

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144513.17 + phase: 0
(319 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
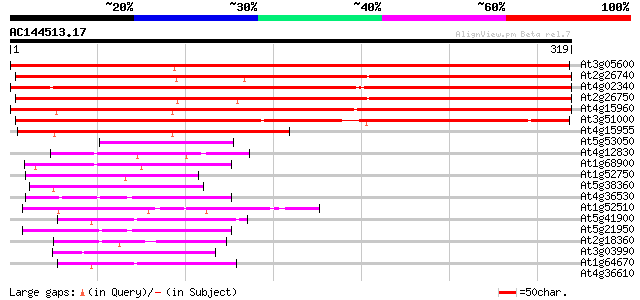
Score E
Sequences producing significant alignments: (bits) Value
At3g05600 putative epoxide hydrolase 427 e-120
At2g26740 epoxide hydrolase (ATsEH) 402 e-112
At4g02340 unknown protein 399 e-111
At2g26750 putative epoxide hydrolase 388 e-108
At4g15960 unknown protein 352 2e-97
At3g51000 epoxide hydrolase-like protein 300 7e-82
At4g15955 unknown protein 207 8e-54
At5g53050 unknown protein 55 4e-08
At4g12830 hydrolase like protein 55 4e-08
At1g68900 hypothetical protein 47 2e-05
At1g52750 alpha/beta fold family protein 47 2e-05
At5g38360 similar to unknown protein (pir||T03994) 45 5e-05
At4g36530 unknown protein 44 1e-04
At1g52510 unknown protein (At1g52510) 42 3e-04
At5g41900 unknown protein 42 4e-04
At5g21950 unknown protein 42 4e-04
At2g18360 unknown protein 42 4e-04
At3g03990 unknown protein 42 6e-04
At1g64670 unknown protein 41 8e-04
At4g36610 unknown protein 40 0.001
>At3g05600 putative epoxide hydrolase
Length = 331
Score = 427 bits (1099), Expect = e-120
Identities = 199/323 (61%), Positives = 250/323 (76%), Gaps = 5/323 (1%)
Query: 1 MERIEHRTVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPD 59
ME I+HR V VNGI MHIAEKG KEGPVVL LHGFP+LWY+WRHQI+ L SLGYRAVAPD
Sbjct: 1 MEGIDHRMVSVNGITMHIAEKGPKEGPVVLLLHGFPDLWYTWRHQISGLSSLGYRAVAPD 60
Query: 60 LRGYGDTDVPSSISSYTCFHVVGDIVSLIDLLG--VEQVFLVGHDMGAIIGWYLCMFRPE 117
LRGYGD+D P S S YTC +VVGD+V+L+D + E+VFLVGHD GAIIGW+LC+FRPE
Sbjct: 61 LRGYGDSDSPESFSEYTCLNVVGDLVALLDSVAGNQEKVFLVGHDWGAIIGWFLCLFRPE 120
Query: 118 RIKAYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNI 177
+I +VCLSVP+ RNPK++ V G +AV+GDDYYICRFQEPG++E ++A ++N+
Sbjct: 121 KINGFVCLSVPYRSRNPKVKPVQGFKAVFGDDYYICRFQEPGKIEGEIASADPRIFLRNL 180
Query: 178 LTTRKTGPPIFPKGE-YGTGFNPDTPD-NLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRN 235
T R GPPI PK +G NP++ + LP W ++ DL ++VSKFEK GFTGGLNYYR
Sbjct: 181 FTGRTLGPPILPKDNPFGEKPNPNSENIELPEWFSKKDLDFYVSKFEKAGFTGGLNYYRA 240
Query: 236 FNLNWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKG 295
+LNWELTAPWTG KI+VPVKF+TG+ DMVYT+ +KEYIHGGGF DVP L+E+++ +
Sbjct: 241 MDLNWELTAPWTGAKIQVPVKFMTGDFDMVYTTPGMKEYIHGGGFAADVPTLQEIVVIED 300
Query: 296 VAHFNNQEAEEEISNYIYEFIMK 318
HF NQE +E++ +I +F K
Sbjct: 301 AGHFVNQEKPQEVTAHINDFFTK 323
>At2g26740 epoxide hydrolase (ATsEH)
Length = 321
Score = 402 bits (1032), Expect = e-112
