

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.8 - phase: 0
(729 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
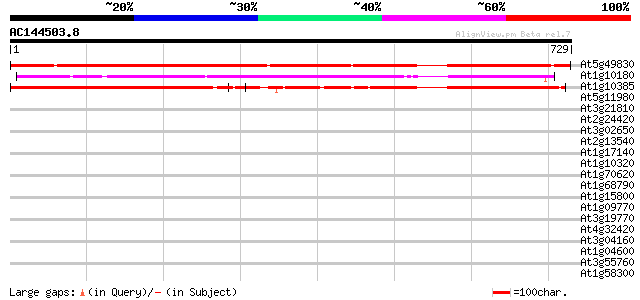
At5g49830 unknown protein 1008 0.0
At1g10180 unknown protein 363 e-100
At1g10385 hypothetical protein 362 e-100
At5g11980 unknown protein 36 0.067
At3g21810 unknown protein 35 0.15
At2g24420 unknown protein 33 0.43
At3g02650 hypothetical protein 33 0.74
At2g13540 nuclear cap-binding protein CBP80 33 0.74
At1g17140 unknown protein 32 1.3
At1g10320 unknown protein 32 1.3
At1g70620 unknown protein 32 1.6
At1g68790 putative nuclear matrix constituent protein 1 (NMCP1) 32 1.6
At1g15800 hypothetical protein 32 1.6
At1g09770 putative DNA-binding protein, Myb 32 1.6
At3g19770 unknown protein 31 2.2
At4g32420 cyclophilin (AtCYP95) 31 2.8
At3g04160 hypothetical protein 31 2.8
At1g04600 putative myosin heavy chain 31 2.8
At3g55760 unknown protein 30 3.7
At1g58300 hypothetical protein 30 3.7
>At5g49830 unknown protein
Length = 752
Score = 1008 bits (2605), Expect = 0.0
Identities = 528/729 (72%), Positives = 611/729 (83%), Gaps = 45/729 (6%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVHIDSLSISDSDGF 60
MRRSVYANY AFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGV+ID +SD
Sbjct: 67 MRRSVYANYPAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVNIDDDKVSDES-- 124
Query: 61 SVNGALDSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLN 120
NG L+ E +SDL+KW EFPD LD LLAERRV+EALAA DEGE +VS+A E +L+
Sbjct: 125 LANGLLNFEDNGLSDLEKWATEFPDHLDALLAERRVDEALAAFDEGEILVSQANEKHTLS 184
Query: 121 PSLLLSLQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLNAH 180
S+L SLQ +I ER+QKLADQLA+AACQPSTRG ELR++++ALK+LGDGP AH++LL+AH
Sbjct: 185 SSVLSSLQFAIAERKQKLADQLAKAACQPSTRGGELRSAIAALKRLGDGPRAHTVLLDAH 244
Query: 181 LQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMWAT 240
QRYQYNMQSLRPS+TSYGGAYTAAL+QLVFSA++QA+SDSL IFG+EPAYSSELV WAT
Sbjct: 245 FQRYQYNMQSLRPSSTSYGGAYTAALSQLVFSAISQASSDSLGIFGKEPAYSSELVTWAT 304
Query: 241 KQTEAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSV 300
KQTEAF+LLVKRHALASSAAAGGLRAAAEC QIALGHCSLLEARGL+LCPVLLK F+P V
Sbjct: 305 KQTEAFSLLVKRHALASSAAAGGLRAAAECAQIALGHCSLLEARGLSLCPVLLKHFKPIV 364
Query: 301 EQALDANLKRIQESTAAMAAADDWVLTYPPNVNRQTGSTTAFQLKLTSSAHRFNLMVQDF 360
EQAL+ANLKRI+E+TAAMAAADDWVLT PP +R ++TAFQ KLTSSAHRFNLMVQDF
Sbjct: 365 EQALEANLKRIEENTAAMAAADDWVLTSPPAGSRH--ASTAFQNKLTSSAHRFNLMVQDF 422
Query: 361 FEDVGPLLSMQLGGQALEGLFQVFNSYVNMLIKALPESMEEEE-SFEDSGNKIVRMAETE 419
FEDVGPLLSMQLG +ALEGLF+VFNSYV++L++ALP S+EEE+ +FE S NKIV+MAETE
