKA 685
Score = 33.1 bits (74), Expect = 0.91
Identities = 26/115 (22%), Positives = 47/115 (40%), Gaps = 5/115 (4%)
Query: 503 CPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSK 562
C +N ++ S A GK+ + + P T + + + + G
Sbjct: 825 CIALSKNDSYVMSAA-GGKVSLFNMMTFKVMTTFMPPPPASTFLAFHPQDNNVIAIGME- 882
Query: 563 DGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMD 617
DS + +N +K G +K+ G+ F T N +++G D+QI FW +D
Sbjct: 883 --DSTIHIYNVRVDEVKSKLKGHQKRITGLA-FSTALNILVSSGADAQICFWSID 934
>At3g16830 unknown protein
Length = 1131
Score = 1048 bits (2709), Expect = 0.0
Identities = 518/685 (75%), Positives = 590/685 (85%), Gaps = 4/685 (0%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFKESVHKLE+ESGFFFN+KYFEEK AGEW+EVEKYLSGF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKESVHKLEQESGFFFNIKYFEEKALAGEWDEVEKYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALDR D+ KAVEIL DLKVF+TFNEELYKEITQLLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRNDRAKAVEILAKDLKVFATFNEELYKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
L NFRENEQLSKYGDTK+AR IM ELKKLIEANPLFR+KL FP+ K+SRLRTLINQSLN
Sbjct: 121 LENFRENEQLSKYGDTKSARSIMYTELKKLIEANPLFREKLAFPSFKASRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLPVAAVAKPAAYTSLGVGAHGPF 240
WQHQLCKNPRPNPDIKTLF+DHSC+PSNG A TPVNLPVAAVA+P+ + LGV GPF
Sbjct: 181 WQHQLCKNPRPNPDIKTLFLDHSCSPSNGARALTPVNLPVAAVARPSNFVPLGVHG-GPF 239
Query: 241 PPAAATA-NANALAGWMANASVSSSVQAAVVTASTIPVPHNQVSILKRPITPSTTPGMVE 299
A A NANALAGWMAN + SSSV + VV AS P+ +QV+ LK P PS + G+++
Sbjct: 240 QSNPAPAPNANALAGWMANPNPSSSVPSGVVAASPFPMQPSQVNELKHPRAPSNSLGLMD 299
Query: 300 YQSADHEQLMKRLRPAPSVEEVSYPSARQASWSLDDLPRTVAMSLHQGSSVTSMDFHPSH 359
YQSADHEQLMKRLR A + EV+YP+ SLDDLPR V ++ QGS V SMDFHPSH
Sbjct: 300 YQSADHEQLMKRLRSAQTSNEVTYPAHSHPPASLDDLPRNVVSTIRQGSVVISMDFHPSH 359
Query: 360 QTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP-SVSRVTWSL 418
TLL VG ++GE++LWE+G RE++V++PFKIW+++ACS+ FQ ++VK+ SV+RV WS
Sbjct: 360 HTLLAVGCSSGEVTLWEVGSREKVVTEPFKIWNMAACSVIFQGSIVKEPSISVTRVAWSP 419
Query: 419 DGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKL 478
DG+ +GV+FTKHLIH+YAY GS +L Q +EIDAH+G VNDLAFAHPNKQ+CVVTCGDDKL
Sbjct: 420 DGNLLGVSFTKHLIHVYAYQGS-DLRQHLEIDAHVGCVNDLAFAHPNKQMCVVTCGDDKL 478
Query: 479 IKVWDLTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDA 538
IKVWDL+G++LF FEGHEAPVYSICPH KENIQFIFSTA+DGKIKAWLYDN+GSRVDYDA
Sbjct: 479 IKVWDLSGKKLFTFEGHEAPVYSICPHQKENIQFIFSTALDGKIKAWLYDNVGSRVDYDA 538
Query: 539 PGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTT 598
PG WCTTMLYSADG+RLFSCGTSK+GDSFLVEWNESEGA+KRTY GFRKKSAGVVQFDTT
Sbjct: 539 PGQWCTTMLYSADGSRLFSCGTSKEGDSFLVEWNESEGALKRTYLGFRKKSAGVVQFDTT 598
Query: 599 QNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKIL 658
+NRFLA GED+QIKFW+MDN N LT EAEGGL LP LRFNK+GNLLAVTTADNGFKIL
Sbjct: 599 RNRFLAVGEDNQIKFWNMDNTNLLTVVEAEGGLPNLPRLRFNKDGNLLAVTTADNGFKIL 658
Query: 659 ANAGGLRSLRTVETPAFEALRSPIE 683
AN GLR+LR E +FEA ++ I+
Sbjct: 659 ANTDGLRTLRAFEARSFEASKASID 683
Score = 30.4 bits (67), Expect = 5.9
Identities = 25/116 (21%), Positives = 46/116 (39%), Gaps = 5/116 (4%)
Query: 503 CPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSK 562
C +N ++ S A GK+ + + P T + + + + G
Sbjct: 836 CIALSKNDSYVMS-ACGGKVSLFNMMTFKVMTTFMPPPPASTFLAFHPQDNNIIAIGME- 893
Query: 563 DGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDN 618
DS + +N +K G +K G+ F T N +++G D+Q+ FW D+
Sbjct: 894 --DSSIHIYNVRVDEVKTKLKGHQKHITGLA-FSTALNILVSSGADAQLFFWTADS 946
>At1g15750 unknown protein (At1g15750)
Length = 1131
Score = 925 bits (2391), Expect = 0.0
Identities = 471/706 (66%), Positives = 552/706 (77%), Gaps = 24/706 (3%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFKE+VHKLE+ESGFFFNMKYFE++V G W+EVEKYLSGF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKETVHKLEQESGFFFNMKYFEDEVHNGNWDEVEKYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALD+ D+PKAV+ILV DLKVFSTFNEEL+KEITQLLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDKHDRPKAVDILVKDLKVFSTFNEELFKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
L NFRENEQLSKYGDTK+AR IML+ELKKLIEANPLFRDKL FPTL++SRLRTLINQSLN
Sbjct: 121 LENFRENEQLSKYGDTKSARAIMLVELKKLIEANPLFRDKLQFPTLRNSRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLP-VAAVAKPAAYTSLGVGAHGP 239
WQHQLCKNPRPNPDIKTLF+DHSC P NG AP+PVN P + + K + L GAHGP
Sbjct: 181 WQHQLCKNPRPNPDIKTLFVDHSCGPPNGARAPSPVNNPLLGGIPKAGGFPPL--GAHGP 238
Query: 240 FPPAAATANANALAGWMANASVSSSVQAAVVTASTIPVPHNQV-SILKRPITPSTTPGMV 298
F P A+ LAGWM S SSV V+A I + + + LK P TP T +
Sbjct: 239 FQPTASPV-PTPLAGWM---SSPSSVPHPAVSAGAIALGGPSIPAALKHPRTPPTNASL- 293
Query: 299 EYQSADHEQLMKRLRPAPSVEEVSY-------------PSARQASWSLDDLPRTVAMSLH 345
+Y SAD E + KR RP +EV+ A + DDLP+TVA +L
Sbjct: 294 DYPSADSEHVSKRTRPMGISDEVNLGVNMLPMSFSGQAHGHSPAFKAPDDLPKTVARTLS 353
Query: 346 QGSSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVV 405
QGSS SMDFHP QTLLLVG+N G+I LWE+G RERLV K FK+WD+S CS+P QAA+V