Identities = 198/322 (61%), Positives = 243/322 (74%), Gaps = 7/322 (2%)
Query: 4 IEHRTVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRG 62
+EHR V NGI +H+A +G +GP+VL LHGFPELWYSWRHQI L + GYRAVAPDLRG
Sbjct: 1 MEHRKVRGNGIDIHVAIQGPSDGPIVLLLHGFPELWYSWRHQIPGLAARGYRAVAPDLRG 60
Query: 63 YGDTDVPSSISSYTCFHVVGDIVSLIDLLGV---EQVFLVGHDMGAIIGWYLCMFRPERI 119
YGD+D P+ ISSYTCF++VGD++++I L E+VF+VGHD GA+I WYLC+FRP+R+
Sbjct: 61 YGDSDAPAEISSYTCFNIVGDLIAVISALTASEDEKVFVVGHDWGALIAWYLCLFRPDRV 120
Query: 120 KAYVCLSVPFLHR--NPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNI 177
KA V LSVPF R +P ++ VD MRA YGDDYYICRFQE G++EA++AEVGT VMK +
Sbjct: 121 KALVNLSVPFSFRPTDPSVKPVDRMRAFYGDDYYICRFQEFGDVEAEIAEVGTERVMKRL 180
Query: 178 LTTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFN 237
LT R GP I PK + G +T LPSWLTE+D+AYFVSKFE+ GF+G +NYYRNFN
Sbjct: 181 LTYRTPGPVIIPKDKSFWGSKGETIP-LPSWLTEEDVAYFVSKFEEKGFSGPVNYYRNFN 239
Query: 238 LNWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVA 297
N EL PW G KI+VP KF+ GELD+VY +KEYIHG FKEDVP LEE ++ +GVA
Sbjct: 240 RNNELLGPWVGSKIQVPTKFVIGELDLVYYMPGVKEYIHGPQFKEDVPLLEEPVVMEGVA 299
Query: 298 HFNNQEAEEEISNYIYEFIMKF 319
HF NQE +EI I +FI KF
Sbjct: 300 HFINQEKPQEILQIILDFISKF 321
>At4g02340 unknown protein
Length = 324
Score = 399 bits (1025), Expect = e-111
Identities = 184/319 (57%), Positives = 235/319 (72%), Gaps = 3/319 (0%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
ME+IEH T+ NGI MH+A G GPV+LF+HGFP+LWYSWRHQ+ + +LGYRA+APDL
Sbjct: 1 MEKIEHTTISTNGINMHVASIGS-GPVILFVHGFPDLWYSWRHQLVSFAALGYRAIAPDL 59
Query: 61 RGYGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIK 120
RGYGD+D P S SYT H+VGD+V L+D LGV++VFLVGHD GAI+ W+LCM RP+R+
Sbjct: 60 RGYGDSDAPPSRESYTILHIVGDLVGLLDSLGVDRVFLVGHDWGAIVAWWLCMIRPDRVN 119
Query: 121 AYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTT 180
A V SV F RNP ++ VD RA++GDDYYICRFQEPGE+E A+V T ++ T+
Sbjct: 120 ALVNTSVVFNPRNPSVKPVDAFRALFGDDYYICRFQEPGEIEEDFAQVDTKKLITRFFTS 179
Query: 181 RKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLNW 240
R PP PK G PD P +LP+WLTE D+ ++ KF + GFTGGLNYYR NL+W
Sbjct: 180 RNPRPPCIPKSVGFRGL-PD-PPSLPAWLTEQDVRFYGDKFSQKGFTGGLNYYRALNLSW 237
Query: 241 ELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAHFN 300
ELTAPWTG++IKVPVKFI G+LD+ Y KEYIH GG K+ VP L+EV++ +GV HF
Sbjct: 238 ELTAPWTGLQIKVPVKFIVGDLDITYNIPGTKEYIHEGGLKKHVPFLQEVVVMEGVGHFL 297
Query: 301 NQEAEEEISNYIYEFIMKF 319
+QE +E++++IY F KF
Sbjct: 298 HQEKPDEVTDHIYGFFKKF 316
