

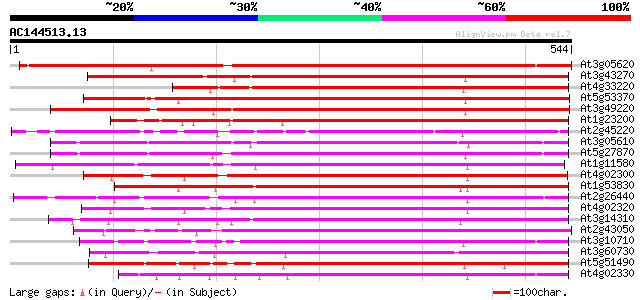
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144513.13 + phase: 0
(544 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g05620 putative pectinesterase 679 0.0
At3g43270 pectinesterase like protein 477 e-135
At4g33220 pectinesterase like protein 432 e-121
At5g53370 pectinesterase 431 e-121
At3g49220 pectinesterase - like protein 423 e-118
At1g23200 putative pectinesterase 399 e-111
At2g45220 pectinesterase like protein 394 e-110
At3g05610 putative pectinesterase 385 e-107
At5g27870 pectin methyl-esterase - like protein 384 e-107
At1g11580 unknown protein 383 e-106
At4g02300 putative pectinesterase 383 e-106
At1g53830 unknown protein 380 e-105
At2g26440 putative pectinesterase 379 e-105
At4g02320 pectinesterase - like protein 377 e-105
At3g14310 pectin methylesterase like protein 376 e-104
At2g43050 putative pectinesterase 373 e-103
At3g10710 putative pectinesterase 372 e-103
At3g60730 pectinesterase - like protein 368 e-102
At5g51490 pectinesterase 367 e-102
At4g02330 unknown protein (At4g02330) 362 e-100
>At3g05620 putative pectinesterase
Length = 543
Score = 679 bits (1752), Expect = 0.0
Identities = 341/546 (62%), Positives = 418/546 (76%), Gaps = 21/546 (3%)
Query: 10 ILILLPSFDQVLSFPDEIPSDIQTQDMQALIAQACMDIENQNSCLTNIHNELTRTGPP-S 68
+L++L S S+ I + + ++L+A+AC I+ C++NI + +G +
Sbjct: 8 LLVMLMSV-HTSSYETTILKPYKEDNFRSLVAKACQFIDAHELCVSNIWTHVKESGHGLN 66
Query: 69 PTSVINAALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEM 128
P SV+ AA++ ++A A+ + + S+ +REQ+AIEDCKEL+ FSV+ELAWS+ EM
Sbjct: 67 PHSVLRAAVKEAHDKAKLAMERIPTVMMLSIRSREQVAIEDCKELVGFSVTELAWSMLEM 126
Query: 129 RRIRAGDR---------TAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQ 179
++ G A GNL+ WLSAA+SNQDTC+EGFEGT+R+ E I GS+ Q
Sbjct: 127 NKLHGGGGIDLDDGSHDAAAAGGNLKTWLSAAMSNQDTCLEGFEGTERKYEELIKGSLRQ 186
Query: 180 VTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELL-KSKPHGKI 238
VTQL+SNVL +YTQLN LPF+ RN ++ + PEW+TE D+ L+ + P
Sbjct: 187 VTQLVSNVLDMYTQLNALPFKASRNESV--------IASPEWLTETDESLMMRHDPSVMH 238
Query: 239 ADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGI 298
+ VVA+DG G+YRTINEA+N AP+HS +R+VIYVKKG+YKENID+KKK TNIM+VGDGI
Sbjct: 239 PNTVVAIDGKGKYRTINEAINEAPNHSTKRYVIYVKKGVYKENIDLKKKKTNIMLVGDGI 298
Query: 299 GQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAF 358
GQTI+T +RNFMQG TTFRTAT AVSG+GFIAKD+TFRNTAGP N QAVALRVDSDQSAF
Sbjct: 299 GQTIITGDRNFMQGLTTFRTATVAVSGRGFIAKDITFRNTAGPQNRQAVALRVDSDQSAF 358
Query: 359 FRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTIT 418
+RCS+EG QDTLYAHSLRQFYR+CEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTIT
Sbjct: 359 YRCSVEGYQDTLYAHSLRQFYRDCEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTIT 418
Query: 419 AQGRKSPHQSTGFTIQDSYVLASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGN 478
AQGRKSP+Q+TGF IQ+SYVLA+QPTYLGRPWK YSRTVY+NTYMS +VQPRGWLEW GN
Sbjct: 419 AQGRKSPNQNTGFVIQNSYVLATQPTYLGRPWKLYSRTVYMNTYMSQLVQPRGWLEWFGN 478
Query: 479 FALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKF 538
FALDTLWYGEY N GPG +GRVKWPGYH++ D A FTV F++G WLP TGV F
Sbjct: 479 FALDTLWYGEYNNIGPGWRSSGRVKWPGYHIM-DKRTALSFTVGSFIDGRRWLPATGVTF 537
Query: 539 TAGLSN 544
TAGL+N
Sbjct: 538 TAGLAN 543
>At3g43270 pectinesterase like protein
Length = 527
Score = 477 bits (1227), Expect = e-135
Identities = 239/480 (49%), Positives = 328/480 (67%), Gaps = 17/480 (3%)
Query: 76 ALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGD 135
A +T ++ A+ ++K + +R AI DC +LLD + EL+W + + D
Sbjct: 50 AAKTVVDAITKAVAIVSKFDKKAGKSRVSNAIVDCVDLLDSAAEELSWIISASQSPNGKD 109
Query: 136 R-TAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQL 194
T +L W+SAALSNQDTC++GFEGT+ ++ ++G +++V + N+L T +
Sbjct: 110 NSTGDVGSDLRTWISAALSNQDTCLDGFEGTNGIIKKIVAGGLSKVGTTVRNLL---TMV 166
Query: 195 NRLPFRPPRNTTLHETSTDESL---EFPEWMTEADQELLKSKPHGKIADAVVALDGSGQY 251
+ P +P +T T +FP W+ D++LL++ + +ADAVVA DG+G +
Sbjct: 167 HSPPSKPKPKPIKAQTMTKAHSGFSKFPSWVKPGDRKLLQTD-NITVADAVVAADGTGNF 225
Query: 252 RTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQ 311
TI++AV AAP +S +R+VI+VK+G+Y EN+++KKK NIMMVGDGI T++T NR+F+
Sbjct: 226 TTISDAVLAAPDYSTKRYVIHVKRGVYVENVEIKKKKWNIMMVGDGIDATVITGNRSFID 285
Query: 312 GWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLY 371
GWTTFR+ATFAVSG+GFIA+D+TF+NTAGP HQAVA+R D+D F+RC++ G QDTLY
Sbjct: 286 GWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVAIRSDTDLGVFYRCAMRGYQDTLY 345
Query: 372 AHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGF 431
AHS+RQF+REC I GT+DFIFG+ AV Q+C+I + LP QK +ITAQGRK P++ TGF
Sbjct: 346 AHSMRQFFRECIITGTVDFIFGDATAVFQSCQIKAKQGLPNQKNSITAQGRKDPNEPTGF 405
Query: 432 TIQDSYVLA---------SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALD 482
TIQ S + A + TYLGRPWK YSRTV++ YMS + P GWLEW GNFALD
Sbjct: 406 TIQFSNIAADTDLLLNLNTTATYLGRPWKLYSRTVFMQNYMSDAINPVGWLEWNGNFALD 465
Query: 483 TLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