Sbjct: 354 QGSSPMSMDFHPIKQTLLLVGTNVGDIGLWEVGSRERLVQKTFKVWDLSKCSMPLQAALV 413
Query: 406 KD-TPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHP 464
K+ SV+RV WS DGS GVA+++H++ +Y+Y+G ++ Q +EIDAH+GGVND++F+ P
Sbjct: 414 KEPVVSVNRVIWSPDGSLFGVAYSRHIVQLYSYHGGEDMRQHLEIDAHVGGVNDISFSTP 473
Query: 465 NKQLCVVTCGDDKLIKVWD-LTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIK 523
NKQLCV+TCGDDK IKVWD TG + FEGHEAPVYS+CPH+KENIQFIFSTA+DGKIK
Sbjct: 474 NKQLCVITCGDDKTIKVWDAATGVKRHTFEGHEAPVYSVCPHYKENIQFIFSTALDGKIK 533
Query: 524 AWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYN 583
AWLYDNMGSRVDYDAPG WCTTM YSADGTRLFSCGTSKDG+SF+VEWNESEGA+KRTY
Sbjct: 534 AWLYDNMGSRVDYDAPGRWCTTMAYSADGTRLFSCGTSKDGESFIVEWNESEGAVKRTYQ 593
Query: 584 GFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEG 643
GF K+S GVVQFDTT+NR+LAAG+D IKFWDMD V LT+ + +GGLQ P +RFNKEG
Sbjct: 594 GFHKRSLGVVQFDTTKNRYLAAGDDFSIKFWDMDAVQLLTAIDGDGGLQASPRIRFNKEG 653
Query: 644 NLLAVTTADNGFKILANAGGLRSLRTVETPAFEALRSPIESAANKA 689
+LLAV+ +N KI+AN+ GLR L T E + E+ + I S A A
Sbjct: 654 SLLAVSGNENVIKIMANSDGLRLLHTFENISSESSKPAINSIAAAA 699
>At3g15880 putative WD-repeat protein
Length = 1160
Score = 890 bits (2299), Expect = 0.0
Identities = 455/724 (62%), Positives = 552/724 (75%), Gaps = 46/724 (6%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFK++VH+LEKESGFFFNM+YFE+ V AGEW++VEKYLSGF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKDTVHRLEKESGFFFNMRYFEDSVTAGEWDDVEKYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALD++D KAV+ILV +LKVFSTFNEEL+KEIT LLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDKKDHAKAVDILVKELKVFSTFNEELFKEITMLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
LTNFRENEQLSKYGDTK+ARGIML ELKKLIEANPLFRDKL FP+LK+SRLRTLINQSLN
Sbjct: 121 LTNFRENEQLSKYGDTKSARGIMLGELKKLIEANPLFRDKLQFPSLKNSRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTP-VNLPVAAVAKPAAYTSLGVGAHGP 239
WQHQLCKNPRPNPDIKTLF+DH+C NG P+P N + +V K + L GAHGP
Sbjct: 181 WQHQLCKNPRPNPDIKTLFVDHTCGHPNGAHTPSPTTNHLMGSVPKVGGFPPL--GAHGP 238
Query: 240 FPPAAATANANALAGWMANASVS------------------SSVQAAVVTASTIPVPHN- 280
F P A +LAGWM N SV +V+ ++ + HN
Sbjct: 239 FQPTPAPL-TTSLAGWMPNPSVQHPTVSAGPIGLGAPNSAVHNVEISLCVFTNCIAYHNL 297
Query: 281 -------QVSILK--RPITPSTTPGMVEYQSADHEQLMKRLRP----------APSVEEV 321
VS+LK RP +P T ++YQ+AD E ++KR RP +V V
Sbjct: 298 FILLFFLAVSMLKRERPRSPPTNSLSMDYQTADSESVLKRPRPFGISDGVNNLPVNVLPV 357
Query: 322 SYP--SARQASWSLDDLPRTVAMSLHQGSSVTSMDFHPSHQTLLLVGSNNGEISLWELGM 379
+YP S A++S DDLP+ V+ L QGS++ SMDFHP QT+LLVG+N G+I++WE+G
Sbjct: 358 TYPGQSHAHATYSTDDLPKNVSRILSQGSAIKSMDFHPVQQTMLLVGTNLGDIAIWEVGS 417
Query: 380 RERLVSKPFKIWDISACSLPFQAAVVKD-TPSVSRVTWSLDGSFVGVAFTKHLIHIYAYN 438
RE+LVS+ FK+WD++ C++ QA++ + T +V+RV WS DG +GVA++KH++HIY+Y+
Sbjct: 418 REKLVSRSFKVWDLATCTVNLQASLASEYTAAVNRVVWSPDGGLLGVAYSKHIVHIYSYH 477
Query: 439 GSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWD-LTGRRLFNFEGHEA 497
G +L +EIDAH G VNDLAF+ PN+QLCVVTCG+DK IKVWD +TG +L FEGHEA
Sbjct: 478 GGEDLRNHLEIDAHAGNVNDLAFSQPNQQLCVVTCGEDKTIKVWDAVTGNKLHTFEGHEA 537
Query: 498 PVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFS 557
PVYS+CPH KENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPG CT+M Y ADGTRLFS
Sbjct: 538 PVYSVCPHQKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGRSCTSMAYCADGTRLFS 597
Query: 558 CGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMD 617
CGTSK+G+SF+VEWNESEGA+KRTY G K+S GVVQFDT +N+FL AG++ Q+KFWDMD
Sbjct: 598 CGTSKEGESFIVEWNESEGAVKRTYLGLGKRSVGVVQFDTMKNKFLVAGDEFQVKFWDMD 657
Query: 618 NVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKILANAGGLRSLRTVETPAFEA 677
+V+ L+ST AEGGL P LR NKEG LLAV+T DNG KILANA G R L ++ ++
Sbjct: 658 SVDLLSSTAAEGGLPSSPCLRINKEGTLLAVSTTDNGIKILANAEGSRILHSMANRGLDS 717
Query: 678 LRSP 681
R+P
Sbjct: 718 SRAP 721
Score = 35.4 bits (80), Expect = 0.18
Identities = 30/125 (24%), Positives = 53/125 (42%), Gaps = 6/125 (4%)
Query: 493 EGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADG 552
EG++ V C +N ++ S A GKI + + + AP T++ +
Sbjct: 866 EGNKEDVVP-CFALSKNDSYVMS-ASGGKISLFNMMTFKTMTTFMAPPPAATSLAFHPQD 923
Query: 553 TRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIK 612
+ + G DS + +N +K G +K+ G+ F N +++G DSQ+
Sbjct: 924 NNIIAIGMD---DSSIQIYNVRVDEVKSKLKGHQKRVTGLA-FSNVLNVLVSSGADSQLC 979
Query: 613 FWDMD 617
W MD
Sbjct: 980 VWSMD 984
>At1g80490 unknown protein
Length = 1073
Score = 421 bits (1083), Expect = e-118
Identities = 230/351 (65%), Positives = 261/351 (73%), Gaps = 12/351 (3%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFKE+VHKLE+ESGFFFNMKYFE++V G W+EVEKYLSGF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKETVHKLEQESGFFFNMKYFEDEVHNGNWDEVEKYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALDR D+PKAV+ILV DLKVFSTFNEEL+KEITQLLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRHDRPKAVDILVKDLKVFSTFNEELFKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