>At2g26750 putative epoxide hydrolase
Length = 320
Score = 388 bits (997), Expect = e-108
Identities = 191/321 (59%), Positives = 241/321 (74%), Gaps = 6/321 (1%)
Query: 4 IEHRTVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRG 62
+EHR V NGI +H+A +G +G +VL LHGFPELWYSWRHQI+ L + GYRAVAPDLRG
Sbjct: 1 MEHRNVRGNGIDIHVAIQGPSDGTIVLLLHGFPELWYSWRHQISGLAARGYRAVAPDLRG 60
Query: 63 YGDTDVPSSISSYTCFHVVGDIVSLIDLLGVE--QVFLVGHDMGAIIGWYLCMFRPERIK 120
YGD+D P+ ISS+TCF++VGD+V++I L E +VF+VGHD GA+I WYLC+FRP+++K
Sbjct: 61 YGDSDAPAEISSFTCFNIVGDLVAVISTLIKEDKKVFVVGHDWGALIAWYLCLFRPDKVK 120
Query: 121 AYVCLSVP--FLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNIL 178
A V LSVP F +P ++ VD MRAVYG+DYY+CRFQE G++EA++AEVGT VMK +L
Sbjct: 121 ALVNLSVPLSFWPTDPSVKPVDRMRAVYGNDYYVCRFQEVGDIEAEIAEVGTERVMKRLL 180
Query: 179 TTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNL 238
T R GP I PK + G +T LPSWLTE+D+AYFVSKF++ GF G +NYYRNFN
Sbjct: 181 TYRTPGPLIIPKDKSFWGSKGETIP-LPSWLTEEDVAYFVSKFKEKGFCGPVNYYRNFNR 239
Query: 239 NWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAH 298
N EL PW G KI+VP KF+ GELD+VY +KEYIHG FKEDVP +EE ++ +GVAH
Sbjct: 240 NNELLGPWVGSKIQVPTKFVIGELDLVYYMPGVKEYIHGPQFKEDVPLIEEPVVMEGVAH 299
Query: 299 FNNQEAEEEISNYIYEFIMKF 319
F NQE +EI I +FI F
Sbjct: 300 FLNQEKPQEILQIILDFISTF 320
>At4g15960 unknown protein
Length = 375
Score = 352 bits (902), Expect = 2e-97
Identities = 167/325 (51%), Positives = 225/325 (68%), Gaps = 7/325 (2%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKEG----PVVLFLHGFPELWYSWRHQIAALGSLGYRAV 56
++ +EH+T++VNGI MH+AEK G P++LFLHGFPELWY+WRHQ+ AL SLGYR +
Sbjct: 51 LDGVEHKTLKVNGINMHVAEKPGSGSGEDPIILFLHGFPELWYTWRHQMVALSSLGYRTI 110
Query: 57 APDLRGYGDTDVPSSISSYTCFHVVGDIVSLIDLL--GVEQVFLVGHDMGAIIGWYLCMF 114
APDLRGYGDT+ P + YT +V+LI + G + V +VGHD GA+I W LC +
Sbjct: 111 APDLRGYGDTEAPEKVEDYTLLKRGRSVVALIVAVTGGDKAVSVVGHDWGAMIAWQLCQY 170
Query: 115 RPERIKAYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVM 174
RPE++KA V +SV F RNP V +R V+GDDYY+CRFQ+ GE+E + ++GT V+
Sbjct: 171 RPEKVKALVNMSVLFSPRNPVRVPVPTLRHVFGDDYYVCRFQKAGEIETEFKKLGTENVL 230
Query: 175 KNILTTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYR 234
K LT + GP PK +Y + + LP WLT++DL Y+V+K+E GFTG +NYYR
Sbjct: 231 KEFLTYKTPGPLNLPKDKYFKR-SENAASALPLWLTQEDLDYYVTKYENKGFTGPINYYR 289
Query: 235 NFNLNWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQK 294
N + NWELTAPWTG KI+VPVKFI G+ D+ Y KEYI+GGGFK DVP L+E ++ K
Sbjct: 290 NIDRNWELTAPWTGAKIRVPVKFIIGDQDLTYNFPGAKEYINGGGFKRDVPLLDETVVLK 349