Sbjct: 423 FEDVGPLLSMQLGSKALEGLFRVFNSYVDVLVRALPGSIEEEDPNFESSCNKIVQMAETE 482
Query: 420 AQQIALLANASLLADELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRHPEQREWRRRLVG 479
A Q+ALLANASLLADELLPRAAMKL SL+Q + D+ RR +RQNR+PEQREW+RRL+
Sbjct: 483 ANQLALLANASLLADELLPRAAMKL-SLDQTGQRTDDLRRPLDRQNRNPEQREWKRRLLS 541
Query: 480 SVDRLKDSFCRQHALSLIFTEDGDSHLTADMYISMERNADEVEWIPSLIFQSVVSWLLIF 539
+VD+LKD+FCRQHAL LIFTE+GDSHL+ADMY++++ N ++V++ PSLIFQ
Sbjct: 542 TVDKLKDAFCRQHALDLIFTEEGDSHLSADMYVNIDENGEDVDFFPSLIFQ--------- 592
Query: 540 KNYRIRQQIDDLSIHIFLIVECLVHSFEELFIKLNRMANIAADMFVGRERFATLLLMRLT 599
ELF KLNRMA++AADMFVGRERFA LLMRLT
Sbjct: 593 ----------------------------ELFAKLNRMASLAADMFVGRERFAISLLMRLT 624
Query: 600 ETVILWISEDQSFWDDIEEGPRPLGPLGLQQFYLDMKFVVCFASNGRYLSRNLQRIVNEI 659
ETVILW+S DQSFWDDIEEGPRPLGPLGL+Q YLDMKFV+CFAS GRYLSRNL R NEI
Sbjct: 625 ETVILWLSGDQSFWDDIEEGPRPLGPLGLRQLYLDMKFVICFASQGRYLSRNLHRGTNEI 684
Query: 660 IRKAMSAFSATGMDPYSDLPEDEWFNEICQDAMERLSGKPKEINGERELSSPTASVSAQS 719
I KA++AF+ATG+DPYS+LPED+WFN+IC DAMERLSGK K NG ++ SPTASVSAQS
Sbjct: 685 ISKALAAFTATGIDPYSELPEDDWFNDICVDAMERLSGKTKGNNG--DVHSPTASVSAQS 742
Query: 720 ISSVRSHNS 728
+SS RSH S
Sbjct: 743 VSSARSHGS 751
>At1g10180 unknown protein
Length = 769
Score = 363 bits (931), Expect = e-100
Identities = 236/709 (33%), Positives = 372/709 (52%), Gaps = 59/709 (8%)
Query: 9 YAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGV--HIDSLSISDSDGFSVNGAL 66
Y AF+R S+E ++E EL +R +S+Q L+ L GV +D + D
Sbjct: 59 YLAFLRISEEAVEMEHELVELRKHISSQGILVQDLMAGVCREMDDWNRLPGDVHDAEVEE 118
Query: 67 DSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPSLLLS 126
D E++D EF + +D+LLAE +V+EAL A+D ER + K ++ S
Sbjct: 119 DPLPNEVTDPKS---EFLEKIDLLLAEHKVDEALEAMDAEERSSPDLKGSVEMS-----S 170
Query: 127 LQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLNAHLQRYQY 186
+S+ ER+ L DQL A QPS AEL+ ++ L +LG GP AH LLL + +
Sbjct: 171 YKSAFMERKAVLEDQLLRIAKQPSICVAELKHALIGLIRLGKGPSAHQLLLKFYATSLRR 230
Query: 187 NMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEE--PAYSSELVMWATKQTE 244
+++ PS + + A L++LVFS ++ A +S A+FG++ PAYS+++V WA ++ E
Sbjct: 231 RIEAFLPSCLTCPNTFPATLSKLVFSNISVATKESAAMFGDDDNPAYSNKVVQWAEREVE 290
Query: 245 AFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQAL 304
LVK +A + S A LRAA+ C+Q L +C +LE +GL L + L LFRP VE+ L
Sbjct: 291 YLVRLVKENA-SPSETASALRAASICLQDCLNYCKVLEPQGLFLSKLFLVLFRPYVEEVL 349
Query: 305 DANLKRIQESTAAMAAADDWVLTYPPNVNRQTGSTTAFQLKLTSSAHRFNLMVQDFFEDV 364
+ N +R + + D+ + + V + A +T + RF +VQD E +
Sbjct: 350 ELNFRRARRVIFDLNETDEGLESPSDFVTILSEFAIASDTMMTDCSIRFMQIVQDILEQL 409
Query: 365 GPLLSMQLGGQALEGLFQVFNSYVNMLIKALPESMEEEESFEDSGNKIVRMAETEAQQIA 424