L NFRENEQLSKYGDTK+AR IML+ELKKLIEANPLFRDKL FPTL++SRLRTLINQSLN
Sbjct: 121 LENFRENEQLSKYGDTKSARAIMLVELKKLIEANPLFRDKLQFPTLRTSRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLPVAAVAKPAAYTSLGVGAHGPF 240
WQHQLCKNPRPNPDIKTLF+DHSC N AP+PVN P+ + P A +GAHGPF
Sbjct: 181 WQHQLCKNPRPNPDIKTLFVDHSCRLPNDARAPSPVNNPLLG-SLPKAEGFPPLGAHGPF 239
Query: 241 PPAAATANANALAGWMANASVSSSVQAAVVTASTIPVPHNQV-SILKRPITPSTTPGMVE 299
P + LAGWM S SSV V+ I + + + LK P TP + V+
Sbjct: 240 QPTPSPV-PTPLAGWM---SSPSSVPHPAVSGGPIALGAPSIQAALKHPRTPPSN-SAVD 294
Query: 300 YQSADHEQLMKRLRPAPSVEEVS-----YPSARQASWSLDDLPRTVAMSLH 345
Y S D + + KR RP +E + S + WS D VA S H
Sbjct: 295 YPSGDSDHVSKRTRPMGISDEAALVKEPVVSVNRVIWSPDGSLFGVAYSRH 345
Score = 418 bits (1074), Expect = e-116
Identities = 195/286 (68%), Positives = 238/286 (83%), Gaps = 2/286 (0%)
Query: 401 QAAVVKD-TPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDL 459
+AA+VK+ SV+RV WS DGS GVA+++H++ +Y+Y+G ++ Q +EIDAH+GGVND+
Sbjct: 315 EAALVKEPVVSVNRVIWSPDGSLFGVAYSRHIVQLYSYHGGEDMRQHLEIDAHVGGVNDI 374
Query: 460 AFAHPNKQLCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAV 518
AF+ PNKQLCV TCGDDK IKVWD TG + + FEGHEAPVYSICPH+KENIQFIFSTA+
Sbjct: 375 AFSTPNKQLCVTTCGDDKTIKVWDAATGVKRYTFEGHEAPVYSICPHYKENIQFIFSTAL 434
Query: 519 DGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAI 578
DGKIKAWLYDNMGSRVDY+APG WCTTM YSADGTRLFSCGTSKDG+SF+VEWNESEGA+
Sbjct: 435 DGKIKAWLYDNMGSRVDYEAPGRWCTTMAYSADGTRLFSCGTSKDGESFIVEWNESEGAV 494
Query: 579 KRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLR 638
KRTY GF K+S GVVQFDTT+NR+LAAG+D IKFWDMD + LT+ +A+GGLQ P +R
Sbjct: 495 KRTYQGFHKRSLGVVQFDTTKNRYLAAGDDFSIKFWDMDTIQLLTAIDADGGLQASPRIR 554
Query: 639 FNKEGNLLAVTTADNGFKILANAGGLRSLRTVETPAFEALRSPIES 684
FNKEG+LLAV+ DN K++AN+ GLR L TVE + E+ + I S
Sbjct: 555 FNKEGSLLAVSANDNMIKVMANSDGLRLLHTVENLSSESSKPAINS 600
>At2g25420 hypothetical protein
Length = 572
Score = 92.0 bits (227), Expect = 2e-18
Identities = 55/178 (30%), Positives = 92/178 (50%), Gaps = 38/178 (21%)
Query: 4 LSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGFTKV 63
L +L+ LILQFL E K+K ++HKLE+E+ FFN+ Y E ++ GE+ + E+YL FT
Sbjct: 93 LKEDLICLILQFLYEAKYKNTLHKLEQETKVFFNLNYLAEVMKLGEYGKAEEYLGAFTNW 152
Query: 64 DDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLTLTN 123
+DN+YS +F E++K L++ + ++ T +
Sbjct: 153 EDNKYSKAMFLELQKLICLQSTE-----------------------------WEVATPSG 183
Query: 124 FRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLNW 181
+N L K + +L K NP+ +D+L FP+++ SRL TL+ Q+++W
Sbjct: 184 SLDNMSLK----IKLHASVAMLAKK-----NPVLKDELKFPSMEKSRLLTLVKQTMDW 232
Score = 35.8 bits (81), Expect = 0.14
Identities = 24/74 (32%), Positives = 36/74 (48%), Gaps = 8/74 (10%)
Query: 103 VFSTFNEELYKEITQLLTLTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLV 162
+ F+EE Y+E L+ + + D R + ++L KL E+NP R KL
Sbjct: 15 ILQFFDEEGYEESLHLIP--------EETCCVDKAPGRAKLCVDLHKLAESNPSLRGKLD 66
Query: 163 FPTLKSSRLRTLIN 176
FP+L S L LI+
Sbjct: 67 FPSLNKSALLALIS 80
Score = 34.3 bits (77), Expect = 0.41
Identities = 14/27 (51%), Positives = 22/27 (80%)
Query: 6 RELVFLILQFLEEEKFKESVHKLEKES 32
+ LVFLILQF +EE ++ES+H + +E+
Sbjct: 9 KNLVFLILQFFDEEGYEESLHLIPEET 35
>At3g49660 putative WD-40 repeat - protein
Length = 317
Score = 58.9 bits (141), Expect = 2e-08
Identities = 68/294 (23%), Positives = 120/294 (40%), Gaps = 54/294 (18%)
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNE-LAQRV-EIDAHIGGVNDLAFAHPNKQ 467
+VS V +S DG + A I Y N N+ +A+ V E H G++D+AF+ +
Sbjct: 26 AVSSVKFSSDGRLLASASADKTIRTYTINTINDPIAEPVQEFTGHENGISDVAFSSDAR- 84
Query: 468 LCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAW- 525
+V+ DDK +K+WD+ TG + GH Y+ C + I S + D ++ W
Sbjct: 85 -FIVSASDDKTLKLWDVETGSLIKTLIGHTN--YAFCVNFNPQSNMIVSGSFDETVRIWD 141
Query: 526 -----------LYDNMGSRVDYDAPGHWCTTMLYSADG-TRLFSCGT------------- 560
+ + + VD++ G + Y DG R++ GT
Sbjct: 142 VTTGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSY--DGLCRIWDSGTGHCVKTLIDDENP 199
Query: 561 -------SKDG--------DSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQN--RFL 603
S +G D+ L WN S +TY G + + N R +
Sbjct: 200 PVSFVRFSPNGKFILVGTLDNTLRLWNISSAKFLKTYTGHVNAQYCISSAFSVTNGKRIV 259
Query: 604 AAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKI 657
+ ED+ + W++++ L + EG + + ++ + NL+A + D +I
Sbjct: 260 SGSEDNCVHMWELNSKKLL--QKLEGHTETVMNVACHPTENLIASGSLDKTVRI 311
>At5g08560 WD-repeat protein-like
Length = 589
Score = 55.1 bits (131), Expect = 2e-07
Identities = 104/553 (18%), Positives = 207/553 (36%), Gaps = 78/553 (14%)
Query: 7 ELVFLILQFLEEEKFKESVHKLEKESGFFFN---MKYFEEKVQAGEWEEVEKYLSGFTKV 63
E V +I + L + ++ LE+ESG + +K F ++V+ G+W++ K L
Sbjct: 67 EFVRIITRALYSLGYDKTGAMLEEESGISLHNSTIKLFLQQVKDGKWDQSVKTLHRIGFP 126
Query: 64 DDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLTLTN 123
D+ F + +QK+LE L + A+ L ++ + +++ + L++ ++
Sbjct: 127 DEKAVKAASFL-LLEQKFLEFLKVEKIADALRTLRNEMAPLRINTKRVHELASSLISPSS 185
Query: 124 FRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLNWQH 183