TL+YGEY N GPG+SL RVKWPGYHV+ ++ A FTV + + G WLP TG+ F AGL
Sbjct: 466 TLYYGEYMNSGPGASLDRRVKWPGYHVLNTSAEANNFTVSQLIQGNLWLPSTGITFIAGL 525
>At4g33220 pectinesterase like protein
Length = 404
Score = 432 bits (1111), Expect = e-121
Identities = 214/405 (52%), Positives = 284/405 (69%), Gaps = 24/405 (5%)
Query: 159 IEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQ------------LNRLPFRPPRNTT 206
+EGF+GT ++S ++GS+ Q+ ++ +L L + + P PP
Sbjct: 1 MEGFDGTSGLVKSLVAGSLDQLYSMLRELLPLVQPEQKPKAVSKPGPIAKGPKAPP-GRK 59
Query: 207 LHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSN 266
L +T DESL+FP+W+ D++LL+S +G+ D VALDG+G + I +A+ AP +S+
Sbjct: 60 LRDTDEDESLQFPDWVRPDDRKLLES--NGRTYDVSVALDGTGNFTKIMDAIKKAPDYSS 117
Query: 267 RRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGK 326
R VIY+KKGLY EN+++KKK NI+M+GDGI T+++ NR+F+ GWTTFR+ATFAVSG+
Sbjct: 118 TRFVIYIKKGLYLENVEIKKKKWNIVMLGDGIDVTVISGNRSFIDGWTTFRSATFAVSGR 177
Query: 327 GFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYG 386
GF+A+D+TF+NTAGP HQAVALR DSD S FFRC++ G QDTLY H++RQFYREC I G
Sbjct: 178 GFLARDITFQNTAGPEKHQAVALRSDSDLSVFFRCAMRGYQDTLYTHTMRQFYRECTITG 237
Query: 387 TIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYV-------- 438
T+DFIFG+G V QNC+I + LP QK TITAQGRK +Q +GF+IQ S +
Sbjct: 238 TVDFIFGDGTVVFQNCQILAKRGLPNQKNTITAQGRKDVNQPSGFSIQFSNISADADLVP 297
Query: 439 -LASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSS 497
L + TYLGRPWK YSRTV+I MS +V+P GWLEW +FALDTL+YGE+ NYGPGS
Sbjct: 298 YLNTTRTYLGRPWKLYSRTVFIRNNMSDVVRPEGWLEWNADFALDTLFYGEFMNYGPGSG 357
Query: 498 LAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
L+ RVKWPGYHV ++ A FTV +F+ G WLP TGV F+ GL
Sbjct: 358 LSSRVKWPGYHVFNNSDQANNFTVSQFIKGNLWLPSTGVTFSDGL 402
>At5g53370 pectinesterase
Length = 587
Score = 431 bits (1108), Expect = e-121
Identities = 224/485 (46%), Positives = 305/485 (62%), Gaps = 17/485 (3%)
Query: 72 VINAALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRI 131
+I+ + T+ + A+ + I+ + R + A + C ELLD SV L +L + +
Sbjct: 106 LIHISFNATLQKFSKALYTSSTITYTQMPPRVRSAYDSCLELLDDSVDALTRALSSVVVV 165
Query: 132 RAGDRTAQYEGNLEAWLSAALSNQDTCIEGF---EGTDRRLESYISGSVTQVTQLISNVL 188
+GD + ++ WLS+A++N DTC +GF EG ++ + G+V +++++SN L
Sbjct: 166 -SGDES---HSDVMTWLSSAMTNHDTCTDGFDEIEGQGGEVKDQVIGAVKDLSEMVSNCL 221
Query: 189 SLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGS 248
+++ + P T+E+ E P W+ D+ELL + AD V+ DGS
Sbjct: 222 AIFAGKVKDLSGVPVVNNRKLLGTEETEELPNWLKREDRELLGTPTSAIQADITVSKDGS 281
Query: 249 GQYRTINEAVNAAPSHSNRRHVIYVKKGLYKE-NIDMKKKMTNIMMVGDGIGQTIVTSNR 307
G ++TI EA+ AP HS+RR VIYVK G Y+E N+ + +K TN+M +GDG G+T++T +
Sbjct: 282 GTFKTIAEAIKKAPEHSSRRFVIYVKAGRYEEENLKVGRKKTNLMFIGDGKGKTVITGGK 341
Query: 308 NFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQ 367
+ TTF TATFA +G GFI +DMTF N AGP HQAVALRV D + +RC+I G Q
Sbjct: 342 SIADDLTTFHTATFAATGAGFIVRDMTFENYAGPAKHQAVALRVGGDHAVVYRCNIIGYQ 401
Query: 368 DTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQ 427
D LY HS RQF+RECEIYGT+DFIFGN A +LQ+C IY R P+ QK+TITAQ RK P+Q
Sbjct: 402 DALYVHSNRQFFRECEIYGTVDFIFGNAAVILQSCNIYARKPMAQQKITITAQNRKDPNQ 461
Query: 428 STGFTIQDSYVLA---------SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGN 478
+TG +I +LA S PTYLGRPWK YSR VY+ + M + PRGWLEW G
Sbjct: 462 NTGISIHACKLLATPDLEASKGSYPTYLGRPWKLYSRVVYMMSDMGDHIDPRGWLEWNGP 521
Query: 479 FALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKF 538
FALD+L+YGEY N G GS + RVKWPGYHVI A FTV +F++G SWLP TGV F
Sbjct: 522 FALDSLYYGEYMNKGLGSGIGQRVKWPGYHVITSTVEASKFTVAQFISGSSWLPSTGVSF 581
Query: 539 TAGLS 543
+GLS
Sbjct: 582 FSGLS 586
>At3g49220 pectinesterase - like protein
Length = 598
Score = 423 bits (1088), Expect = e-118
Identities = 228/518 (44%), Positives = 317/518 (61%), Gaps = 21/518 (4%)
Query: 40 IAQACMDIENQNSCLTNIHNELTRTGPPSPTSVINAALRTTINEAIGAINNMTKISTFSV 99
I++AC C+ ++ + S +I+ + T++ A+ + +S +
Sbjct: 87 ISKACELTRFPELCVDSLMDFPGSLAASSSKDLIHVTVNMTLHHFSHALYSSASLSFVDM 146
Query: 100 NNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCI 159
R + A + C ELLD SV L+ +L + A + ++ WLSAAL+N DTC
Sbjct: 147 PPRARSAYDSCVELLDDSVDALSRALSSVVSSSAKPQ------DVTTWLSAALTNHDTCT 200
Query: 160 EGFEGTDRR-LESYISGSVTQVTQLISNVLSLYTQLNR---LPFRPPRNTTLHETSTDES 215
EGF+G D ++ +++ ++ +++L+SN L++++ + P +N L E
Sbjct: 201 EGFDGVDDGGVKDHMTAALQNLSELVSNCLAIFSASHDGDDFAGVPIQNRRLLGVEEREE 260
Query: 216 LEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKK 275
+FP WM ++E+L+ AD +V+ DG+G +TI+EA+ AP +S RR +IYVK
Sbjct: 261 -KFPRWMRPKEREILEMPVSQIQADIIVSKDGNGTCKTISEAIKKAPQNSTRRIIIYVKA 319
Query: 276 GLYKEN-IDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMT 334
G Y+EN + + +K N+M VGDG G+T+++ ++ TTF TA+FA +G GFIA+D+T
Sbjct: 320 GRYEENNLKVGRKKINLMFVGDGKGKTVISGGKSIFDNITTFHTASFAATGAGFIARDIT 379
Query: 335 FRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGN 394
F N AGP HQAVALR+ +D + +RC+I G QDTLY HS RQF+REC+IYGT+DFIFGN
Sbjct: 380 FENWAGPAKHQAVALRIGADHAVIYRCNIIGYQDTLYVHSNRQFFRECDIYGTVDFIFGN 439
