

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144513.16 + phase: 0
(319 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
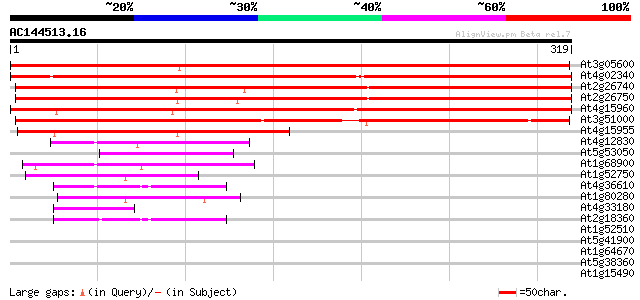
Score E
Sequences producing significant alignments: (bits) Value
At3g05600 putative epoxide hydrolase 424 e-119
At4g02340 unknown protein 399 e-112
At2g26740 epoxide hydrolase (ATsEH) 395 e-110
At2g26750 putative epoxide hydrolase 380 e-106
At4g15960 unknown protein 351 3e-97
At3g51000 epoxide hydrolase-like protein 307 6e-84
At4g15955 unknown protein 206 1e-53
At4g12830 hydrolase like protein 56 3e-08
At5g53050 unknown protein 47 1e-05
At1g68900 hypothetical protein 44 9e-05
At1g52750 alpha/beta fold family protein 44 2e-04
At4g36610 unknown protein 42 6e-04
At1g80280 unknown protein 42 6e-04
At4g33180 putative protein 41 8e-04
At2g18360 unknown protein 41 8e-04
At1g52510 unknown protein (At1g52510) 41 0.001
At5g41900 unknown protein 40 0.001
At1g64670 unknown protein 40 0.002
At5g38360 similar to unknown protein (pir||T03994) 39 0.003
At1g15490 unknown protein 38 0.006
>At3g05600 putative epoxide hydrolase
Length = 331
Score = 424 bits (1091), Expect = e-119
Identities = 198/323 (61%), Positives = 248/323 (76%), Gaps = 5/323 (1%)
Query: 1 MEGIEHRRVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPD 59
MEGI+HR V VNGI MHIAEKG KEGPVVL LHGFP+LWY+WRHQI+ L SLGYRAVAPD
Sbjct: 1 MEGIDHRMVSVNGITMHIAEKGPKEGPVVLLLHGFPDLWYTWRHQISGLSSLGYRAVAPD 60
Query: 60 LRGYGDTEAPSSISSYTGFHIVGDLVALIDLLGVDQ--VFLVAHDWGAIIGWYLCMFRPE 117
LRGYGD+++P S S YT ++VGDLVAL+D + +Q VFLV HDWGAIIGW+LC+FRPE
Sbjct: 61 LRGYGDSDSPESFSEYTCLNVVGDLVALLDSVAGNQEKVFLVGHDWGAIIGWFLCLFRPE 120
Query: 118 RIKAYVCLSVPFTRRNPKIRTVDGMRAAYGDDYYISRFQEPGKMEAQMAEVGTAYVMKST 177
+I +VCLSVP+ RNPK++ V G +A +GDDYYI RFQEPGK+E ++A +++
Sbjct: 121 KINGFVCLSVPYRSRNPKVKPVQGFKAVFGDDYYICRFQEPGKIEGEIASADPRIFLRNL 180
Query: 178 LTTRKTGPPIFPKGE-FGTGFNPDTPD-KLPSWLTEDDLAYFVSKFEKTGFVGGLNYYRN 235
T R GPPI PK FG NP++ + +LP W ++ DL ++VSKFEK GF GGLNYYR
Sbjct: 181 FTGRTLGPPILPKDNPFGEKPNPNSENIELPEWFSKKDLDFYVSKFEKAGFTGGLNYYRA 240