F + + ++ +R +L EL+ L+ A+ + +K RL L+ SL+ Q
Sbjct: 186 FISHTTSTPGKESVNSRSKVLEELQTLLPASVIIPEK---------RLECLVENSLHIQR 236
Query: 184 QLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLPVAAVAKPAAYTSLGVGAHGPFPPA 243
C +L+ DH C P + + L +G +
Sbjct: 237 DSCVFHNTLDSDLSLYSDHQCGKHQIPSQTAQI-----LESHTDEVWFLQFSHNGKY--L 289
Query: 244 AATANANALAGWMANASVSSSVQAAVVTASTIPVPHNQVSILKRPITPSTTPGMVEYQSA 303
A+++ W +A I + H V K I +P + +
Sbjct: 290 ASSSKDQTAIIWEISAD------------GHISLKHTLVGHHKPVIAILWSPDDRQVLTC 337
Query: 304 DHEQLMKRLRPAPSVEEVSYPSARQASWSLDDLPRTVAMSLHQGSSVTSMDFHPSHQTLL 363
E++++R W +D V M G S S ++P Q +
Sbjct: 338 GAEEVIRR-------------------WDVDS-GDCVHMYEKGGISPISCGWYPDGQG-I 376
Query: 364 LVGSNNGEISLWELGMRERLVSKPFKI-------------WDISAC-----SLPFQAA-- 403
+ G + I +W+L RE+ K + W +S C SL + A
Sbjct: 377 IAGMTDRSICMWDLDGREKECWKGQRTQKVSDIAMTDDGKWLVSVCKDSVISLFDREATV 436
Query: 404 --VVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAF 461
++++ ++ + S D ++ V I ++ G ++ R + + F
Sbjct: 437 ERLIEEEDMITSFSLSNDNKYILVNLLNQEIRLWNIEGDPKIVSRYKGHKRSRFIIRSCF 496
Query: 462 AHPNKQLCVVTCGDDKLIKVWD-LTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDG 520
KQ + + +D + +W TG+ + GH V + N+ + S + DG
Sbjct: 497 G-GYKQAFIASGSEDSQVYIWHRSTGKLIVELPGHAGAVNCV-SWSPTNLHMLASASDDG 554
Query: 521 KIKAWLYDNMGSR 533
I+ W D + +
Sbjct: 555 TIRIWGLDRINQQ 567
>At2g43770 putative splicing factor
Length = 343
Score = 53.1 bits (126), Expect = 8e-07
Identities = 53/238 (22%), Positives = 95/238 (39%), Gaps = 15/238 (6%)
Query: 425 VAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDL 484
+A H I+ + + + + H + DL + Q +V+ DK ++ WD+
Sbjct: 68 IASGSHDREIFLWRVHGDCKNFMVLKGHKNAILDLHWTSDGSQ--IVSASPDKTVRAWDV 125
Query: 485 -TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWC 543
TG+++ H + V S CP + I S + DG K W G+ + +
Sbjct: 126 ETGKQIKKMAEHSSFVNSCCP-TRRGPPLIISGSDDGTAKLWDMRQRGA-IQTFPDKYQI 183
Query: 544 TTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFL 603
T + +S ++F+ G D + W+ +G T G + G + + L
Sbjct: 184 TAVSFSDAADKIFTGGVDND----VKVWDLRKGEATMTLEGHQDTITG-MSLSPDGSYLL 238
Query: 604 AAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKILANA 661
G D+++ WDM P +G H N E NLL + + +G K+ A +
Sbjct: 239 TNGMDNKLCVWDMRPYAP--QNRCVKIFEGHQH---NFEKNLLKCSWSPDGTKVTAGS 291
Score = 36.6 bits (83), Expect = 0.082
Identities = 54/277 (19%), Positives = 102/277 (36%), Gaps = 39/277 (14%)
Query: 348 SSVTSMDFHPSHQTLLLVGSNNGEISLWEL--GMRERLVSKPFKIWDISACSLPFQAAVV 405
S+V +M F+P+ TL+ GS++ EI LW + + +V K K
Sbjct: 54 SAVYTMKFNPAG-TLIASGSHDREIFLWRVHGDCKNFMVLKGHK---------------- 96
Query: 406 KDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPN 465
++ + W+ DGS + A + + ++ + E H VN
Sbjct: 97 ---NAILDLHWTSDGSQIVSASPDKTVRAWDVETGKQIKKMAE---HSSFVNSCCPTRRG 150
Query: 466 KQLCVVTCGDDKLIKVWDLTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAW 525
L +++ DD K+WD+ R + + ++ + IF+ VD +K W
Sbjct: 151 PPL-IISGSDDGTAKLWDMRQRGAIQTFPDKYQITAV--SFSDAADKIFTGGVDNDVKVW 207
Query: 526 LYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWN----ESEGAIKRT 581
+ + + T M S DG+ L + G D+ L W+ + +
Sbjct: 208 DLRKGEATMTLEGHQDTITGMSLSPDGSYLLTNGM----DNKLCVWDMRPYAPQNRCVKI 263
Query: 582 YNGFR---KKSAGVVQFDTTQNRFLAAGEDSQIKFWD 615
+ G + +K+ + + A D + WD
Sbjct: 264 FEGHQHNFEKNLLKCSWSPDGTKVTAGSSDRMVHIWD 300
Score = 32.3 bits (72), Expect = 1.5
Identities = 37/178 (20%), Positives = 64/178 (35%), Gaps = 31/178 (17%)
Query: 362 LLLVGSNNGEISLWELGMRERL-------------------------VSKPFKIWDISAC 396
L++ GS++G LW++ R + V K+WD+
Sbjct: 153 LIISGSDDGTAKLWDMRQRGAIQTFPDKYQITAVSFSDAADKIFTGGVDNDVKVWDLRKG 212
Query: 397 SLPFQAAVVKDTPSVSRVTWSLDGSFV---GVAFTKHLIHIYAYNGSNELAQRVEIDAHI 453
+DT ++ ++ S DGS++ G+ + + Y N + E H
Sbjct: 213 EATMTLEGHQDT--ITGMSLSPDGSYLLTNGMDNKLCVWDMRPYAPQNRCVKIFEGHQHN 270
Query: 454 GGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRR-LFNFEGHEAPVYSICPHHKENI 510
N L + V D+++ +WD T RR ++ GH V H E I
Sbjct: 271 FEKNLLKCSWSPDGTKVTAGSSDRMVHIWDTTSRRTIYKLPGHTGSVNECVFHPTEPI 328
>At1g10580 putative pre-mRNA splicing factor
Length = 616
Score = 51.2 bits (121), Expect = 3e-06
Identities = 44/206 (21%), Positives = 90/206 (43%), Gaps = 13/206 (6%)
Query: 452 HIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDL--TGRRLFNFEGHEAPVYSICPHHKEN 509
H GV+ + F P + +++ G D +K+WD+ +G+ + + GH V IC +
Sbjct: 281 HTKGVSAIRF-FPKQGHLLLSAGMDCKVKIWDVYNSGKCMRTYMGHAKAVRDIC-FSNDG 338
Query: 510 IQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGH--WCTTMLYSADGTRLFSCGTSKDGDSF 567
+F+ + D IK W + G + + G + + D + G S D
Sbjct: 339 SKFL-TAGYDKNIKYWDTET-GQVISTFSTGKIPYVVKLNPDDDKQNILLAGMS---DKK 393
Query: 568 LVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEA 627
+V+W+ + G + + Y+ + + F RF+ + +D ++ W+ + + +
Sbjct: 394 IVQWDINTGEVTQEYDQ-HLGAVNTITFVDNNRRFVTSSDDKSLRVWEF-GIPVVIKYIS 451
Query: 628 EGGLQGLPHLRFNKEGNLLAVTTADN 653
E + +P + + GN LA + DN