Query: 395 GAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLA---------SQPTY 445
A VLQNC IY R P+ QK TITAQ RK P+Q+TG +I S VLA S TY
Sbjct: 440 AAVVLQNCSIYARKPMDFQKNTITAQNRKDPNQNTGISIHASRVLAASDLQATNGSTQTY 499
Query: 446 LGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWP 505
LGRPWK +SRTVY+ +Y+ V RGWLEW FALDTL+YGEY N GPGS L RV WP
Sbjct: 500 LGRPWKLFSRTVYMMSYIGGHVHTRGWLEWNTTFALDTLYYGEYLNSGPGSGLGQRVSWP 559
Query: 506 GYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGLS 543
GY VI + A FTV F+ G SWLP TGV F AGLS
Sbjct: 560 GYRVINSTAEANRFTVAEFIYGSSWLPSTGVSFLAGLS 597
>At1g23200 putative pectinesterase
Length = 554
Score = 399 bits (1026), Expect = e-111
Identities = 222/469 (47%), Positives = 284/469 (60%), Gaps = 33/469 (7%)
Query: 98 SVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDT 157
S++ A+ DC EL + ++ +L S R G ++ ++ LSAA++NQDT
Sbjct: 95 SLHKHATSALFDCLELYEDTIDQLNHS-----RRSYGQYSSPHDRQTS--LSAAIANQDT 147
Query: 158 CIEGFEGTD--------------RRLESYISGS--VTQVTQLISNVLSLYTQLNRLPFRP 201
C GF R L IS S VT+ V Y F
Sbjct: 148 CRNGFRDFKLTSSYSKYFPVQFHRNLTKSISNSLAVTKAAAEAEAVAEKYPSTGFTKFSK 207
Query: 202 PRNTTLHETS------TDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTIN 255
R++ + +DE +FP W +D++LL+ AD VVA DGSG Y +I
Sbjct: 208 QRSSAGGGSHRRLLLFSDE--KFPSWFPLSDRKLLEDSKTTAKADLVVAKDGSGHYTSIQ 265
Query: 256 EAVNAAPS--HSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGW 313
+AVNAA N+R VIYVK G+Y+EN+ +KK + N+M++GDGI TIVT NRN G
Sbjct: 266 QAVNAAAKLPRRNQRLVIYVKAGVYRENVVIKKSIKNVMVIGDGIDSTIVTGNRNVQDGT 325
Query: 314 TTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAH 373
TTFR+ATFAVSG GFIA+ +TF NTAGP HQAVALR SD S F+ CS +G QDTLY H
Sbjct: 326 TTFRSATFAVSGNGFIAQGITFENTAGPEKHQAVALRSSSDFSVFYACSFKGYQDTLYLH 385
Query: 374 SLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTI 433
S RQF R C IYGT+DFIFG+ A+LQNC IY R P+ QK TITAQ RK P ++TGF I
Sbjct: 386 SSRQFLRNCNIYGTVDFIFGDATAILQNCNIYARKPMSGQKNTITAQSRKEPDETTGFVI 445
Query: 434 QDSYVLASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYG 493
Q S V + TYLGRPW+ +SRTV++ + ++V P GWL W G+FAL TL+YGEY N G
Sbjct: 446 QSSTVATASETYLGRPWRSHSRTVFMKCNLGALVSPAGWLPWSGSFALSTLYYGEYGNTG 505
Query: 494 PGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
G+S++GRVKWPGYHVIK + A FTV+ FL+G W+ TGV GL
Sbjct: 506 AGASVSGRVKWPGYHVIKTVTEAEKFTVENFLDGNYWITATGVPVNDGL 554
>At2g45220 pectinesterase like protein
Length = 511
Score = 394 bits (1013), Expect = e-110
Identities = 235/553 (42%), Positives = 327/553 (58%), Gaps = 59/553 (10%)
Query: 2 ALLVNFLFILILLPSFDQVLSFPDEIPSDIQTQDMQALIAQACMDIENQNSCLTNIHNEL 61
A ++NF+ + IL+ S S +D++A C N C + +
Sbjct: 6 AYIINFVILCILVAS----------TVSGYNQKDVKAW----CSQTPNPKPCEYFLTHNS 51
Query: 62 TRTGPPSPTSVINAALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSEL 121
S + + +++ ++ AI A + + + RE+ A EDC +L D +VS++
Sbjct: 52 NNEPIKSESEFLKISMKLVLDRAILAKTHAFTLGPKCRDTREKAAWEDCIKLYDLTVSKI 111
Query: 122 AWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQV- 180
++ ++ AQ WLS AL+N DTC GF LE ++ V +
Sbjct: 112 NETMDP--NVKCSKLDAQ------TWLSTALTNLDTCRAGF------LELGVTDIVLPLM 157
Query: 181 TQLISNVLSLYTQLNRLPFR--PPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKI 238
+ +SN+L +N++PF PP E FP W+ D++LL+S
Sbjct: 158 SNNVSNLLCNTLAINKVPFNYTPP-----------EKDGFPSWVKPGDRKLLQSSTPKD- 205
Query: 239 ADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGI 298
+AVVA DGSG ++TI EA++AA R VIYVK+G+Y EN++++KK N+M+ GDGI
Sbjct: 206 -NAVVAKDGSGNFKTIKEAIDAASGSG--RFVIYVKQGVYSENLEIRKK--NVMLRGDGI 260
Query: 299 GQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAF 358
G+TI+T +++ G TTF +AT A G GFIA+ +TFRNTAG N QAVALR SD S F
Sbjct: 261 GKTIITGSKSVGGGTTTFNSATVAAVGDGFIARGITFRNTAGASNEQAVALRSGSDLSVF 320
Query: 359 FRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTIT 418
++CS E QDTLY HS RQFYR+C++YGT+DFIFGN AAVLQNC I+ R P + TIT
Sbjct: 321 YQCSFEAYQDTLYVHSNRQFYRDCDVYGTVDFIFGNAAAVLQNCNIFARRPRS-KTNTIT 379
Query: 419 AQGRKSPHQSTGFTIQDSY---------VLASQPTYLGRPWKEYSRTVYINTYMSSMVQP 469
AQGR P+Q+TG I +S VL S TYLGRPW++YSRTV++ T + S++ P
Sbjct: 380 AQGRSDPNQNTGIIIHNSRVTAASDLRPVLGSTKTYLGRPWRQYSRTVFMKTSLDSLIDP 439
Query: 470 RGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGS 529
RGWLEW GNFAL TL+Y E++N GPG+S +GRV WPG+ V+ AS A FTV FL GGS
Sbjct: 440 RGWLEWDGNFALKTLFYAEFQNTGPGASTSGRVTWPGFRVLGSASEASKFTVGTFLAGGS 499
Query: 530 WLPRTGVKFTAGL 542
W+P + V FT+GL
Sbjct: 500 WIP-SSVPFTSGL 511
>At3g05610 putative pectinesterase
Length = 669
Score = 385 bits (989), Expect = e-107
Identities = 207/521 (39%), Positives = 301/521 (57%), Gaps = 31/521 (5%)
Query: 40 IAQACMDIENQNSCL-TNIHNELTRTGPPSPTSVINAALRTTINEAIGAINNMTKISTFS 98
+ C + + +C T I N T P ++ A T+ + A I
Sbjct: 58 VKDVCAPTDYRKTCEDTLIKNGKNTT---DPMELVKTAFNVTMKQITDAAKKSQTIMELQ 114
Query: 99 VNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTC 158
++R ++A++ CKEL+D+++ EL+ S E+ + + NL WLSAA+S+++TC
Sbjct: 115 KDSRTRMALDQCKELMDYALDELSNSFEELGKFEF-HLLDEALINLRIWLSAAISHEETC 173