Query: 236 LNLNWELMAPWNGVKIKVPVKFITGDLDIVYTSPKVKEYIHGGGFKEDVPNLEEVIVQKG 295
++LNWEL APW G KI+VPVKF+TGD D+VYT+P +KEYIHGGGF DVP L+E++V +
Sbjct: 241 MDLNWELTAPWTGAKIQVPVKFMTGDFDMVYTTPGMKEYIHGGGFAADVPTLQEIVVIED 300
Query: 296 VAHFNNQEAAEEISNHIYEFIKK 318
HF NQE +E++ HI +F K
Sbjct: 301 AGHFVNQEKPQEVTAHINDFFTK 323
>At4g02340 unknown protein
Length = 324
Score = 399 bits (1026), Expect = e-112
Identities = 188/319 (58%), Positives = 231/319 (71%), Gaps = 3/319 (0%)
Query: 1 MEGIEHRRVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
ME IEH + NGI MH+A G GPV+LF+HGFP+LWYSWRHQ+ + +LGYRA+APDL
Sbjct: 1 MEKIEHTTISTNGINMHVASIGS-GPVILFVHGFPDLWYSWRHQLVSFAALGYRAIAPDL 59
Query: 61 RGYGDTEAPSSISSYTGFHIVGDLVALIDLLGVDQVFLVAHDWGAIIGWYLCMFRPERIK 120
RGYGD++AP S SYT HIVGDLV L+D LGVD+VFLV HDWGAI+ W+LCM RP+R+
Sbjct: 60 RGYGDSDAPPSRESYTILHIVGDLVGLLDSLGVDRVFLVGHDWGAIVAWWLCMIRPDRVN 119
Query: 121 AYVCLSVPFTRRNPKIRTVDGMRAAYGDDYYISRFQEPGKMEAQMAEVGTAYVMKSTLTT 180
A V SV F RNP ++ VD RA +GDDYYI RFQEPG++E A+V T ++ T+
Sbjct: 120 ALVNTSVVFNPRNPSVKPVDAFRALFGDDYYICRFQEPGEIEEDFAQVDTKKLITRFFTS 179
Query: 181 RKTGPPIFPKGEFGTGFNPDTPDKLPSWLTEDDLAYFVSKFEKTGFVGGLNYYRNLNLNW 240
R PP PK G PD P LP+WLTE D+ ++ KF + GF GGLNYYR LNL+W
Sbjct: 180 RNPRPPCIPKSVGFRGL-PD-PPSLPAWLTEQDVRFYGDKFSQKGFTGGLNYYRALNLSW 237
Query: 241 ELMAPWNGVKIKVPVKFITGDLDIVYTSPKVKEYIHGGGFKEDVPNLEEVIVQKGVAHFN 300
EL APW G++IKVPVKFI GDLDI Y P KEYIH GG K+ VP L+EV+V +GV HF
Sbjct: 238 ELTAPWTGLQIKVPVKFIVGDLDITYNIPGTKEYIHEGGLKKHVPFLQEVVVMEGVGHFL 297
Query: 301 NQEAAEEISNHIYEFIKKF 319
+QE +E+++HIY F KKF
Sbjct: 298 HQEKPDEVTDHIYGFFKKF 316
>At2g26740 epoxide hydrolase (ATsEH)
Length = 321
Score = 395 bits (1015), Expect = e-110
Identities = 199/322 (61%), Positives = 242/322 (74%), Gaps = 7/322 (2%)
Query: 4 IEHRRVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRG 62
+EHR+V NGI +H+A +G +GP+VL LHGFPELWYSWRHQI L + GYRAVAPDLRG
Sbjct: 1 MEHRKVRGNGIDIHVAIQGPSDGPIVLLLHGFPELWYSWRHQIPGLAARGYRAVAPDLRG 60
Query: 63 YGDTEAPSSISSYTGFHIVGDLVALIDLLGV---DQVFLVAHDWGAIIGWYLCMFRPERI 119
YGD++AP+ ISSYT F+IVGDL+A+I L ++VF+V HDWGA+I WYLC+FRP+R+