Sbjct: 452 EPHMHSMPSISVHPNGNWLAAQSLDN 477
Score = 41.2 bits (95), Expect = 0.003
Identities = 41/174 (23%), Positives = 68/174 (38%), Gaps = 15/174 (8%)
Query: 448 EIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRLFNF--EGHEAPVYSICPH 505
E D H+G VN + F N++ VT DDK ++VW+ + + E H + SI H
Sbjct: 407 EYDQHLGAVNTITFVDNNRRF--VTSSDDKSLRVWEFGIPVVIKYISEPHMHSMPSISVH 464
Query: 506 HKENIQFIFSTAVDGKIKAW-----LYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGT 560
N ++ + ++D +I + N R + + +S DG +
Sbjct: 465 PNGN--WLAAQSLDNQILIYSTRERFQLNKKKRFAGHIVAGYACQVNFSPDGRFVM---- 518
Query: 561 SKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFW 614
S DG+ W+ + RT G Q++ G D IK+W
Sbjct: 519 SGDGEGKCWFWDWKSCKVFRTLKCHNGVCIGAEWHPLEQSKVATCGWDGLIKYW 572
>At3g18130 protein kinase C-receptor/G-protein, putative
Length = 326
Score = 50.4 bits (119), Expect = 5e-06
Identities = 63/273 (23%), Positives = 117/273 (42%), Gaps = 30/273 (10%)
Query: 350 VTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP 409
VT++ + +++ S + I LW+L +K K + ++ L + V+D
Sbjct: 18 VTAIATPIDNSDIIVTASRDKSIILWKL-------TKDDKSYGVAQRRLTGHSHFVEDVV 70
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLC 469
S ++L GS+ G L + E +R H V +AF+ N+Q
Sbjct: 71 LSSDGQFALSGSWDGELRLWDL-------ATGETTRRFV--GHTKDVLSVAFSTDNRQ-- 119
Query: 470 VVTCGDDKLIKVWDLTGRRLFNF---EGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWL 526
+V+ D+ IK+W+ G + +GH+ V + + I S + D +K W
Sbjct: 120 IVSASRDRTIKLWNTLGECKYTISEGDGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWN 179
Query: 527 YDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFR 586
N R + T+ S DG+ S G KDG ++ W+ +EG K+ Y+
Sbjct: 180 LQNCKLRNSLVGHSGYLNTVAVSPDGSLCASGG--KDG--VILLWDLAEG--KKLYS--L 231
Query: 587 KKSAGVVQFDTTQNRF-LAAGEDSQIKFWDMDN 618
+ + + + NR+ L A ++ I+ WD+++
Sbjct: 232 EAGSIIHSLCFSPNRYWLCAATENSIRIWDLES 264
Score = 35.8 bits (81), Expect = 0.14
Identities = 33/132 (25%), Positives = 61/132 (46%), Gaps = 26/132 (19%)
Query: 446 RVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLT-GRRLFNFEGHEAPVYSICP 504
R + H G +N +A + P+ LC + G D +I +WDL G++L++ E ++S+C
Sbjct: 186 RNSLVGHSGYLNTVAVS-PDGSLCA-SGGKDGVILLWDLAEGKKLYSLEAGSI-IHSLCF 242
Query: 505 HHKENIQFIFSTAVDGKIKAWLYDNMGS----RVDYDAPGH---------------WCTT 545
++ A + I+ W ++ +VD + +CT+
Sbjct: 243 SPN---RYWLCAATENSIRIWDLESKSVVEDLKVDLKSEAEKNEGGVGTGNQKKVIYCTS 299
Query: 546 MLYSADGTRLFS 557
+ +SADG+ LFS
Sbjct: 300 LNWSADGSTLFS 311
Score = 35.8 bits (81), Expect = 0.14
Identities = 27/100 (27%), Positives = 46/100 (46%), Gaps = 5/100 (5%)
Query: 541 HWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQN 600
H+ ++ S+DG F+ S DG+ L W+ + G R + G K V F T
Sbjct: 64 HFVEDVVLSSDGQ--FALSGSWDGELRL--WDLATGETTRRFVGHTKDVLSVA-FSTDNR 118
Query: 601 RFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFN 640
+ ++A D IK W+ T +E +G + + +RF+
Sbjct: 119 QIVSASRDRTIKLWNTLGECKYTISEGDGHKEWVSCVRFS 158
Score = 32.7 bits (73), Expect = 1.2
Identities = 38/155 (24%), Positives = 58/155 (36%), Gaps = 8/155 (5%)
Query: 476 DKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRV 534
D +++WDL TG F GH V S+ +N Q + S + D IK W
Sbjct: 84 DGELRLWDLATGETTRRFVGHTKDVLSVA-FSTDNRQIV-SASRDRTIKLWNTLGECKYT 141
Query: 535 DYDAPGH--WCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGV 592
+ GH W + + +S + L S D + WN ++ + G
Sbjct: 142 ISEGDGHKEWVSCVRFSPN--TLVPTIVSASWDKTVKVWNLQNCKLRNSLVG-HSGYLNT 198
Query: 593 VQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEA 627
V + + G+D I WD+ L S EA
Sbjct: 199 VAVSPDGSLCASGGKDGVILLWDLAEGKKLYSLEA 233
>At5g08390 katanin p80 subunit - like protein
Length = 823
Score = 49.3 bits (116), Expect = 1e-05
Identities = 71/287 (24%), Positives = 106/287 (36%), Gaps = 74/287 (25%)
Query: 337 PRTVAMSLHQGSSVTSMDFHPSHQTLLLVGSNNGEISLWEL----------GMRERLVSK 386
P + S + S+ F S + L+ G+ +G I LW+L G R VS
Sbjct: 142 PNAILSLYGHSSGIDSVTFDAS-EGLVAAGAASGTIKLWDLEEAKVVRTLTGHRSNCVSV 200
Query: 387 PFKIWDISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQR 446
F PF + + W + K IH Y
Sbjct: 201 NFH---------PFGEFFASGSLDTNLKIWDIR--------KKGCIHTYK---------- 233
Query: 447 VEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLT-GRRLFNFEGHEAPVYSICPH 505
H GVN L F + +V+ G+D ++KVWDLT G+ L F+ HE + S+ H
Sbjct: 234 ----GHTRGVNVLRFTPDGRW--IVSGGEDNVVKVWDLTAGKLLHEFKSHEGKIQSLDFH 287
Query: 506 HKENIQFIFST-AVDGKIKAW---LYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTS 561
E F+ +T + D +K W ++ +GS + G C T +
Sbjct: 288 PHE---FLLATGSADKTVKFWDLETFELIGSG-GTETTGVRCLTF--------------N 329
Query: 562 KDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGED 608
DG S L ES +KRT S G Q ++ + +G D
Sbjct: 330 PDGKSVLCGLQES---LKRT----EPMSGGATQSNSHPEKTSGSGRD 369
Score = 48.9 bits (115), Expect = 2e-05
Identities = 54/213 (25%), Positives = 93/213 (43%), Gaps = 17/213 (7%)
Query: 448 EIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHH 506
E AH VN L + ++ +VT G+D + +W + + + GH + + S+