Query: 295 GVAHFNNQEAEEEISNYIYEFIMKF 319
G+ HF ++E + I+ +I+ F KF
Sbjct: 350 GLGHFLHEENPDVINQHIHNFFHKF 374
>At3g51000 epoxide hydrolase-like protein
Length = 323
Score = 300 bits (768), Expect = 7e-82
Identities = 152/325 (46%), Positives = 205/325 (62%), Gaps = 21/325 (6%)
Query: 4 IEHRTVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRG 62
+ + ++ NGI +++AEKG +EGP+VL LHGFPE WYSWRHQI L S GY VAPDLRG
Sbjct: 5 VREKKIKTNGIWLNVAEKGDEEGPLVLLLHGFPETWYSWRHQIDFLSSHGYHVVAPDLRG 64
Query: 63 YGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAY 122
YGD+D S SYT H+V D++ L+D G Q F+ GHD GAIIGW LC+FRP+R+K +
Sbjct: 65 YGDSDSLPSHESYTVSHLVADVIGLLDHYGTTQAFVAGHDWGAIIGWCLCLFRPDRVKGF 124
Query: 123 VCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTTRK 182
+ LSVP+ R+PK++ D + ++GD YI +FQ+PG EA A+ VMK L +
Sbjct: 125 ISLSVPYFPRDPKLKPSDFFK-IFGDGLYITQFQKPGRAEAAFAKHDCLSVMKKFLLITR 183
Query: 183 TGPPIFPKGEYGTGFNPDT--------PDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYR 234
T + P PDT P +P W+TE+++ + KF+++GFTG LNYYR
Sbjct: 184 TDYLVAP---------PDTEIIDHLEIPSTIPDWITEEEIQVYAEKFQRSGFTGPLNYYR 234
Query: 235 NFNLNWELTAPWTGVKIKVPVKFITGELDMVYTSFN-LKEYIHGGGFKEDVPNLEEVIIQ 293
+ ++NWE+ APW KI VP KFI G+ D+ Y N EY+ G FK VPNLE V+I+
Sbjct: 235 SMDMNWEILAPWQDSKIVVPTKFIAGDKDIGYEGPNGTMEYVKGEVFKIVVPNLEIVVIE 294
Query: 294 KGVAHFNNQEAEEEISNYIYEFIMK 318
G HF QE E++S I F+ K
Sbjct: 295 GG-HHFIQQEKSEQVSQEILSFLNK 318
>At4g15955 unknown protein
Length = 178
Score = 207 bits (526), Expect = 8e-54
Identities = 98/165 (59%), Positives = 119/165 (71%), Gaps = 10/165 (6%)
Query: 5 EHRTVEVNGIKMHIAEKGKE--------GPVVLFLHGFPELWYSWRHQIAALGSLGYRAV 56
+H V+VNGI MH+AEK PV+LFLHGFPELWY+WRHQ+ AL SLGYR +
Sbjct: 6 DHSFVKVNGITMHVAEKSPSVAGNGAIRPPVILFLHGFPELWYTWRHQMVALSSLGYRTI 65
Query: 57 APDLRGYGDTDVPSSISSYTCFHVVGDIVSLIDLL--GVEQVFLVGHDMGAIIGWYLCMF 114
APDLRGYGDTD P S+ +YT HVVGD++ LID + E+VF+VGHD GAII W+LC+F
Sbjct: 66 APDLRGYGDTDAPESVDAYTSLHVVGDLIGLIDAVVGDREKVFVVGHDWGAIIAWHLCLF 125
Query: 115 RPERIKAYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPG 159
RP+R+KA V +SV F NPK + +A YGDDYYICRFQ G
Sbjct: 126 RPDRVKALVNMSVVFDPWNPKRKPTSTFKAFYGDDYYICRFQNMG 170
>At5g53050 unknown protein
Length = 396
Score = 55.5 bits (132), Expect = 4e-08
Identities = 29/76 (38%), Positives = 42/76 (55%)
Query: 52 GYRAVAPDLRGYGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYL 111
G A D RG G + VP+ S YT + D +SL+D LG ++ ++GH MGA+I L
Sbjct: 78 GIEVCAFDNRGMGRSSVPTHKSEYTTTIMANDSISLLDHLGWKKAHIIGHSMGAMIACKL 137