Query: 159 IEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLEF 218
+EGF+GT + ++ +L N L++ ++++ + E F
Sbjct: 174 LEGFQGTQGNAGETMKKALKTAIELTHNGLAIISEMSNFVGQMQIPGLNSRRLLAEG--F 231
Query: 219 PEWMTEADQELLKS-------KPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVI 271
P W+ + ++LL++ KP D VVA DGSGQY+TINEA+ P N V+
Sbjct: 232 PSWVDQRGRKLLQAAAAYSDVKP-----DIVVAQDGSGQYKTINEALQFVPKKRNTTFVV 286
Query: 272 YVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAK 331
++K GLYKE + + K M++++ +GDG +TI++ N+N+ G TT+RTAT A+ G FIAK
Sbjct: 287 HIKAGLYKEYVQVNKTMSHLVFIGDGPDKTIISGNKNYKDGITTYRTATVAIVGNYFIAK 346
Query: 332 DMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFI 391
++ F NTAG + HQAVA+RV SD+S FF C +G QDTLY HS RQF+R+C I GTIDF+
Sbjct: 347 NIGFENTAGAIKHQAVAVRVQSDESIFFNCRFDGYQDTLYTHSHRQFFRDCTISGTIDFL 406
Query: 392 FGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLASQP-------- 443
FG+ AAV QNC + R PLP Q ITA GRK P +STGF Q +A +P
Sbjct: 407 FGDAAAVFQNCTLLVRKPLPNQACPITAHGRKDPRESTGFVFQ-GCTIAGEPDYLAVKET 465
Query: 444 --TYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGR 501
YLGRPWKEYSRT+ +NT++ VQP+GW WLG+F L TL+Y E +N GPGS+LA R
Sbjct: 466 SKAYLGRPWKEYSRTIIMNTFIPDFVQPQGWQPWLGDFGLKTLFYSEVQNTGPGSALANR 525
Query: 502 VKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
V W G + + FT +++ G W+P GV +T GL
Sbjct: 526 VTWAGIKTLSEEDIL-KFTPAQYIQGDDWIPGKGVPYTTGL 565
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 384 bits (987), Expect = e-107
Identities = 196/518 (37%), Positives = 295/518 (56%), Gaps = 27/518 (5%)
Query: 40 IAQACMDIENQNSCLTNIHNELTRTGPPSPTSVINAALRTTINEAIGAINNMTKISTFSV 99
I C + + +C + + T P ++ A T+ + +
Sbjct: 57 IKDVCAPTDYKETCEDTLRKDAKDTS--DPLELVKTAFNATMKQISDVAKKSQTMIELQK 114
Query: 100 NNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCI 159
+ R ++A++ CKEL+D+++ EL+ S E+ + + + L WLSA +S++ TC+
Sbjct: 115 DPRAKMALDQCKELMDYAIGELSKSFEELGKFEF-HKVDEALVKLRIWLSATISHEQTCL 173
Query: 160 EGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQLN------RLPFRPPRNTTLHETSTD 213
+GF+GT I ++ QL N L++ T+++ ++P R
Sbjct: 174 DGFQGTQGNAGETIKKALKTAVQLTHNGLAMVTEMSNYLGQMQIPEMNSRRLL------- 226
Query: 214 ESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYV 273
S EFP WM + LL + D VVA DGSGQY+TINEA+N P N V+++
Sbjct: 227 -SQEFPSWMDARARRLLNAPMSEVKPDIVVAQDGSGQYKTINEALNFVPKKKNTTFVVHI 285
Query: 274 KKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDM 333
K+G+YKE + + + MT+++ +GDG +T+++ ++++ G TT++TAT A+ G FIAK++
Sbjct: 286 KEGIYKEYVQVNRSMTHLVFIGDGPDKTVISGSKSYKDGITTYKTATVAIVGDHFIAKNI 345
Query: 334 TFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFG 393
F NTAG + HQAVA+RV +D+S F+ C +G QDTLYAHS RQFYR+C I GTIDF+FG
Sbjct: 346 AFENTAGAIKHQAVAIRVLADESIFYNCKFDGYQDTLYAHSHRQFYRDCTISGTIDFLFG 405
Query: 394 NGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLA---------SQPT 444
+ AAV QNC + R PL Q ITA GRK P +STGF +Q ++ T
Sbjct: 406 DAAAVFQNCTLLVRKPLLNQACPITAHGRKDPRESTGFVLQGCTIVGEPDYLAVKEQSKT 465
Query: 445 YLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKW 504
YLGRPWKEYSRT+ +NT++ V P GW WLG F L+TL+Y E +N GPG+++ RV W
Sbjct: 466 YLGRPWKEYSRTIIMNTFIPDFVPPEGWQPWLGEFGLNTLFYSEVQNTGPGAAITKRVTW 525
Query: 505 PGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
PG + D FT +++ G +W+P GV + GL
Sbjct: 526 PGIKKLSDEEIL-KFTPAQYIQGDAWIPGKGVPYILGL 562
>At1g11580 unknown protein
Length = 557
Score = 383 bits (984), Expect = e-106
Identities = 212/547 (38%), Positives = 309/547 (55%), Gaps = 26/547 (4%)
Query: 6 NFLFILILLPSFDQVLSFPDEIPSDIQTQDMQALI--AQACMDIENQNSCLTNIHNELTR 63
N +L + V F ++ S + +L+ +Q C +Q+SC + T
Sbjct: 19 NLCLVLSFVAILGSVAFFTAQLISVNTNNNDDSLLTTSQICHGAHDQDSCQALLSEFTTL 78
Query: 64 T-GPPSPTSVINAALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELA 122
+ + +++ L+ ++ + +++ S R++ DC+E++D S +
Sbjct: 79 SLSKLNRLDLLHVFLKNSVWRLESTMTMVSEARIRSNGVRDKAGFADCEEMMDVSKDRM- 137
Query: 123 WSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVTQ 182
+ M +R G+ + N+ WLS+ L+N TC+E + + + +
Sbjct: 138 --MSSMEELRGGNYNLESYSNVHTWLSSVLTNYMTCLESISDVSVNSKQIVKPQLEDLVS 195
Query: 183 LISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGK--IAD 240
L+++ + LP R + S FP W+T D++LL+S P A+
Sbjct: 196 RARVALAIFVSV--LPARDDLKMII-------SNRFPSWLTALDRKLLESSPKTLKVTAN 246
Query: 241 AVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQ 300
VVA DG+G+++T+NEAV AAP +SN R+VIYVKKG+YKE ID+ KK N+M+VGDG
Sbjct: 247 VVVAKDGTGKFKTVNEAVAAAPENSNTRYVIYVKKGVYKETIDIGKKKKNLMLVGDGKDA 306
Query: 301 TIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFR 360
TI+T + N + G TTFR+AT A +G GF+A+D+ F+NTAGP HQAVALRV +DQ+ R
Sbjct: 307 TIITGSLNVIDGSTTFRSATVAANGDGFMAQDIWFQNTAGPAKHQAVALRVSADQTVINR 366
Query: 361 CSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQ 420
C I+ QDTLY H+LRQFYR+ I GT+DFIFGN A V QNC I R P QK +TAQ
Sbjct: 367 CRIDAYQDTLYTHTLRQFYRDSYITGTVDFIFGNSAVVFQNCDIVARNPGAGQKNMLTAQ 426
Query: 421 GRKSPHQSTGFTIQDSYVLASQ---------PTYLGRPWKEYSRTVYINTYMSSMVQPRG 471