Sbjct: 104 EFVAHSAAVNCLKIGRKSSRV-LVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDA 162
Query: 507 KENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHW--CTTMLYSADGTRLFSCGTSKDG 564
E + + + A G IK W D ++V GH C ++ + G F+ G+
Sbjct: 163 SEGL--VAAGAASGTIKLW--DLEEAKVVRTLTGHRSNCVSVNFHPFG-EFFASGSL--- 214
Query: 565 DSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTS 624
D+ L W+ + TY G + V++F ++ GED+ +K WD+ L
Sbjct: 215 DTNLKIWDIRKKGCIHTYKG-HTRGVNVLRFTPDGRWIVSGGEDNVVKVWDLTAGKLLHE 273
Query: 625 TEA-EGGLQGLPHLRFNKEGNLLAVTTADNGFK 656
++ EG +Q L F+ LLA +AD K
Sbjct: 274 FKSHEGKIQSLD---FHPHEFLLATGSADKTVK 303
Score = 45.1 bits (105), Expect = 2e-04
Identities = 47/217 (21%), Positives = 86/217 (38%), Gaps = 19/217 (8%)
Query: 430 HLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRL 489
H ++++A N + + + H G++ + F + V IK+WDL ++
Sbjct: 132 HKVNLWAIGKPNAI---LSLYGHSSGIDSVTF--DASEGLVAAGAASGTIKLWDLEEAKV 186
Query: 490 FN-FEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLY 548
GH + S+ H +F S ++D +K W G Y + +
Sbjct: 187 VRTLTGHRSNCVSVNFHPFG--EFFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRF 244
Query: 549 SADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAA--G 606
+ DG + S G D+ + W+ + G + + F+ + D + FL A
Sbjct: 245 TPDGRWIVSGGE----DNVVKVWDLTAGKL---LHEFKSHEGKIQSLDFHPHEFLLATGS 297
Query: 607 EDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEG 643
D +KFWD++ + S E G+ L FN +G
Sbjct: 298 ADKTVKFWDLETFELIGSGGTE--TTGVRCLTFNPDG 332
>At1g48630 unknown protein
Length = 326
Score = 49.3 bits (116), Expect = 1e-05
Identities = 62/273 (22%), Positives = 117/273 (42%), Gaps = 30/273 (10%)
Query: 350 VTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP 409
VT++ + +++ S + I LW+L +K K + ++ + + V+D
Sbjct: 18 VTAIATPVDNSDVIVTSSRDKSIILWKL-------TKEDKSYGVAQRRMTGHSHFVQDVV 70
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLC 469
S ++L GS+ G L + E +R H V +AF+ N+Q
Sbjct: 71 LSSDGQFALSGSWDGELRLWDL-------ATGESTRRFV--GHTKDVLSVAFSTDNRQ-- 119
Query: 470 VVTCGDDKLIKVWDLTGRRLFNF---EGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWL 526
+V+ D+ IK+W+ G + +GH+ V + + I S + D +K W
Sbjct: 120 IVSASRDRTIKLWNTLGECKYTISEADGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWN 179
Query: 527 YDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFR 586
N R + T+ S DG+ S G KDG ++ W+ +EG K+ Y+
Sbjct: 180 LQNCKLRNTLAGHSGYLNTVAVSPDGSLCASGG--KDG--VILLWDLAEG--KKLYS--L 231
Query: 587 KKSAGVVQFDTTQNRF-LAAGEDSQIKFWDMDN 618
+ + + + NR+ L A ++ I+ WD+++
Sbjct: 232 EAGSIIHSLCFSPNRYWLCAATENSIRIWDLES 264
Score = 37.0 bits (84), Expect = 0.063
Identities = 28/100 (28%), Positives = 47/100 (47%), Gaps = 5/100 (5%)
Query: 541 HWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQN 600
H+ ++ S+DG F+ S DG+ L W+ + G R + G K V F T
Sbjct: 64 HFVQDVVLSSDGQ--FALSGSWDGELRL--WDLATGESTRRFVGHTKDVLSVA-FSTDNR 118
Query: 601 RFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFN 640
+ ++A D IK W+ T +EA+G + + +RF+
Sbjct: 119 QIVSASRDRTIKLWNTLGECKYTISEADGHKEWVSCVRFS 158
Score = 36.6 bits (83), Expect = 0.082
Identities = 34/132 (25%), Positives = 60/132 (44%), Gaps = 26/132 (19%)
Query: 446 RVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLT-GRRLFNFEGHEAPVYSICP 504
R + H G +N +A + P+ LC + G D +I +WDL G++L++ E ++S+C
Sbjct: 186 RNTLAGHSGYLNTVAVS-PDGSLCA-SGGKDGVILLWDLAEGKKLYSLEAGSI-IHSLCF 242
Query: 505 HHKENIQFIFSTAVDGKIKAWLYDNMGS----RVDYDAPGH---------------WCTT 545
++ A + I+ W ++ +VD A +CT+
Sbjct: 243 SPN---RYWLCAATENSIRIWDLESKSVVEDLKVDLKAEAEKTDGSTGIGNKTKVIYCTS 299
Query: 546 MLYSADGTRLFS 557
+ +SADG LFS
Sbjct: 300 LNWSADGNTLFS 311
Score = 36.2 bits (82), Expect = 0.11
Identities = 40/155 (25%), Positives = 59/155 (37%), Gaps = 8/155 (5%)
Query: 476 DKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRV 534
D +++WDL TG F GH V S+ +N Q + S + D IK W
Sbjct: 84 DGELRLWDLATGESTRRFVGHTKDVLSVA-FSTDNRQIV-SASRDRTIKLWNTLGECKYT 141
Query: 535 DYDAPGH--WCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGV 592
+A GH W + + +S + L S D + WN ++ T G
Sbjct: 142 ISEADGHKEWVSCVRFSPN--TLVPTIVSASWDKTVKVWNLQNCKLRNTLAG-HSGYLNT 198
Query: 593 VQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEA 627
V + + G+D I WD+ L S EA
Sbjct: 199 VAVSPDGSLCASGGKDGVILLWDLAEGKKLYSLEA 233
>At1g11160 hypothetical protein
Length = 974
Score = 48.9 bits (115), Expect = 2e-05
Identities = 51/184 (27%), Positives = 83/184 (44%), Gaps = 37/184 (20%)
Query: 348 SSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSL----PFQAA 403
S V S+ F+ S + L+L G+++G I LW+L E + + F S CS PF
Sbjct: 8 SPVDSVAFN-SEEVLVLAGASSGVIKLWDL--EESKMVRAF-TGHRSNCSAVEFHPFGEF 63
Query: 404 VVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAH 463
+ + + W T+ I Y G H G++ + F+
Sbjct: 64 LASGSSDTNLRVWD----------TRKKGCIQTYKG------------HTRGISTIEFSP 101
Query: 464 PNKQLCVVTCGDDKLIKVWDLT-GRRLFNFEGHEAPVYSICPHHKENIQFIFST-AVDGK 521