Query: 112 CMFRPERIKAYVCLSV 127
PER+ + L+V
Sbjct: 138 AAMAPERVLSLALLNV 153
>At4g12830 hydrolase like protein
Length = 393
Score = 55.5 bits (132), Expect = 4e-08
Identities = 39/117 (33%), Positives = 61/117 (51%), Gaps = 7/117 (5%)
Query: 24 EGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPSS--ISSYTCFHVV 81
+ P V+ +HGFP YS+R I L S YRA+A D G+G +D P + +YT V
Sbjct: 132 DSPPVILIHGFPSQAYSYRKTIPVL-SKNYRAIAFDWLGFGFSDKPQAGYGFNYTMDEFV 190
Query: 82 GDIVSLIDLLGVEQVFLV--GHDMGAIIGWYLCMFRPERIKAYVCLSVPFLHRNPKI 136
+ S ID + +V LV G+ A++ + RP++IK + L+ P + K+
Sbjct: 191 SSLESFIDEVTTSKVSLVVQGYFSAAVVKY--ARNRPDKIKNLILLNPPLTPEHAKL 245
>At1g68900 hypothetical protein
Length = 656
Score = 46.6 bits (109), Expect = 2e-05
Identities = 36/128 (28%), Positives = 63/128 (49%), Gaps = 11/128 (8%)
Query: 9 VEVNG----IKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYG 64
V+V+G I++H + EG V LFLHGF W + + S R ++ D+ G+G
Sbjct: 360 VDVDGFSHFIRVHDVGENAEGSVALFLHGFLGTGEEWIPIMTGI-SGSARCISVDIPGHG 418
Query: 65 DTDVPSSIS------SYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPER 118
+ V S S +++ + + LI+ + +V +VG+ MGA I Y+ + +
Sbjct: 419 RSRVQSHASETQTSPTFSMEMIAEALYKLIEQITPGKVTIVGYSMGARIALYMALRFSNK 478
Query: 119 IKAYVCLS 126
I+ V +S
Sbjct: 479 IEGAVVVS 486
>At1g52750 alpha/beta fold family protein
Length = 633
Score = 46.6 bits (109), Expect = 2e-05
Identities = 32/107 (29%), Positives = 50/107 (45%), Gaps = 9/107 (8%)
Query: 10 EVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALG-SLGYRAVAPDLRGYG---- 64
++N IK+ + E ++ +HGF +SWRH + L LG R VA D G+G
Sbjct: 338 QINSIKLGDDMEKDENTGIVLVHGFGGGVFSWRHVMGELSLQLGCRVVAYDRPGWGLTSR 397
Query: 65 ----DTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAII 107
D + + + Y V ++S +G V LVGHD G ++
Sbjct: 398 LIRKDWEKRNLANPYKLESQVDLLLSFCSEMGFSSVILVGHDDGGLL 444
>At5g38360 similar to unknown protein (pir||T03994)
Length = 242
Score = 45.1 bits (105), Expect = 5e-05
Identities = 32/106 (30%), Positives = 48/106 (45%), Gaps = 7/106 (6%)
Query: 12 NGIKMHIAEKGK-----EGPVVLFLHGFPELWYSWRHQ-IAALGSLGYRAVAPDLRGYGD 65
+G+KM + E+ K E P ++F+HG + W + S G+ + A L G G+
Sbjct: 48 SGLKMEVIEQRKSKSERENPPLVFVHGSYHAAWCWAENWLPFFSSSGFDSYAVSLLGQGE 107
Query: 66 TDVPSSISSYTCFHVVGDIVSLIDL-LGVEQVFLVGHDMGAIIGWY 110
+D P + T DI I+ LG LVGH G +I Y
Sbjct: 108 SDEPLGTVAGTLQTHASDIADFIESNLGSSPPVLVGHSFGGLIVQY 153
>At4g36530 unknown protein
Length = 378
Score = 43.9 bits (102), Expect = 1e-04