Sbjct: 61 YGDSDAPAEISSYTCFNIVGDLIAVISALTASEDEKVFVVGHDWGALIAWYLCLFRPDRV 120
Query: 120 KAYVCLSVPFTRR--NPKIRTVDGMRAAYGDDYYISRFQEPGKMEAQMAEVGTAYVMKST 177
KA V LSVPF+ R +P ++ VD MRA YGDDYYI RFQE G +EA++AEVGT VMK
Sbjct: 121 KALVNLSVPFSFRPTDPSVKPVDRMRAFYGDDYYICRFQEFGDVEAEIAEVGTERVMKRL 180
Query: 178 LTTRKTGPPIFPKGEFGTGFNPDTPDKLPSWLTEDDLAYFVSKFEKTGFVGGLNYYRNLN 237
LT R GP I PK + G +T LPSWLTE+D+AYFVSKFE+ GF G +NYYRN N
Sbjct: 181 LTYRTPGPVIIPKDKSFWGSKGETIP-LPSWLTEEDVAYFVSKFEEKGFSGPVNYYRNFN 239
Query: 238 LNWELMAPWNGVKIKVPVKFITGDLDIVYTSPKVKEYIHGGGFKEDVPNLEEVIVQKGVA 297
N EL+ PW G KI+VP KF+ G+LD+VY P VKEYIHG FKEDVP LEE +V +GVA
Sbjct: 240 RNNELLGPWVGSKIQVPTKFVIGELDLVYYMPGVKEYIHGPQFKEDVPLLEEPVVMEGVA 299
Query: 298 HFNNQEAAEEISNHIYEFIKKF 319
HF NQE +EI I +FI KF
Sbjct: 300 HFINQEKPQEILQIILDFISKF 321
>At2g26750 putative epoxide hydrolase
Length = 320
Score = 380 bits (976), Expect = e-106
Identities = 191/321 (59%), Positives = 238/321 (73%), Gaps = 6/321 (1%)
Query: 4 IEHRRVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRG 62
+EHR V NGI +H+A +G +G +VL LHGFPELWYSWRHQI+ L + GYRAVAPDLRG
Sbjct: 1 MEHRNVRGNGIDIHVAIQGPSDGTIVLLLHGFPELWYSWRHQISGLAARGYRAVAPDLRG 60
Query: 63 YGDTEAPSSISSYTGFHIVGDLVALIDLLGVD--QVFLVAHDWGAIIGWYLCMFRPERIK 120
YGD++AP+ ISS+T F+IVGDLVA+I L + +VF+V HDWGA+I WYLC+FRP+++K
Sbjct: 61 YGDSDAPAEISSFTCFNIVGDLVAVISTLIKEDKKVFVVGHDWGALIAWYLCLFRPDKVK 120
Query: 121 AYVCLSVP--FTRRNPKIRTVDGMRAAYGDDYYISRFQEPGKMEAQMAEVGTAYVMKSTL 178
A V LSVP F +P ++ VD MRA YG+DYY+ RFQE G +EA++AEVGT VMK L
Sbjct: 121 ALVNLSVPLSFWPTDPSVKPVDRMRAVYGNDYYVCRFQEVGDIEAEIAEVGTERVMKRLL 180
Query: 179 TTRKTGPPIFPKGEFGTGFNPDTPDKLPSWLTEDDLAYFVSKFEKTGFVGGLNYYRNLNL 238
T R GP I PK + G +T LPSWLTE+D+AYFVSKF++ GF G +NYYRN N
Sbjct: 181 TYRTPGPLIIPKDKSFWGSKGETIP-LPSWLTEEDVAYFVSKFKEKGFCGPVNYYRNFNR 239
Query: 239 NWELMAPWNGVKIKVPVKFITGDLDIVYTSPKVKEYIHGGGFKEDVPNLEEVIVQKGVAH 298
N EL+ PW G KI+VP KF+ G+LD+VY P VKEYIHG FKEDVP +EE +V +GVAH
Sbjct: 240 NNELLGPWVGSKIQVPTKFVIGELDLVYYMPGVKEYIHGPQFKEDVPLIEEPVVMEGVAH 299
Query: 299 FNNQEAAEEISNHIYEFIKKF 319
F NQE +EI I +FI F
Sbjct: 300 FLNQEKPQEILQIILDFISTF 320