+ VV+ G D ++KVWDLT G+ L F+ HE P+ S+ H ++F+ +T + D
Sbjct: 102 DGR--WVVSGGLDNVVKVWDLTAGKLLHEFKCHEGPIRSLDFH---PLEFLLATGSADRT 156
Query: 522 IKAW 525
+K W
Sbjct: 157 VKFW 160
Score = 47.8 bits (112), Expect = 4e-05
Identities = 48/209 (22%), Positives = 90/209 (42%), Gaps = 17/209 (8%)
Query: 452 HIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRLFN-FEGHEAPVYSICPHHKENI 510
H V+ +AF ++++ V+ +IK+WDL ++ F GH + ++ H
Sbjct: 6 HTSPVDSVAFN--SEEVLVLAGASSGVIKLWDLEESKMVRAFTGHRSNCSAVEFHPFG-- 61
Query: 511 QFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVE 570
+F+ S + D ++ W G Y +T+ +S DG + S G D+ +
Sbjct: 62 EFLASGSSDTNLRVWDTRKKGCIQTYKGHTRGISTIEFSPDGRWVVSGGL----DNVVKV 117
Query: 571 WNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAA--GEDSQIKFWDMDNVNPLTSTEAE 628
W+ + G + + F+ + D FL A D +KFWD++ + +T E
Sbjct: 118 WDLTAGKL---LHEFKCHEGPIRSLDFHPLEFLLATGSADRTVKFWDLETFELIGTTRPE 174
Query: 629 GGLQGLPHLRFNKEGNLLAVTTADNGFKI 657
G+ + F+ +G L D+G K+
Sbjct: 175 A--TGVRAIAFHPDGQTL-FCGLDDGLKV 200
>At5g23430 unknown protein
Length = 837
Score = 48.5 bits (114), Expect = 2e-05
Identities = 54/213 (25%), Positives = 94/213 (43%), Gaps = 17/213 (7%)
Query: 448 EIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHH 506
E AH VN L + ++ +VT G+D + +W + + + GH + + S+
Sbjct: 11 EFVAHSAAVNCLKIGRKSSRV-LVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDA 69
Query: 507 KENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHW--CTTMLYSADGTRLFSCGTSKDG 564
E + + + A G IK W D +++ GH C ++ + G F+ G+
Sbjct: 70 SEVL--VAAGAASGTIKLW--DLEEAKIVRTLTGHRSNCISVDFHPFG-EFFASGSL--- 121
Query: 565 DSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTS 624
D+ L W+ + TY G + V++F ++ GED+ +K WD+ LT
Sbjct: 122 DTNLKIWDIRKKGCIHTYKG-HTRGVNVLRFTPDGRWVVSGGEDNIVKVWDLTAGKLLTE 180
Query: 625 TEA-EGGLQGLPHLRFNKEGNLLAVTTADNGFK 656
++ EG +Q L F+ LLA +AD K
Sbjct: 181 FKSHEGQIQSLD---FHPHEFLLATGSADRTVK 210
Score = 46.2 bits (108), Expect = 1e-04
Identities = 47/217 (21%), Positives = 86/217 (38%), Gaps = 19/217 (8%)
Query: 430 HLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRL 489
H ++++A N + + + H G++ + F ++ V IK+WDL ++
Sbjct: 39 HKVNLWAIGKPNAI---LSLYGHSSGIDSVTF--DASEVLVAAGAASGTIKLWDLEEAKI 93
Query: 490 FN-FEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLY 548
GH + S+ H +F S ++D +K W G Y + +
Sbjct: 94 VRTLTGHRSNCISVDFHPFG--EFFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRF 151
Query: 549 SADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAA--G 606
+ DG + S G D+ + W+ + G + F+ + D + FL A
Sbjct: 152 TPDGRWVVSGGE----DNIVKVWDLTAGKL---LTEFKSHEGQIQSLDFHPHEFLLATGS 204
Query: 607 EDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEG 643
D +KFWD++ + S E G+ L FN +G
Sbjct: 205 ADRTVKFWDLETFELIGSGGPE--TAGVRCLSFNPDG 239
>At1g79990 coatomer protein like complex, subunit beta 2 (beta
prime)
Length = 920
Score = 48.5 bits (114), Expect = 2e-05
Identities = 64/272 (23%), Positives = 106/272 (38%), Gaps = 40/272 (14%)
Query: 350 VTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP 409
V S+D HP+ + +L +G + +W + + + K F + ++ S F
Sbjct: 18 VKSVDLHPT-EPWILASLYSGTLCIWNY--QTQTMVKSFDVTELPVRSAKF--------- 65
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLC 469
++R W + G+ I +Y YN +++ +AH + +A HP
Sbjct: 66 -IARKQWVVAGA------DDMFIRVYNYNTMDKIKV---FEAHADYIRCVA-VHPTLPY- 113
Query: 470 VVTCGDDKLIKVWDLTGRRLFN--FEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLY 527
V++ DD LIK+WD L FEGH V + + K+ F S ++D IK W
Sbjct: 114 VLSSSDDMLIKLWDWEKGWLCTQIFEGHSHYVMQVTFNPKDTNTFA-SASLDRTIKIW-- 170
Query: 528 DNMGS-----RVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTY 582
N+GS +D G C D L T D + V W+ + +T
Sbjct: 171 -NLGSPDPNFTLDAHLKGVNCVDYFTGGDKPYLI---TGSDDHTAKV-WDYQTKSCVQTL 225
Query: 583 NGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFW 614
G + V F + ED ++ W
Sbjct: 226 EG-HTHNVSAVSFHPELPIIITGSEDGTVRIW 256
>At3g15980 putative coatomer complex subunit
Length = 921
Score = 48.1 bits (113), Expect = 3e-05
Identities = 58/267 (21%), Positives = 105/267 (38%), Gaps = 30/267 (11%)
Query: 350 VTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP 409
V S+D HP+ + +L +G + +W + + ++K F++ ++ S F
Sbjct: 21 VKSVDLHPT-EPWILASLYSGTVCIWNY--QTQTITKSFEVTELPVRSAKF--------- 68
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLC 469
+ R W + G+ I +Y YN +++ +AH + +A HP
Sbjct: 69 -IPRKQWVVAGA------DDMYIRVYNYNTMDKVKV---FEAHSDYIRCVA-VHPTLPY- 116
Query: 470 VVTCGDDKLIKVWDLTGRRLFN--FEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLY 527
V++ DD LIK+WD FEGH V + + K+ F S ++D IK W
Sbjct: 117 VLSSSDDMLIKLWDWENGWACTQIFEGHSHYVMQVVFNPKDTNTFA-SASLDRTIKIWNL 175
Query: 528 DNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRK 587
+ DA + Y G + + S D + + W+ + +T +G
Sbjct: 176 GSPDPNFTLDAHQKGVNCVDYFTGGDKPYLITGSDDHTAKV--WDYQTKSCVQTLDG-HT 232
Query: 588 KSAGVVQFDTTQNRFLAAGEDSQIKFW 614
+ V F + ED ++ W
Sbjct: 233 HNVSAVCFHPELPIIITGSEDGTVRIW 259