Identities = 29/117 (24%), Positives = 53/117 (44%), Gaps = 4/117 (3%)
Query: 10 EVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVP 69
E G K+H +G+ P+V +HGF + WR+ I L Y+ A DL G+G +D
Sbjct: 85 EWRGHKIHYVVQGEGSPLV-LIHGFGASVFHWRYNIPELAK-KYKVYALDLLGFGWSD-- 140
Query: 70 SSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYVCLS 126
++ Y ++ + + E +VG+ +G + + PE++ L+
Sbjct: 141 KALIEYDAMVWTDQVIDFMKEVVKEPAVVVGNSLGGFTALSVAVGLPEQVTGVALLN 197
>At1g52510 unknown protein (At1g52510)
Length = 380
Score = 42.4 bits (98), Expect = 3e-04
Identities = 40/180 (22%), Positives = 81/180 (44%), Gaps = 19/180 (10%)
Query: 8 TVEVNGIKMHIAEKGKEGP---VVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYG 64
T++ ++ + E G + ++F+HG P +S+R ++ L G+ APD G+G
Sbjct: 105 TIKSGKLRWFVRETGSKESRRGTIVFVHGAPTQSFSYRTVMSELSDAGFHCFAPDWIGFG 164
Query: 65 DTDVPSSISSYTC----FHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWY---LCMFRPE 117
+D P + +H D L+++L V+ F + G ++G Y + P
Sbjct: 165 FSDKPQPGYGFNYTEKEYHEAFD--KLLEVLEVKSPFFL-VVQGFLVGSYGLTWALKNPS 221
Query: 118 RIKAYVCLSVPFLHRNPKIRTVDGMR-AVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKN 176
+++ L+ P +P +R ++G+ + C + + + E G+ YV+KN
Sbjct: 222 KVEKLAILNSPLTVSSPVPGLFKQLRIPLFGE--FTC---QNAILAERFIEGGSPYVLKN 276
>At5g41900 unknown protein
Length = 471
Score = 42.0 bits (97), Expect = 4e-04
Identities = 33/113 (29%), Positives = 53/113 (46%), Gaps = 7/113 (6%)
Query: 28 VLFLHGFPELWYSWRHQI----AALGSLGYRAVAPDLRGYGDTDVPSSISSYTC-FHVVG 82
V+F+HGF W + + YR +A DL GYG + P+ S YT H+
Sbjct: 193 VVFIHGFVSSSAFWTETLFPNFSDSAKSNYRFIAVDLLGYGRSPKPND-SLYTLREHLEM 251
Query: 83 DIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYVCLSVPFLHRNPK 135
S+I ++ +V H +G I+ L + P IK+ L+ P+ ++ PK
Sbjct: 252 IEKSVISKFKLKTFHIVAHSLGCILALALAVKHPGAIKSLTLLAPPY-YKVPK 303
>At5g21950 unknown protein
Length = 220
Score = 42.0 bits (97), Expect = 4e-04
Identities = 31/120 (25%), Positives = 55/120 (45%), Gaps = 4/120 (3%)
Query: 8 TVEVNGIKMHIAEKGKEGPVVLFLHGF-PELWYSWRHQIAALGSLGYRAVAPDLRGYGDT 66
T++ G + + + P +L LHGF P + W HQ+ L +R PDL +G +
Sbjct: 37 TIQFWGPPPSSSSENTQKPSLLLLHGFGPSAVWQWSHQVKPLSHF-FRLYVPDLVFFGGS 95
Query: 67 DVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYVCLS 126
SS + + + L++ L VE+ +VG G + + + PE+++ V S
Sbjct: 96 S--SSGENRSEMFQALCMGKLMEKLEVERFSVVGTSYGGFVAYNMAKMFPEKVEKVVLAS 153
>At2g18360 unknown protein
Length = 313
Score = 42.0 bits (97), Expect = 4e-04
Identities = 31/103 (30%), Positives = 48/103 (46%), Gaps = 12/103 (11%)