>At4g15960 unknown protein
Length = 375
Score = 351 bits (901), Expect = 3e-97
Identities = 167/325 (51%), Positives = 223/325 (68%), Gaps = 7/325 (2%)
Query: 1 MEGIEHRRVEVNGIKMHIAEKGKEG----PVVLFLHGFPELWYSWRHQIAALGSLGYRAV 56
++G+EH+ ++VNGI MH+AEK G P++LFLHGFPELWY+WRHQ+ AL SLGYR +
Sbjct: 51 LDGVEHKTLKVNGINMHVAEKPGSGSGEDPIILFLHGFPELWYTWRHQMVALSSLGYRTI 110
Query: 57 APDLRGYGDTEAPSSISSYTGFHIVGDLVALIDLL--GVDQVFLVAHDWGAIIGWYLCMF 114
APDLRGYGDTEAP + YT +VALI + G V +V HDWGA+I W LC +
Sbjct: 111 APDLRGYGDTEAPEKVEDYTLLKRGRSVVALIVAVTGGDKAVSVVGHDWGAMIAWQLCQY 170
Query: 115 RPERIKAYVCLSVPFTRRNPKIRTVDGMRAAYGDDYYISRFQEPGKMEAQMAEVGTAYVM 174
RPE++KA V +SV F+ RNP V +R +GDDYY+ RFQ+ G++E + ++GT V+
Sbjct: 171 RPEKVKALVNMSVLFSPRNPVRVPVPTLRHVFGDDYYVCRFQKAGEIETEFKKLGTENVL 230
Query: 175 KSTLTTRKTGPPIFPKGEFGTGFNPDTPDKLPSWLTEDDLAYFVSKFEKTGFVGGLNYYR 234
K LT + GP PK ++ + + LP WLT++DL Y+V+K+E GF G +NYYR
Sbjct: 231 KEFLTYKTPGPLNLPKDKYFKR-SENAASALPLWLTQEDLDYYVTKYENKGFTGPINYYR 289
Query: 235 NLNLNWELMAPWNGVKIKVPVKFITGDLDIVYTSPKVKEYIHGGGFKEDVPNLEEVIVQK 294
N++ NWEL APW G KI+VPVKFI GD D+ Y P KEYI+GGGFK DVP L+E +V K
Sbjct: 290 NIDRNWELTAPWTGAKIRVPVKFIIGDQDLTYNFPGAKEYINGGGFKRDVPLLDETVVLK 349
Query: 295 GVAHFNNQEAAEEISNHIYEFIKKF 319
G+ HF ++E + I+ HI+ F KF
Sbjct: 350 GLGHFLHEENPDVINQHIHNFFHKF 374
>At3g51000 epoxide hydrolase-like protein
Length = 323
Score = 307 bits (786), Expect = 6e-84
Identities = 151/325 (46%), Positives = 210/325 (64%), Gaps = 21/325 (6%)
Query: 4 IEHRRVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRG 62
+ ++++ NGI +++AEKG +EGP+VL LHGFPE WYSWRHQI L S GY VAPDLRG
Sbjct: 5 VREKKIKTNGIWLNVAEKGDEEGPLVLLLHGFPETWYSWRHQIDFLSSHGYHVVAPDLRG 64
Query: 63 YGDTEAPSSISSYTGFHIVGDLVALIDLLGVDQVFLVAHDWGAIIGWYLCMFRPERIKAY 122
YGD+++ S SYT H+V D++ L+D G Q F+ HDWGAIIGW LC+FRP+R+K +
Sbjct: 65 YGDSDSLPSHESYTVSHLVADVIGLLDHYGTTQAFVAGHDWGAIIGWCLCLFRPDRVKGF 124
Query: 123 VCLSVPFTRRNPKIRTVDGMRAAYGDDYYISRFQEPGKMEAQMAEVGTAYVMKSTLTTRK 182
+ LSVP+ R+PK++ D + +GD YI++FQ+PG+ EA A+ VMK L +
Sbjct: 125 ISLSVPYFPRDPKLKPSDFFK-IFGDGLYITQFQKPGRAEAAFAKHDCLSVMKKFLLITR 183