GR+ +Q+T +IQ + AS T+LGRPWK YSRTV + +++ + + P G
Sbjct: 427 GREDQNQNTAISIQKCKITASSDLAPVKGSVKTFLGRPWKLYSRTVIMQSFIDNHIDPAG 486
Query: 472 WLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWL 531
W W G FAL TL+YGEY N GPG+ + RV W G+ VIKD+ A FTV + + GG WL
Sbjct: 487 WFPWDGEFALSTLYYGEYANTGPGADTSKRVNWKGFKVIKDSKEAEQFTVAKLIQGGLWL 546
Query: 532 PRTGVKF 538
TGV F
Sbjct: 547 KPTGVTF 553
>At4g02300 putative pectinesterase
Length = 532
Score = 383 bits (983), Expect = e-106
Identities = 203/491 (41%), Positives = 300/491 (60%), Gaps = 36/491 (7%)
Query: 72 VINAALRTTINEAIGAINNMTKISTF---SVNNREQLAIEDCKELLDFSVSELAWSLGEM 128
+I A L TI + A +N + + T ++ + E+ A EDC LLD ++S+L ++ ++
Sbjct: 56 LIIADLNLTILKVNLASSNFSDLQTRLFPNLTHYERCAFEDCLGLLDDTISDLETAVSDL 115
Query: 129 RRIRAGDRTAQYEGN-LEAWLSAALSNQDTCIEGFEGTDRR--------LESYISGSVTQ 179
R ++ E N + L+ ++ QDTC++GF +D L + +
Sbjct: 116 R-------SSSLEFNDISMLLTNVMTYQDTCLDGFSTSDNENNNDMTYELPENLKEIILD 168
Query: 180 VTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIA 239
++ +SN L + ++R P S++ +E+P W++E DQ LL++
Sbjct: 169 ISNNLSNSLHMLQVISRKKPSPK--------SSEVDVEYPSWLSENDQRLLEAPVQETNY 220
Query: 240 DAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIG 299
+ VA+DG+G + TIN+AV AAP+ S R +IY+K G Y EN+++ KK T IM +GDGIG
Sbjct: 221 NLSVAIDGTGNFTTINDAVFAAPNMSETRFIIYIKGGEYFENVELPKKKTMIMFIGDGIG 280
Query: 300 QTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFF 359
+T++ +NR+ + GW+TF+T T V GKG+IAKD++F N+AGP QAVA R SD SAF+
Sbjct: 281 KTVIKANRSRIDGWSTFQTPTVGVKGKGYIAKDISFVNSAGPAKAQAVAFRSGSDHSAFY 340
Query: 360 RCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITA 419
RC +G QDTLY HS +QFYREC+IYGTIDFIFGN A V QN +Y R P P K+ TA
Sbjct: 341 RCEFDGYQDTLYVHSAKQFYRECDIYGTIDFIFGNAAVVFQNSSLYARKPNPGHKIAFTA 400
Query: 420 QGRKSPHQSTGFTIQDSYVLASQ---------PTYLGRPWKEYSRTVYINTYMSSMVQPR 470
Q R Q TG +I + +LA+ YLGRPW++YSRTV I +++ ++ P
Sbjct: 401 QSRNQSDQPTGISILNCRILAAPDLIPVKENFKAYLGRPWRKYSRTVIIKSFIDDLIHPA 460
Query: 471 GWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSW 530
GWLE +FAL+TL+YGEY N GPG+++A RV WPG+ I++ + A FTV F++G +W
Sbjct: 461 GWLEGKKDFALETLYYGEYMNEGPGANMAKRVTWPGFRRIENQTEATQFTVGPFIDGSTW 520
Query: 531 LPRTGVKFTAG 541
L TG+ F+ G
Sbjct: 521 LNSTGIPFSLG 531
>At1g53830 unknown protein
Length = 587
Score = 380 bits (976), Expect = e-105
Identities = 207/465 (44%), Positives = 288/465 (61%), Gaps = 26/465 (5%)
Query: 102 REQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEG 161
RE A+ DC E +D ++ EL ++ ++ + ++ +L+ +S+A++NQ TC++G
Sbjct: 125 REVTALHDCLETIDETLDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDG 184
Query: 162 F--EGTDRRLESYISGSVTQVTQLISNVLSLYTQLNRLP------------FRPPRNTTL 207
F + DR++ + V + SN L++ + F N L
Sbjct: 185 FSYDDADRKVRKALLKGQVHVEHMCSNALAMIKNMTETDIANFELRDKSSTFTNNNNRKL 244
Query: 208 HETSTD-ESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSN 266
E + D +S +P+W++ D+ LL+ ADA VA DGSG + T+ AV AAP SN
Sbjct: 245 KEVTGDLDSDGWPKWLSVGDRRLLQGSTIK--ADATVADDGSGDFTTVAAAVAAAPEKSN 302
Query: 267 RRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGK 326
+R VI++K G+Y+EN+++ KK TNIM +GDG G+TI+T +RN + G TTF +AT A G+
Sbjct: 303 KRFVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFHSATVAAVGE 362
Query: 327 GFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYG 386
F+A+D+TF+NTAGP HQAVALRV SD SAF++C + QDTLY HS RQF+ +C I G
Sbjct: 363 RFLARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITG 422
Query: 387 TIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQD------SYVLA 440
T+DFIFGN AAVLQ+C I R P QK +TAQGR P+Q+TG IQ+ S +LA
Sbjct: 423 TVDFIFGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDLLA 482
Query: 441 SQ---PTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSS 497
+ PTYLGRPWKEYSRTV + + +S +++P GW EW G+FALDTL Y EY N G G+
Sbjct: 483 VKGTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSFALDTLTYREYLNRGGGAG 542
Query: 498 LAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
A RVKW GY VI + A FT +F+ GG WL TG F+ L
Sbjct: 543 TANRVKWKGYKVITSDTEAQPFTAGQFIGGGGWLASTGFPFSLSL 587
>At2g26440 putative pectinesterase
Length = 547
Score = 379 bits (972), Expect = e-105
Identities = 218/560 (38%), Positives = 315/560 (55%), Gaps = 41/560 (7%)
Query: 4 LVNFLFILILLPSFDQVLSFPDEIPSDIQTQDMQALIAQACMDIENQNSCLTNIHNELTR 63
L + LF+L PS P P + C + ++C T++ ++
Sbjct: 8 LSSLLFLLFFTPSVFSYSYQPSLNPHETSATSF-------CKNTPYPDACFTSLKLSISI 60
Query: 64 TGPPSPTSVINAALRTTINEAIGAINNMTKISTFSVN--NREQLAIEDCKELLDFSVSEL 121
P+ S + L+T ++EA G + ++ + S N ++ +++DCK+L + S L
Sbjct: 61 NISPNILSFLLQTLQTALSEA-GKLTDLLSGAGVSNNLVEGQRGSLQDCKDLHHITSSFL 119
Query: 122 AWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVT 181
S+ +I+ G ++ + A+LSAAL+N+ TC+EG E L+ + S T
Sbjct: 120 KRSIS---KIQDGVNDSRKLADARAYLSAALTNKITCLEGLESASGPLKPKLVTSFTTTY 176
Query: 182 QLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLE-----FPEWMTEADQELLKSKPHG 236
+ ISN LS + R TT +T + FP+W+ + D L+ G