L+ + G L + Q+++ Y++ LIKALP +E+ E N ++ AET+++Q+A
Sbjct: 410 THLVVLHFGESVLTRILQLYDKYIDFLIKALPGHSDEDGLPELQDNTVLARAETDSEQLA 469
Query: 425 LLANASLLADELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRHPEQREWRRRLVGSVDRL 484
LL A + DELLPR+ +K+ L + + + PE +EW+R +V + D+L
Sbjct: 470 LLGAAFTILDELLPRSLVKVWKLQIENGGGEGENSAALNSSAAPELKEWKRHMVQAFDKL 529
Query: 485 KDSFCRQHALSLIFTEDGDSHLTADMYISMERNADEVEWIPSLIFQSVVSWLLIFKNYRI 544
++ FC Q LS I++ +G + L A +Y++ D++ +PSL FQ+
Sbjct: 530 RNYFCLQFVLSFIYSREGLTRLDALIYLT--ETPDDLH-LPSLPFQA------------- 573
Query: 545 RQQIDDLSIHIFLIVECLVHSFEELFIKLNRMANIAADMFVGRERFATLLLMRLTETVIL 604
LF KL ++A IA D+ +G+E+ +LL RLTETVI+
Sbjct: 574 ------------------------LFSKLQQLAIIAGDVLLGKEKLQKILLARLTETVII 609
Query: 605 WISEDQSFWDDIEEGPRPLGPLGLQQFYLDMKFVVCFASNGRYLSRNLQRIVNEIIRKAM 664
W+S +Q FW E+ PL P GLQQ LDM F V A Y + +Q + +I +A+
Sbjct: 610 WLSNEQEFWSAFEDESNPLQPSGLQQLILDMNFTVEIARFAGYPFKVVQNHASVVINRAI 669
Query: 665 SAFSATGMDPYSDLPEDEWFNEICQDAMERL------SGKPKEINGERE 707
+ FS G++P S LP+ EWF E + A+ RL + +P+E E E
Sbjct: 670 NIFSERGINPQSSLPKTEWFTEAAKSAINRLLMGSEDASEPEEYECEEE 718
>At1g10385 hypothetical protein
Length = 660
Score = 362 bits (928), Expect = e-100
Identities = 215/443 (48%), Positives = 287/443 (64%), Gaps = 59/443 (13%)
Query: 285 GLALCPVLLKLFRPSVEQALDANLKRIQESTAAMAAADDWVLTYPPNVNRQTGSTTAFQL 344
G+A L +L ++ QA + + E A + W + + + A L
Sbjct: 271 GVAFAAALSQLVFSTIAQAASDSQAVVGEDPAYTSELVTWAV--------KQAESFALLL 322
Query: 345 K---LTSSAHRFNLMVQDFFEDVGPL-LSMQLGGQALEGLFQVFNSYVNMLIKALPESME 400
K L SSA +L ++F ED GPL ++QL G AL+G+ QVFNSYV++LI ALP S E
Sbjct: 323 KRHTLASSAAAGSL--REFLEDAGPLDEALQLDGIALDGVLQVFNSYVDLLINALPGSAE 380
Query: 401 EEESFEDSGNKIVRMAETEAQQIALLANASLLADELLPRAAMKLSSLNQDPYKDDNRRRT 460
EE+ ++IV++AETE+QQ ALL NA LLADEL+PR+A ++ L Q + RR +
Sbjct: 381 NEEN---PVHRIVKVAETESQQTALLVNALLLADELIPRSASRI--LPQGTSQSTPRRGS 435
Query: 461 TERQNRHPEQREWRRRLVGSVDRLKDSFCRQHALSLIFTEDGDSHLTADMYISMERNADE 520
++RQNR PEQREW+++L SVDRL+DSFCRQHAL LIFTE+G+ L++++YI M+ +E
Sbjct: 436 SDRQNR-PEQREWKKKLQRSVDRLRDSFCRQHALELIFTEEGEVRLSSEIYILMDETTEE 494
Query: 521 VEWIPSLIFQSVVSWLLIFKNYRIRQQIDDLSIHIFLIVECLVHSFEELFIKLNRMANIA 580
EW PS IFQ ELF KL R+A I
Sbjct: 495 PEWFPSPIFQ-------------------------------------ELFAKLTRIAMIV 517
Query: 581 ADMFVGRERFATLLLMRLTETVILWISEDQSFWDDIEEGPRPLGPLGLQQFYLDMKFVVC 640
+DMFVGRERFAT+LLMRLTETVILWIS+DQSFW+++E G +PLGPLGLQQFYLDM+FV+
Sbjct: 518 SDMFVGRERFATILLMRLTETVILWISDDQSFWEEMETGDKPLGPLGLQQFYLDMEFVMI 577
Query: 641 FASNGRYLSRNLQRIVNEIIRKAMSAFSATGMDPYSDLPEDEWFNEICQDAMERLSGKPK 700