Query: 183 TGPPIFPKGEFGTGFNPDT--------PDKLPSWLTEDDLAYFVSKFEKTGFVGGLNYYR 234
T + P PDT P +P W+TE+++ + KF+++GF G LNYYR
Sbjct: 184 TDYLVAP---------PDTEIIDHLEIPSTIPDWITEEEIQVYAEKFQRSGFTGPLNYYR 234
Query: 235 NLNLNWELMAPWNGVKIKVPVKFITGDLDIVYTSPK-VKEYIHGGGFKEDVPNLEEVIVQ 293
++++NWE++APW KI VP KFI GD DI Y P EY+ G FK VPNLE V+++
Sbjct: 235 SMDMNWEILAPWQDSKIVVPTKFIAGDKDIGYEGPNGTMEYVKGEVFKIVVPNLEIVVIE 294
Query: 294 KGVAHFNNQEAAEEISNHIYEFIKK 318
G HF QE +E++S I F+ K
Sbjct: 295 GG-HHFIQQEKSEQVSQEILSFLNK 318
>At4g15955 unknown protein
Length = 178
Score = 206 bits (524), Expect = 1e-53
Identities = 97/165 (58%), Positives = 119/165 (71%), Gaps = 10/165 (6%)
Query: 5 EHRRVEVNGIKMHIAEKGKE--------GPVVLFLHGFPELWYSWRHQIAALGSLGYRAV 56
+H V+VNGI MH+AEK PV+LFLHGFPELWY+WRHQ+ AL SLGYR +
Sbjct: 6 DHSFVKVNGITMHVAEKSPSVAGNGAIRPPVILFLHGFPELWYTWRHQMVALSSLGYRTI 65
Query: 57 APDLRGYGDTEAPSSISSYTGFHIVGDLVALIDLLGVD--QVFLVAHDWGAIIGWYLCMF 114
APDLRGYGDT+AP S+ +YT H+VGDL+ LID + D +VF+V HDWGAII W+LC+F
Sbjct: 66 APDLRGYGDTDAPESVDAYTSLHVVGDLIGLIDAVVGDREKVFVVGHDWGAIIAWHLCLF 125
Query: 115 RPERIKAYVCLSVPFTRRNPKIRTVDGMRAAYGDDYYISRFQEPG 159
RP+R+KA V +SV F NPK + +A YGDDYYI RFQ G
Sbjct: 126 RPDRVKALVNMSVVFDPWNPKRKPTSTFKAFYGDDYYICRFQNMG 170
>At4g12830 hydrolase like protein
Length = 393
Score = 55.8 bits (133), Expect = 3e-08
Identities = 37/115 (32%), Positives = 58/115 (50%), Gaps = 3/115 (2%)
Query: 24 EGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTEAPSS--ISSYTGFHIV 81
+ P V+ +HGFP YS+R I L S YRA+A D G+G ++ P + +YT V
Sbjct: 132 DSPPVILIHGFPSQAYSYRKTIPVL-SKNYRAIAFDWLGFGFSDKPQAGYGFNYTMDEFV 190
Query: 82 GDLVALIDLLGVDQVFLVAHDWGAIIGWYLCMFRPERIKAYVCLSVPFTRRNPKI 136
L + ID + +V LV + + RP++IK + L+ P T + K+
Sbjct: 191 SSLESFIDEVTTSKVSLVVQGYFSAAVVKYARNRPDKIKNLILLNPPLTPEHAKL 245
>At5g53050 unknown protein
Length = 396
Score = 47.0 bits (110), Expect = 1e-05
Identities = 25/76 (32%), Positives = 38/76 (49%)
Query: 52 GYRAVAPDLRGYGDTEAPSSISSYTGFHIVGDLVALIDLLGVDQVFLVAHDWGAIIGWYL 111
G A D RG G + P+ S YT + D ++L+D LG + ++ H GA+I L
Sbjct: 78 GIEVCAFDNRGMGRSSVPTHKSEYTTTIMANDSISLLDHLGWKKAHIIGHSMGAMIACKL 137