Sbjct: 177 KHISNSLSALPK--------QRRTTNPKTGGNTKNRRLLGLFPDWVYKKDHRFLEDSSDG 228
Query: 237 -----KIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNI 291
VVA DG+G + TINEA++ AP+ SN R +IYVK+G+Y ENID+ TNI
Sbjct: 229 YDEYDPSESLVVAADGTGNFSTINEAISFAPNMSNDRVLIYVKEGVYDENIDIPIYKTNI 288
Query: 292 MMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRV 351
+++GDG T +T NR+ GWTTFR+AT AVSG+GF+A+D+ NTAGP HQAVALRV
Sbjct: 289 VLIGDGSDVTFITGNRSVGDGWTTFRSATLAVSGEGFLARDIMITNTAGPEKHQAVALRV 348
Query: 352 DSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLP 411
++D A +RC I+G QDTLY HS RQFYREC+IYGTID+IFGN A V Q C I +++P+P
Sbjct: 349 NADFVALYRCVIDGYQDTLYTHSFRQFYRECDIYGTIDYIFGNAAVVFQGCNIVSKLPMP 408
Query: 412 LQKVTITAQGRKSPHQSTGFTIQDSYVLASQ---------PTYLGRPWKEYSRTVYINTY 462
Q ITAQ R + + TG ++Q+ +LAS+ +YLGRPW+E+SRTV + +Y
Sbjct: 409 GQFTVITAQSRDTQDEDTGISMQNCSILASEDLFNSSNKVKSYLGRPWREFSRTVVMESY 468
Query: 463 MSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQ 522
+ + GW +W G ALDTL+YGEY N GPGS RV WPG+H++ A FT
Sbjct: 469 IDEFIDGSGWSKWNGGEALDTLYYGEYNNNGPGSETVKRVNWPGFHIMGYEDAFN-FTAT 527
Query: 523 RFLNGGSWLPRTGVKFTAGL 542
F+ G WL T + G+
Sbjct: 528 EFITGDGWLGSTSFPYDNGI 547
>At4g02320 pectinesterase - like protein
Length = 518
Score = 377 bits (969), Expect = e-105
Identities = 200/497 (40%), Positives = 299/497 (59%), Gaps = 40/497 (8%)
Query: 70 TSVINAALRTTINEAIGAINNMTKIST---FSVNNREQLAIEDCKELLDFSVSELAWSLG 126
T ++ AL TI+ + +N + + ++++R+ A +DC ELLD +V +L ++
Sbjct: 38 TELVVTALNQTISNVNLSSSNFSDLLQRLGSNLSHRDLCAFDDCLELLDDTVFDLTTAIS 97
Query: 127 EMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRR------------LESYIS 174
++R + N++ LSAA++N TC++GF +D + +
Sbjct: 98 KLRS------HSPELHNVKMLLSAAMTNTRTCLDGFASSDNDENLNNNDNKTYGVAESLK 151
Query: 175 GSVTQVTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKP 234
S+ ++ +S+ L++ L +P P E + FP W++ +D+ LL+
Sbjct: 152 ESLFNISSHVSDSLAM---LENIPGHIPGKVK-------EDVGFPMWVSGSDRNLLQDPV 201
Query: 235 HGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMV 294
+ VVA +G+G Y TI EA++AAP+ S R VIY+K G Y ENI++ ++ T IM +
Sbjct: 202 DETKVNLVVAQNGTGNYTTIGEAISAAPNSSETRFVIYIKCGEYFENIEIPREKTMIMFI 261
Query: 295 GDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSD 354
GDGIG+T++ +NR++ GWT F +AT V G GFIAKD++F N AGP HQAVALR SD
Sbjct: 262 GDGIGRTVIKANRSYADGWTAFHSATVGVRGSGFIAKDLSFVNYAGPEKHQAVALRSSSD 321
Query: 355 QSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQK 414
SA++RCS E QDT+Y HS +QFYREC+IYGT+DFIFG+ + V QNC +Y R P P QK
Sbjct: 322 LSAYYRCSFESYQDTIYVHSHKQFYRECDIYGTVDFIFGDASVVFQNCSLYARRPNPNQK 381
Query: 415 VTITAQGRKSPHQSTGFTIQDSYVLASQ---------PTYLGRPWKEYSRTVYINTYMSS 465
+ TAQGR++ + TG +I S +LA+ YLGRPW+ YSRTV + +++
Sbjct: 382 IIYTAQGRENSREPTGISIISSRILAAPDLIPVQANFKAYLGRPWQLYSRTVIMKSFIDD 441
Query: 466 MVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFL 525
+V P GWL+W +FAL+TL+YGEY N GPGS++ RV+WPG+ I+ A F+V F+
Sbjct: 442 LVDPAGWLKWKDDFALETLYYGEYMNEGPGSNMTNRVQWPGFKRIETVEEASQFSVGPFI 501
Query: 526 NGGSWLPRTGVKFTAGL 542
+G WL T + FT L
Sbjct: 502 DGNKWLNSTRIPFTLDL 518
>At3g14310 pectin methylesterase like protein
Length = 592
Score = 376 bits (965), Expect = e-104
Identities = 213/543 (39%), Positives = 310/543 (56%), Gaps = 45/543 (8%)
Query: 38 ALIAQACMDIENQNSCLTNIHN----ELTRTGPPSPTSVINAALRTTINEAIGAINNMTK 93
A++ +C C++ + ELT S VI A++ TI + K
Sbjct: 57 AVLRSSCSSTRYPELCISAVVTAGGVELT-----SQKDVIEASVNLTITAVEHNYFTVKK 111
Query: 94 I--STFSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAA 151
+ + RE+ A+ DC E +D ++ EL ++ ++ ++ G+L+ +S+A
Sbjct: 112 LIKKRKGLTPREKTALHDCLETIDETLDELHETVEDLHLYPTKKTLREHAGDLKTLISSA 171
Query: 152 LSNQDTCIEGF--EGTDRRLESYISGSVTQVTQLISNVLSLYTQLNRLPFR--------P 201
++NQ+TC++GF + D+++ + V + SN L++ +
Sbjct: 172 ITNQETCLDGFSHDDADKQVRKALLKGQIHVEHMCSNALAMIKNMTDTDIANFEQKAKIT 231
Query: 202 PRNTTLHETSTD-------------ESLEFPEWMTEADQELLKSKPHGKIADAVVALDGS 248
N L E + + +S +P W++ D+ LL+ G ADA VA DGS
Sbjct: 232 SNNRKLKEENQETTVAVDIAGAGELDSEGWPTWLSAGDRRLLQGS--GVKADATVAADGS 289
Query: 249 GQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRN 308
G ++T+ AV AAP +SN+R+VI++K G+Y+EN+++ KK NIM +GDG +TI+T +RN
Sbjct: 290 GTFKTVAAAVAAAPENSNKRYVIHIKAGVYRENVEVAKKKKNIMFMGDGRTRTIITGSRN 349
Query: 309 FMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQD 368
+ G TTF +AT A G+ F+A+D+TF+NTAGP HQAVALRV SD SAF+ C + QD
Sbjct: 350 VVDGSTTFHSATVAAVGERFLARDITFQNTAGPSKHQAVALRVGSDFSAFYNCDMLAYQD 409
Query: 369 TLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQS 428
TLY HS RQF+ +C I GT+DFIFGN A VLQ+C I+ R P QK +TAQGR P+Q+
Sbjct: 410 TLYVHSNRQFFVKCLIAGTVDFIFGNAAVVLQDCDIHARRPNSGQKNMVTAQGRTDPNQN 469
Query: 429 TGFTIQD---------SYVLASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNF 479
TG IQ V S PTYLGRPWKEYS+TV + + +S +++P GW EW G F
Sbjct: 470 TGIVIQKCRIGATSDLQSVKGSFPTYLGRPWKEYSQTVIMQSAISDVIRPEGWSEWTGTF 529
Query: 480 ALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFT 539