FAS GRYLSRNL +++ II +A+ A SATG+DPYS LPE+EWF E+ Q A++ L GK
Sbjct: 578 FASQGRYLSRNLHQVIKNIIARAVEAVSATGLDPYSTLPEEEWFAEVAQIAIKMLMGKGN 637
Query: 701 -EINGERELSSPTASVSAQSISS 722
+GER+++SP+ S SA+S +S
Sbjct: 638 FGGHGERDVTSPSVS-SAKSYTS 659
Score = 339 bits (869), Expect = 4e-93
Identities = 185/306 (60%), Positives = 236/306 (76%), Gaps = 4/306 (1%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVHIDSLSISDSDGF 60
MR+SVYANYAAFIRTSKEIS LEG+L S+RNLLS QA L+HGLADGVHI SL D+D
Sbjct: 73 MRKSVYANYAAFIRTSKEISALEGQLLSMRNLLSAQAALVHGLADGVHISSLCADDADDL 132
Query: 61 SVNGALDSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLN 120
D ++K++S+++ W+VEF D L+VLLAE+RVEE++AAL+EG RV EA E ++L+
Sbjct: 133 RDEDLYDMDNKQLSNIENWVVEFFDRLEVLLAEKRVEESMAALEEGRRVAVEAHEKRTLS 192
Query: 121 PSLLLSLQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLNAH 180
P+ LLSL ++I E+RQ+LADQLAEA QPSTRG ELR++V +LKKLGDG AH+LLL ++
Sbjct: 193 PTTLLSLNNAIKEKRQELADQLAEAISQPSTRGGELRSAVLSLKKLGDGSRAHTLLLRSY 252
Query: 181 LQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMWAT 240
+R Q N+QSLR SNTSYG A+ AAL+QLVFS +AQAASDS A+ GE+PAY+SELV WA
Sbjct: 253 ERRLQANIQSLRASNTSYGVAFAAALSQLVFSTIAQAASDSQAVVGEDPAYTSELVTWAV 312
Query: 241 KQTEAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSV 300
KQ E+FALL+KRH LASSAAAG LR E ++ A L+ G+AL V L++F V
Sbjct: 313 KQAESFALLLKRHTLASSAAAGSLR---EFLEDAGPLDEALQLDGIALDGV-LQVFNSYV 368
Query: 301 EQALDA 306
+ ++A
Sbjct: 369 DLLINA 374
>At5g11980 unknown protein
Length = 569
Score = 36.2 bits (82), Expect = 0.067
Identities = 40/148 (27%), Positives = 67/148 (45%), Gaps = 16/148 (10%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGV--HIDSLSISDSD 58
M+ NY AFI + + + E+SSI L + + L G IDS + +
Sbjct: 58 MQEVAVGNYRAFITAADALLAIRQEVSSIDKHLESLIGEVPKLTSGCTEFIDSAE-NILE 116
Query: 59 GFSVNGALDSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKS 118
+N AL + H + D L+E P L+D + +EAL + E VS ++ +
Sbjct: 117 KRKMNQALLANHSTLLD----LLEIPQLMDTCVRNGNFDEAL----DLEAFVS---KLAT 165
Query: 119 LNPSL--LLSLQSSITERRQKLADQLAE 144
L+P L + +L + + + Q L QL +
Sbjct: 166 LHPKLPVIQALAAEVRQTTQSLLSQLLQ 193
>At3g21810 unknown protein
Length = 388
Score = 35.0 bits (79), Expect = 0.15
Identities = 28/106 (26%), Positives = 46/106 (42%), Gaps = 2/106 (1%)
Query: 9 YAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVHIDSLSISDSDGFSVNGALDS 68
Y F+R ++ E L + N LST G V +D +S +++G ++ A D
Sbjct: 166 YNRFLRAQDDLKRSEARLQKLGNQLSTYLAGSEGNNRDVGLDIVSDEETNGRNLRTACD- 224
Query: 69 EHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAK 114
H E+ + L +D + VE+ L E E+V +E K
Sbjct: 225 PHNELQNTSS-LSRKKHYVDQYTTKEPVEDGLIGRGEEEKVENEKK 269