Query: 112 CMFRPERIKAYVCLSV 127
PER+ + L+V
Sbjct: 138 AAMAPERVLSLALLNV 153
>At1g68900 hypothetical protein
Length = 656
Score = 44.3 bits (103), Expect = 9e-05
Identities = 38/142 (26%), Positives = 66/142 (45%), Gaps = 11/142 (7%)
Query: 8 RVEVNG----IKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGY 63
RV+V+G I++H + EG V LFLHGF W + + S R ++ D+ G+
Sbjct: 359 RVDVDGFSHFIRVHDVGENAEGSVALFLHGFLGTGEEWIPIMTGI-SGSARCISVDIPGH 417
Query: 64 GDTEAPSSIS------SYTGFHIVGDLVALIDLLGVDQVFLVAHDWGAIIGWYLCMFRPE 117
G + S S +++ I L LI+ + +V +V + GA I Y+ +
Sbjct: 418 GRSRVQSHASETQTSPTFSMEMIAEALYKLIEQITPGKVTIVGYSMGARIALYMALRFSN 477
Query: 118 RIKAYVCLSVPFTRRNPKIRTV 139
+I+ V +S ++P R +
Sbjct: 478 KIEGAVVVSGSPGLKDPVARKI 499
>At1g52750 alpha/beta fold family protein
Length = 633
Score = 43.5 bits (101), Expect = 2e-04
Identities = 32/107 (29%), Positives = 49/107 (44%), Gaps = 9/107 (8%)
Query: 10 EVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALG-SLGYRAVAPDLRGYG---- 64
++N IK+ + E ++ +HGF +SWRH + L LG R VA D G+G
Sbjct: 338 QINSIKLGDDMEKDENTGIVLVHGFGGGVFSWRHVMGELSLQLGCRVVAYDRPGWGLTSR 397
Query: 65 ----DTEAPSSISSYTGFHIVGDLVALIDLLGVDQVFLVAHDWGAII 107
D E + + Y V L++ +G V LV HD G ++
Sbjct: 398 LIRKDWEKRNLANPYKLESQVDLLLSFCSEMGFSSVILVGHDDGGLL 444
>At4g36610 unknown protein
Length = 317
Score = 41.6 bits (96), Expect = 6e-04
Identities = 32/99 (32%), Positives = 50/99 (50%), Gaps = 4/99 (4%)
Query: 26 PVVLFLHGFP-ELWYSWRHQIAALGSLGYRAVAPDLRGYGDTEAPSSISSYTGFHIVGDL 84
PVVL +HGF E +W+ Q+ AL S Y PDL +G + +S S F L
Sbjct: 61 PVVLLIHGFAGEGIVTWQFQVGAL-SKKYSVYIPDLLFFGGSYTDNSDRS-PAFQ-ADCL 117
Query: 85 VALIDLLGVDQVFLVAHDWGAIIGWYLCMFRPERIKAYV 123
V + +LGVD+ V +G ++ + + P+ ++A V
Sbjct: 118 VKGLRILGVDKFVPVGFSYGGMVAFKIAEAYPDMVRAIV 156
>At1g80280 unknown protein
Length = 647
Score = 41.6 bits (96), Expect = 6e-04
Identities = 37/118 (31%), Positives = 53/118 (44%), Gaps = 14/118 (11%)
Query: 28 VLFLHGFPELWYSWRHQIAALG-SLGYRAVAPDLRGYG--------DTEAPSSISSYTGF 78
V+ +HGF +SWRH +++L LG A D G+G D E + YT
Sbjct: 374 VVLVHGFGGGVFSWRHVMSSLAHQLGCVVTAFDRPGWGLTARPHKKDLEEREMPNPYTLD 433