AL+TL Y EY N G G+ A RVKW G+ VI A+ A +T +F+ GG WL TG F+
Sbjct: 530 ALNTLTYREYSNTGAGAGTANRVKWRGFKVITAAAEAQKYTAGQFIGGGGWLSSTGFPFS 589
Query: 540 AGL 542
GL
Sbjct: 590 LGL 592
>At2g43050 putative pectinesterase
Length = 518
Score = 373 bits (958), Expect = e-103
Identities = 215/493 (43%), Positives = 295/493 (59%), Gaps = 36/493 (7%)
Query: 63 RTGPPSPTSV-INAALRTTINEAIGAIN---NMTKISTFSVNNREQLAIEDCKELLDFSV 118
R+ P S T A ++ ++N A+ A + N+T +S +V I DC ELLD ++
Sbjct: 51 RSSPSSKTKQGFLATVQESMNHALLARSLAFNLT-LSHRTVQTHTFDPIHDCLELLDDTL 109
Query: 119 SELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVT 178
L+ RI A + E ++ WLSAAL+NQDTC + + + ESY G
Sbjct: 110 DMLS-------RIHADND----EEDVHTWLSAALTNQDTCEQSLQ---EKSESYKHGLAM 155
Query: 179 Q-----VTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSK 233
+T L+++ L L+ + + H + FP ++ ++Q L
Sbjct: 156 DFVARNLTGLLTSSLDLFVSVK----------SKHRKLLSKQEYFPTFVPSSEQRRLLEA 205
Query: 234 PHGKI-ADAVVALDGSGQYRTINEAV-NAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNI 291
P ++ DAVVA DGSG ++TI EA+ + + + S R IY+K G Y ENI++ K N+
Sbjct: 206 PVEELNVDAVVAPDGSGTHKTIGEALLSTSLASSGGRTKIYLKAGTYHENINIPTKQKNV 265
Query: 292 MMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRV 351
M+VGDG G+T++ +R+ GWTT++TAT A G+GFIA+DMTF N AGP + QAVALRV
Sbjct: 266 MLVGDGKGKTVIVGSRSNRGGWTTYKTATVAAMGEGFIARDMTFVNNAGPKSEQAVALRV 325
Query: 352 DSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLP 411
+D+S RCS+EG QD+LY HS RQFYRE +I GT+DFIFGN A V Q+C I R PLP
Sbjct: 326 GADKSVVHRCSVEGYQDSLYTHSKRQFYRETDITGTVDFIFGNSAVVFQSCNIAARKPLP 385
Query: 412 LQKVTITAQGRKSPHQSTGFTIQDSYVLASQPTYLGRPWKEYSRTVYINTYMSSMVQPRG 471
Q+ +TAQGR +P Q+TG IQ+ + A TYLGRPWKEYSRTV + +++ + P G
Sbjct: 386 GQRNFVTAQGRSNPGQNTGIAIQNCRITAESMTYLGRPWKEYSRTVVMQSFIGGSIHPSG 445
Query: 472 WLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWL 531
W W G F L +L+YGEY N GPGSS++GRVKW G H + A FTV F++G WL
Sbjct: 446 WSPWSGGFGLKSLFYGEYGNSGPGSSVSGRVKWSGCHPSLTVTEAEKFTVASFIDGNIWL 505
Query: 532 PRTGVKFTAGLSN 544
P TGV F GL N
Sbjct: 506 PSTGVSFDPGLVN 518
>At3g10710 putative pectinesterase
Length = 561
Score = 372 bits (954), Expect = e-103
Identities = 212/484 (43%), Positives = 293/484 (59%), Gaps = 28/484 (5%)
Query: 68 SPTSVINAALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGE 127
+P + A++ TI E AIN + NN + + C ELLD ++ L +L
Sbjct: 97 NPEELFRYAVKITIAEVSKAINAFSSSLGDEKNN---ITMNACAELLDLTIDNLNNTLTS 153
Query: 128 MRRIRAGDRTA-QYEGNLEAWLSAALSNQDTCIEGFEGTDRRL-ESYISGSVTQVTQLIS 185
GD T + +L WLS+A + Q TC+E R ES++ S T+L S
Sbjct: 154 SSN---GDVTVPELVDDLRTWLSSAGTYQRTCVETLAPDMRPFGESHLKNS----TELTS 206
Query: 186 NVLSLYTQLNRLP--FRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVV 243
N L++ T L ++ F+ R T+ D ++F A + LL+S K+AD VV
Sbjct: 207 NALAIITWLGKIADSFKLRRRLL---TTADVEVDF-----HAGRRLLQSTDLRKVADIVV 258
Query: 244 ALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIV 303
A DGSG+YRTI A+ P S +R +IYVKKG+Y EN+ ++KKM N+++VGDG ++IV
Sbjct: 259 AKDGSGKYRTIKRALQDVPEKSEKRTIIYVKKGVYFENVKVEKKMWNVIVVGDGESKSIV 318
Query: 304 TSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSI 363
+ N + G TF+TATFAV GKGF+A+DM F NTAGP HQAVAL V +D +AF+RC++
Sbjct: 319 SGRLNVIDGTPTFKTATFAVFGKGFMARDMGFINTAGPSKHQAVALMVSADLTAFYRCTM 378
Query: 364 EGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRK 423
QDTLY H+ RQFYREC I GT+DFIFGN A+VLQ+C+I R P+ Q+ TITAQGR
Sbjct: 379 NAYQDTLYVHAQRQFYRECTIIGTVDFIFGNSASVLQSCRILPRRPMKGQQNTITAQGRT 438
Query: 424 SPHQSTGFTIQDSYV-----LASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGN 478
P+ +TG +I + L T+LGRPWK +S TV +++Y+ + +GWL W G+
Sbjct: 439 DPNMNTGISIHRCNISPLGDLTDVMTFLGRPWKNFSTTVIMDSYLHGFIDRKGWLPWTGD 498
Query: 479 FALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKF 538
A DT++YGEY+N GPG+S RVKW G + A FTV+ F++GG WLP T V F
Sbjct: 499 SAPDTIFYGEYKNTGPGASTKNRVKWKGLRFL-STKEANRFTVKPFIDGGRWLPATKVPF 557
Query: 539 TAGL 542
+GL
Sbjct: 558 RSGL 561
>At3g60730 pectinesterase - like protein
Length = 496
Score = 368 bits (945), Expect = e-102
Identities = 203/474 (42%), Positives = 277/474 (57%), Gaps = 24/474 (5%)
Query: 78 RTTINEAIGAINNM--TKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGD 135
RT + EA + +M T+ ++ + +L + +C++L D S + L+ + + D
Sbjct: 38 RTAVVEAKTSFGSMAVTEATSEVAGSYYKLGLSECEKLYDESEARLSKLVVDHENFTVED 97
Query: 136 RTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQLN 195
+ WLS L+N TC++G + + + +VT V + L+ Y +
Sbjct: 98 --------VRTWLSGVLANHHTCLDGLIQQRQGHKPLVHSNVTFV---LHEALAFYKKSR 146
Query: 196 RL----PFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQY 251
P RP T S P + L+ P AD VVA DGS +
Sbjct: 147 ARQGHGPTRPKHRPTRPNHGPGRSHHGPSRPNQNGGMLVSWNPTSSRADFVVARDGSATH 206
Query: 252 RTINEAVNAAPSHSN---RRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRN 308
RTIN+A+ A R +IY+K G+Y E I++ + M NIM+VGDG+ +TIVT+NRN
Sbjct: 207 RTINQALAAVSRMGKSRLNRVIIYIKAGVYNEKIEIDRHMKNIMLVGDGMDRTIVTNNRN 266
Query: 309 FMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQD 368