>At1g52360 coatomer complex subunit, putative
Length = 926
Score = 47.8 bits (112), Expect = 4e-05
Identities = 59/267 (22%), Positives = 106/267 (39%), Gaps = 30/267 (11%)
Query: 350 VTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP 409
V S+D HP+ + +L +G + +W + ++++K F++ ++ S F
Sbjct: 18 VKSVDLHPT-EPWILASLYSGTLCIWNY--QTQVMAKSFEVTELPVRSAKF--------- 65
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLC 469
V+R W + G+ I +Y YN +++ +AH + +A HP
Sbjct: 66 -VARKQWVVAGA------DDMYIRVYNYNTMDKVKV---FEAHSDYIRCVA-VHPTLPY- 113
Query: 470 VVTCGDDKLIKVWDLTGRRLFN--FEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLY 527
V++ DD LIK+WD FEGH V + + K+ F S ++D IK W
Sbjct: 114 VLSSSDDMLIKLWDWEKGWACTQIFEGHSHYVMQVTFNPKDTNTFA-SASLDRTIKIWNL 172
Query: 528 DNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRK 587
+ DA + Y G + + S D + + W+ + +T G
Sbjct: 173 GSPDPNFTLDAHQKGVNCVDYFTGGDKPYLITGSDDHTAKV--WDYQTKSCVQTLEG-HT 229
Query: 588 KSAGVVQFDTTQNRFLAAGEDSQIKFW 614
+ V F + ED ++ W
Sbjct: 230 HNVSAVCFHPELPIIITGSEDGTVRIW 256
>At5g67320 unknown protein
Length = 613
Score = 47.4 bits (111), Expect = 5e-05
Identities = 24/64 (37%), Positives = 36/64 (55%), Gaps = 3/64 (4%)
Query: 463 HPNKQLCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGK 521
+PNKQL + + D +K+WD G+ L +F GH PVYS+ N ++I S ++D
Sbjct: 507 NPNKQLTLASASFDSTVKLWDAELGKMLCSFNGHREPVYSLA--FSPNGEYIASGSLDKS 564
Query: 522 IKAW 525
I W
Sbjct: 565 IHIW 568
Score = 33.9 bits (76), Expect = 0.53
Identities = 52/238 (21%), Positives = 96/238 (39%), Gaps = 40/238 (16%)
Query: 392 DISACSL-PFQAAVVKDTPSVSRVTWSL-DGSFVGVAFTKH---LIHIYAYNGSNELAQR 446
++ AC+ P + + + + WS+ +GSF V ++ LI +A SNE ++
Sbjct: 267 EVCACAWSPSASLLASGSGDATARIWSIPEGSFKAVHTGRNINALILKHAKGKSNEKSK- 325
Query: 447 VEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRLFNFEGHEAPVYSICPHH 506
V L + L +C D ++W L G + H+ P++S+ +
Sbjct: 326 --------DVTTLDWNGEGTLLATGSC--DGQARIWTLNGELISTLSKHKGPIFSLKWNK 375
Query: 507 KENIQFIFS---TAV--DGKIKAWLYD---NMGSRVDYDAPGHWCTTMLYSADGTRLFSC 558
K + S TAV D K + W + G +D D W + ++ T
Sbjct: 376 KGDYLLTGSVDRTAVVWDVKAEEWKQQFEFHSGPTLDVD----WRNNVSFATSST----- 426
Query: 559 GTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDM 616
D +L + E+ A +T+ G + V++D T + + +DS K W++
Sbjct: 427 ----DSMIYLCKIGETRPA--KTFTG-HQGEVNCVKWDPTGSLLASCSDDSTAKIWNI 477
Score = 33.1 bits (74), Expect = 0.91
Identities = 24/95 (25%), Positives = 43/95 (45%), Gaps = 6/95 (6%)
Query: 565 DSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGE-DSQIKFWDMDNVNPLT 623
DS + W+ G + ++NG R+ + + ++A+G D I W + +
Sbjct: 520 DSTVKLWDAELGKMLCSFNGHREPVYSLAF--SPNGEYIASGSLDKSIHIWSIKEGKIVK 577
Query: 624 STEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKIL 658
+ GG+ + +NKEGN +A ADN +L
Sbjct: 578 TYTGNGGIF---EVCWNKEGNKIAACFADNSVCVL 609
>At1g61210
Length = 282
Score = 47.0 bits (110), Expect = 6e-05
Identities = 38/178 (21%), Positives = 74/178 (41%), Gaps = 10/178 (5%)
Query: 452 HIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRLFN-FEGHEAPVYSICPHHKENI 510
H V+ +AF + ++ V+ +IK+WD+ ++ F GH + ++ H
Sbjct: 87 HTSAVDSVAF--DSAEVLVLAGASSGVIKLWDVEEAKMVRAFTGHRSNCSAVEFHPFG-- 142
Query: 511 QFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVE 570
+F+ S + D +K W G Y +T+ ++ DG + S G D+ +
Sbjct: 143 EFLASGSSDANLKIWDIRKKGCIQTYKGHSRGISTIRFTPDGRWVVSGGL----DNVVKV 198
Query: 571 WNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAE 628
W+ + G + + F + + F + D +KFWD++ + ST E
Sbjct: 199 WDLTAGKLLHEFK-FHEGPIRSLDFHPLEFLLATGSADRTVKFWDLETFELIGSTRPE 255
Score = 45.1 bits (105), Expect = 2e-04
Identities = 46/181 (25%), Positives = 80/181 (43%), Gaps = 31/181 (17%)
Query: 348 SSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSL-PFQAAVVK 406
S+V S+ F S + L+L G+++G I LW++ + + + + SA PF +
Sbjct: 89 SAVDSVAFD-SAEVLVLAGASSGVIKLWDVEEAKMVRAFTGHRSNCSAVEFHPFGEFLAS 147
Query: 407 DTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNK 466
+ + W + I Y G H G++ + F +
Sbjct: 148 GSSDANLKIWDIRKKGC----------IQTYKG------------HSRGISTIRFTPDGR 185
Query: 467 QLCVVTCGDDKLIKVWDLT-GRRLFNFEGHEAPVYSICPHHKENIQFIFST-AVDGKIKA 524
VV+ G D ++KVWDLT G+ L F+ HE P+ S+ H ++F+ +T + D +K
Sbjct: 186 W--VVSGGLDNVVKVWDLTAGKLLHEFKFHEGPIRSLDFH---PLEFLLATGSADRTVKF 240
Query: 525 W 525
W
Sbjct: 241 W 241
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.333 0.143 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,150,579
Number of Sequences: 26719
Number of extensions: 1026015
Number of successful extensions: 4540
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 107
Number of HSP's that attempted gapping in prelim test: 4193
Number of HSP's gapped (non-prelim): 387
length of query: 1115
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1005
effective length of database: 8,379,506
effective search space: 8421403530
effective search space used: 8421403530
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144513.3