Query: 79 HIVGDLVALIDLLGVDQVFLVAHDWGAIIGW-----YLCMFRPERIKAYVCLSVPFTR 131
+ V L+A +G V LV HD G ++ L P ++K V L+V TR
Sbjct: 434 NQVDMLLAFCHEMGFASVVLVGHDDGGLLALKAAQRLLETKDPIKVKGVVLLNVSLTR 491
>At4g33180 putative protein
Length = 111
Score = 41.2 bits (95), Expect = 8e-04
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 1/47 (2%)
Query: 26 PVVLFLHGF-PELWYSWRHQIAALGSLGYRAVAPDLRGYGDTEAPSS 71
PV+L LHGF P + WR Q+ A +R +PDL +GD+ + S+
Sbjct: 56 PVMLLLHGFGPSSMWQWRRQMQAFSPSAFRVYSPDLVFFGDSTSSST 102
>At2g18360 unknown protein
Length = 313
Score = 41.2 bits (95), Expect = 8e-04
Identities = 29/99 (29%), Positives = 52/99 (52%), Gaps = 4/99 (4%)
Query: 26 PVVLFLHGFP-ELWYSWRHQIAALGSLGYRAVAPDLRGYGDTEAPSSISSYTGFHIVGDL 84
PV+LF+HGF E +W+ Q+ +L Y PDL +G + + ++ S F L
Sbjct: 63 PVLLFIHGFAAEGIVTWQFQVGSLAKK-YSVYIPDLLFFGGSYSDNADRS-PAFQ-AHCL 119
Query: 85 VALIDLLGVDQVFLVAHDWGAIIGWYLCMFRPERIKAYV 123
V + +LG+++ LV +G ++ + + PE ++A V
Sbjct: 120 VKSLRILGIEKFTLVGFSYGGMVAFKIAEEYPEMVQAMV 158
>At1g52510 unknown protein (At1g52510)
Length = 380
Score = 40.8 bits (94), Expect = 0.001
Identities = 31/121 (25%), Positives = 56/121 (45%), Gaps = 7/121 (5%)
Query: 19 AEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYG--DTEAPSSISSYT 76
+++ + G +V F+HG P +S+R ++ L G+ APD G+G D P +YT
Sbjct: 120 SKESRRGTIV-FVHGAPTQSFSYRTVMSELSDAGFHCFAPDWIGFGFSDKPQPGYGFNYT 178
Query: 77 GFHIVGDLVALIDLLGVDQVFLVAHDWGAIIGWY---LCMFRPERIKAYVCLSVPFTRRN 133
L+++L V F + G ++G Y + P +++ L+ P T +
Sbjct: 179 EKEYHEAFDKLLEVLEVKSPFFLVVQ-GFLVGSYGLTWALKNPSKVEKLAILNSPLTVSS 237
Query: 134 P 134
P
Sbjct: 238 P 238
>At5g41900 unknown protein
Length = 471
Score = 40.4 bits (93), Expect = 0.001
Identities = 28/106 (26%), Positives = 45/106 (42%), Gaps = 4/106 (3%)
Query: 28 VLFLHGFPELWYSWRHQI----AALGSLGYRAVAPDLRGYGDTEAPSSISSYTGFHIVGD 83
V+F+HGF W + + YR +A DL GYG + P+ H+
Sbjct: 193 VVFIHGFVSSSAFWTETLFPNFSDSAKSNYRFIAVDLLGYGRSPKPNDSLYTLREHLEMI 252
Query: 84 LVALIDLLGVDQVFLVAHDWGAIIGWYLCMFRPERIKAYVCLSVPF 129
++I + +VAH G I+ L + P IK+ L+ P+
Sbjct: 253 EKSVISKFKLKTFHIVAHSLGCILALALAVKHPGAIKSLTLLAPPY 298
>At1g64670 unknown protein