Query: 26 PVVLFLHGFP-ELWYSWRHQIAALGSLGYRAVAPDLR----GYGDTDVPSSISSYTCFHV 80
PV+LF+HGF E +W+ Q+ +L Y PDL Y D S C
Sbjct: 63 PVLLFIHGFAAEGIVTWQFQVGSLAKK-YSVYIPDLLFFGGSYSDNADRSPAFQAHC--- 118
Query: 81 VGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYV 123
+V + +LG+E+ LVG G ++ + + PE ++A V
Sbjct: 119 ---LVKSLRILGIEKFTLVGFSYGGMVAFKIAEEYPEMVQAMV 158
>At3g03990 unknown protein
Length = 267
Score = 41.6 bits (96), Expect = 6e-04
Identities = 32/96 (33%), Positives = 47/96 (48%), Gaps = 4/96 (4%)
Query: 25 GPVVLFL-HGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPS-SISSYTCFH-VV 81
G +LFL HGF +W H I + YR V DL G + + YT V
Sbjct: 18 GDRILFLAHGFGTDQSAW-HLILPYFTQNYRVVLYDLVCAGSVNPDYFDFNRYTTLDPYV 76
Query: 82 GDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPE 117
D+++++D LG++ VGH + A+IG + RPE
Sbjct: 77 DDLLNIVDSLGIQNCAYVGHSVSAMIGIIASIRRPE 112
>At1g64670 unknown protein
Length = 469
Score = 41.2 bits (95), Expect = 8e-04
Identities = 32/107 (29%), Positives = 49/107 (44%), Gaps = 6/107 (5%)
Query: 28 VLFLHGFPELWYSWRHQI----AALGSLGYRAVAPDLRGYGDTDVPSSISSYTC-FHVVG 82
V+F+HGF W + + YR +A DL GYG + P+ S YT H+
Sbjct: 185 VVFIHGFLSSSTFWTETLFPNFSDSAKSNYRFLAVDLLGYGKSPKPND-SLYTLKEHLEM 243
Query: 83 DIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYVCLSVPF 129
S+I ++ LV H +G I+ L + P IK+ L+ P+
Sbjct: 244 IERSVISQFRLKTFHLVAHSLGCILALALAVKHPGAIKSLTLLAPPY 290
>At4g36610 unknown protein
Length = 317
Score = 40.4 bits (93), Expect = 0.001
Identities = 30/99 (30%), Positives = 49/99 (49%), Gaps = 4/99 (4%)
Query: 26 PVVLFLHGFP-ELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPSSISSYTCFHVVGDI 84
PVVL +HGF E +W+ Q+ AL S Y PDL +G + +S S +
Sbjct: 61 PVVLLIHGFAGEGIVTWQFQVGAL-SKKYSVYIPDLLFFGGSYTDNSDRSPA--FQADCL 117
Query: 85 VSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYV 123
V + +LGV++ VG G ++ + + P+ ++A V
Sbjct: 118 VKGLRILGVDKFVPVGFSYGGMVAFKIAEAYPDMVRAIV 156
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.142 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,121,008
Number of Sequences: 26719
Number of extensions: 379670
Number of successful extensions: 783
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 724
Number of HSP's gapped (non-prelim): 59
length of query: 319
length of database: 11,318,596
effective HSP length: 99
effective length of query: 220
effective length of database: 8,673,415
effective search space: 1908151300
effective search space used: 1908151300
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144513.17