G TT+ +ATF VSG GF A+D+TF NTAGP HQAVALRV SD S F+RCS +G QD
Sbjct: 267 VPDGSTTYGSATFGVSGDGFWARDITFENTAGPHKHQAVALRVSSDLSLFYRCSFKGYQD 326
Query: 369 TLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQS 428
TL+ HSLRQFYR+C IYGTIDFIFG+ AAV QNC I+ R P+ Q ITAQGR PH +
Sbjct: 327 TLFTHSLRQFYRDCHIYGTIDFIFGDAAAVFQNCDIFVRRPMDHQGNMITAQGRDDPHTN 386
Query: 429 TGFTIQDSYVLASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGE 488
+ F V +YLGRPWK+YSRTV++ T + ++ PRGW EW G++AL TL+YGE
Sbjct: 387 SEF----EAVKGRFKSYLGRPWKKYSRTVFLKTDIDELIDPRGWREWSGSYALSTLYYGE 442
Query: 489 YRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
+ N G G+ RV WPG+HV++ A FTV RF+ G SW+P TGV F+AG+
Sbjct: 443 FMNTGAGAGTGRRVNWPGFHVLRGEEEASPFTVSRFIQGDSWIPITGVPFSAGV 496
>At5g51490 pectinesterase
Length = 536
Score = 367 bits (943), Expect = e-102
Identities = 207/481 (43%), Positives = 293/481 (60%), Gaps = 30/481 (6%)
Query: 77 LRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDR 136
+ ++ AI A +T ++++Q + DC +L ++ +L +L + +AG
Sbjct: 71 VEAAMDRAISARAELTNSGKNCTDSKKQAVLADCIDLYGDTIMQLNRTLHGVSP-KAGAA 129
Query: 137 TAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSV--TQVTQLISNVLSLYTQL 194
+ + + + WLS AL+N +TC G +D + +I+ V T+++ LISN L++ L
Sbjct: 130 KSCTDFDAQTWLSTALTNTETCRRG--SSDLNVTDFITPIVSNTKISHLISNCLAVNGAL 187
Query: 195 NRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTI 254
L NTT ++ FP W++ D+ LL++ A+ VVA DGSG + T+
Sbjct: 188 --LTAGNKGNTTANQKG------FPTWLSRKDKRLLRAVR----ANLVVAKDGSGHFNTV 235
Query: 255 NEAVNAAPSH--SNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQG 312
A++ A ++ R VIYVK+G+Y+ENI+++ +IM+VGDG+ TI+T R+ G
Sbjct: 236 QAAIDVAGRRKVTSGRFVIYVKRGIYQENINVRLNNDDIMLVGDGMRSTIITGGRSVQGG 295
Query: 313 WTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYA 372
+TT+ +AT + G FIAK +TFRNTAGP QAVALR SD S F++CSIEG QDTL
Sbjct: 296 YTTYNSATAGIEGLHFIAKGITFRNTAGPAKGQAVALRSSSDLSIFYKCSIEGYQDTLMV 355
Query: 373 HSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFT 432
HS RQFYREC IYGT+DFIFGN AAV QNC I R PL Q ITAQGR P Q+TG +
Sbjct: 356 HSQRQFYRECYIYGTVDFIFGNAAAVFQNCLILPRRPLKGQANVITAQGRADPFQNTGIS 415
Query: 433 IQDSYVL---------ASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGN--FAL 481
I +S +L + TY+GRPW ++SRTV + TY+ ++V P GW W+ F L
Sbjct: 416 IHNSRILPAPDLKPVVGTVKTYMGRPWMKFSRTVVLQTYLDNVVSPVGWSPWIEGSVFGL 475
Query: 482 DTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAG 541
DTL+Y EY+N GP SS RV W G+HV+ AS A FTV +F+ G +WLPRTG+ FT+G
Sbjct: 476 DTLFYAEYKNTGPASSTRWRVSWKGFHVLGRASDASAFTVGKFIAGTAWLPRTGIPFTSG 535
Query: 542 L 542
L
Sbjct: 536 L 536
>At4g02330 unknown protein (At4g02330)
Length = 573
Score = 362 bits (930), Expect = e-100
Identities = 202/478 (42%), Positives = 281/478 (58%), Gaps = 43/478 (8%)
Query: 106 AIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYE--GNLEAWLSAALSNQDTCIEGFE 163
A++DC+ L + L S E I +T + ++ LSAAL+N+ TC++G
Sbjct: 96 ALQDCRYLASLTTDYLITSF-ETVNITTSSKTLSFSKADEIQTLLSAALTNEQTCLDGIN 154
Query: 164 ---GTDRRLESYISGSVTQVTQLISNVLSLYT---------QLNRLPFRPPRNTTLHETS 211
+ + + ++ + T+L S L+L+T Q+ + P+NT H
Sbjct: 155 TAASSSWTIRNGVALPLINDTKLFSVSLALFTKGWVPKKKKQVASYSWAHPKNTHSHTKP 214
Query: 212 TDESLE--FPEWMTEADQELLKSKPHGKIAD-------------AVVALDGSGQYRTINE 256
P MTE + + +S K+AD V +G+G + TI E
Sbjct: 215 FRHFRNGALPLKMTEHTRAVYESLSRRKLADDDNDVNTVLVSDIVTVNQNGTGNFTTITE 274
Query: 257 AVNAAPSHSNRR---HVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGW 313
AVN+AP+ ++ VIYV G+Y+EN+ + K +MM+GDGI +T+VT NRN + GW
Sbjct: 275 AVNSAPNKTDGTAGYFVIYVTSGVYEENVVIAKNKRYLMMIGDGINRTVVTGNRNVVDGW 334
Query: 314 TTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAH 373
TTF +ATFAV+ F+A +MTFRNTAGP HQAVA+R +D S F+ CS E QDTLY H
Sbjct: 335 TTFNSATFAVTSPNFVAVNMTFRNTAGPEKHQAVAMRSSADLSIFYSCSFEAYQDTLYTH 394
Query: 374 SLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTI 433
SLRQFYREC+IYGT+DFIFGN A V Q+C +Y R P+ Q ITAQGR P+Q+TG +I
Sbjct: 395 SLRQFYRECDIYGTVDFIFGNAAVVFQDCNLYPRQPMQNQFNAITAQGRTDPNQNTGISI 454
Query: 434 QDSYVLASQ---------PTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTL 484
+ + + TYLGRPWKEYSRTV++ +Y+ +V+P GW EW G+FAL TL
Sbjct: 455 HNCTIKPADDLVSSNYTVKTYLGRPWKEYSRTVFMQSYIDEVVEPVGWREWNGDFALSTL 514
Query: 485 WYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
+Y EY N G GSS RV WPGYHVI +++ A FTV+ FL G W+ ++GV + +GL
Sbjct: 515 YYAEYNNTGSGSSTTDRVVWPGYHVI-NSTDANNFTVENFLLGDGWMVQSGVPYISGL 571
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,816,174
Number of Sequences: 26719
Number of extensions: 488081
Number of successful extensions: 1489
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 1161
Number of HSP's gapped (non-prelim): 113
length of query: 544
length of database: 11,318,596
effective HSP length: 104
effective length of query: 440
effective length of database: 8,539,820
effective search space: 3757520800
effective search space used: 3757520800
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144513.13