Length = 469
Score = 40.0 bits (92), Expect = 0.002
Identities = 29/106 (27%), Positives = 45/106 (42%), Gaps = 4/106 (3%)
Query: 28 VLFLHGFPELWYSWRHQI----AALGSLGYRAVAPDLRGYGDTEAPSSISSYTGFHIVGD 83
V+F+HGF W + + YR +A DL GYG + P+ H+
Sbjct: 185 VVFIHGFLSSSTFWTETLFPNFSDSAKSNYRFLAVDLLGYGKSPKPNDSLYTLKEHLEMI 244
Query: 84 LVALIDLLGVDQVFLVAHDWGAIIGWYLCMFRPERIKAYVCLSVPF 129
++I + LVAH G I+ L + P IK+ L+ P+
Sbjct: 245 ERSVISQFRLKTFHLVAHSLGCILALALAVKHPGAIKSLTLLAPPY 290
>At5g38360 similar to unknown protein (pir||T03994)
Length = 242
Score = 39.3 bits (90), Expect = 0.003
Identities = 29/106 (27%), Positives = 48/106 (44%), Gaps = 7/106 (6%)
Query: 12 NGIKMHIAEKGK-----EGPVVLFLHGFPELWYSWRHQ-IAALGSLGYRAVAPDLRGYGD 65
+G+KM + E+ K E P ++F+HG + W + S G+ + A L G G+
Sbjct: 48 SGLKMEVIEQRKSKSERENPPLVFVHGSYHAAWCWAENWLPFFSSSGFDSYAVSLLGQGE 107
Query: 66 TEAPSSISSYTGFHIVGDLVALIDL-LGVDQVFLVAHDWGAIIGWY 110
++ P + T D+ I+ LG LV H +G +I Y
Sbjct: 108 SDEPLGTVAGTLQTHASDIADFIESNLGSSPPVLVGHSFGGLIVQY 153
>At1g15490 unknown protein
Length = 648
Score = 38.1 bits (87), Expect = 0.006
Identities = 30/99 (30%), Positives = 46/99 (46%), Gaps = 11/99 (11%)
Query: 18 IAEKGKEGPVVLFLHGFPELWYSWRHQIAALG-SLGYRAVAPDLRGYG--------DTEA 68
+ E G+ G V+ +HGF +SWRH + +L LG A D G+G D E
Sbjct: 361 LGESGQFG--VVLVHGFGGGVFSWRHVMGSLAQQLGCVVTAFDRPGWGLTARPHKNDLEE 418
Query: 69 PSSISSYTGFHIVGDLVALIDLLGVDQVFLVAHDWGAII 107
++ Y+ + V L+A +G V V HD G ++
Sbjct: 419 RQLLNPYSLENQVEMLIAFCYEMGFSSVVFVGHDDGGLL 457
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.141 0.440
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,966,618
Number of Sequences: 26719
Number of extensions: 365666
Number of successful extensions: 755
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 695
Number of HSP's gapped (non-prelim): 53
length of query: 319
length of database: 11,318,596
effective HSP length: 99
effective length of query: 220
effective length of database: 8,673,415
effective search space: 1908151300
effective search space used: 1908151300
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144513.16