>At2g24420 unknown protein
Length = 440
Score = 33.5 bits (75), Expect = 0.43
Identities = 20/99 (20%), Positives = 42/99 (42%)
Query: 93 ERRVEEALAALDEGERVVSEAKEMKSLNPSLLLSLQSSITERRQKLADQLAEAACQPSTR 152
E ++++ L E +V+E +++ + SL++ ++ R+K + E + R
Sbjct: 57 ESQIDDKTKELKGREELVTEKEKLLQERQDKVASLETEVSSLRKKGSSDSVELLSKAQAR 116
Query: 153 GAELRASVSALKKLGDGPHAHSLLLNAHLQRYQYNMQSL 191
EL V LKK + + L+ A + + L
Sbjct: 117 ATELEKQVEVLKKFLEQKNKEKELIEAQTSETEKKLNEL 155
>At3g02650 hypothetical protein
Length = 1077
Score = 32.7 bits (73), Expect = 0.74
Identities = 27/89 (30%), Positives = 40/89 (44%), Gaps = 10/89 (11%)
Query: 339 TTAFQLKLTSSAHRFNLMV-QDFFEDVGPLLSMQLGGQALEGLFQVFNSYVNMLIKALPE 397
T FQL+ T SA L V +D+ PL+ Q L+ FQ N ++ L + +
Sbjct: 183 TLEFQLESTESADDLLLSVLTGLCKDLSPLVIKFDDDQILKAQFQECNHILDFLYQMIVP 242
Query: 398 SMEEEESFED---------SGNKIVRMAE 417
+EEEE ED GN+ +R +
Sbjct: 243 RIEEEELEEDDEENRADENGGNRAIRFMD 271
>At2g13540 nuclear cap-binding protein CBP80
Length = 848
Score = 32.7 bits (73), Expect = 0.74
Identities = 27/102 (26%), Positives = 50/102 (48%), Gaps = 5/102 (4%)
Query: 32 LLSTQATLIHGLADGVHIDSLSISDSDGFSVNGALDSEHKEISDLDKWLVEFPDLLDVLL 91
L+S QA ++ + ++D +SD + AL+ + ISDL K + +VL+
Sbjct: 604 LVSNQA-IVRWVFSPENVDQFHVSDQPWEILGNALNKTYNRISDLRKDISNITK--NVLV 660
Query: 92 AERRVEEALAALDEGERVVS--EAKEMKSLNPSLLLSLQSSI 131
AE+ A L+ E +S E + + NP+ + L+S++
Sbjct: 661 AEKASANARVELEAAESKLSLVEGEPVLGENPAKMKRLKSTV 702
>At1g17140 unknown protein
Length = 344
Score = 32.0 bits (71), Expect = 1.3
Identities = 25/98 (25%), Positives = 48/98 (48%), Gaps = 3/98 (3%)
Query: 97 EEALAALDEGERVVSEAKEMKSLNPSL--LLSLQSSITERRQKLADQLAEAACQPSTRGA 154
EE + DE E++V++ E+K L L + S+ + + L +QL+++A + S A
Sbjct: 153 EELRSGNDEAEKLVAKEDEIKMLKARLYDMEKEHESLGKENESLKNQLSDSASEISNVKA 212
Query: 155 ELRASVSALKKLGDGPHAHSLLLNAHLQRYQYNMQSLR 192
VS + ++G+ S AHL+ +M+ +
Sbjct: 213 NEDEMVSKVSRIGE-ELEESRAKTAHLKEKLESMEEAK 249
>At1g10320 unknown protein
Length = 757
Score = 32.0 bits (71), Expect = 1.3
Identities = 21/90 (23%), Positives = 38/90 (41%), Gaps = 9/90 (10%)
Query: 397 ESMEEEESFEDSGNKIVRMAETEAQQIALLANASLLADELLPRAAMKLSSLNQDP----- 451
E+ E+ESFE+S K M+ E ++ + E+ + + + DP
Sbjct: 15 EAAGEKESFEESKEKAAEMSRKEKRKAMKKLKRKQVRKEIAAKEREEAKAKLNDPAEQER 74
Query: 452 ----YKDDNRRRTTERQNRHPEQREWRRRL 477
++D RRR E ++ +R WR +
Sbjct: 75 LKAIEEEDARRREKELKDFEESERAWREAM 104
>At1g70620 unknown protein
Length = 897
Score = 31.6 bits (70), Expect = 1.6
Identities = 23/64 (35%), Positives = 35/64 (53%), Gaps = 3/64 (4%)
Query: 439 RAAMKLSSLNQDPYKDDNRRRTT-ERQNRHPEQREWRRRLVGSVDRLKDSFCRQHALSLI 497
R+ K SS + D DD++R+++ +R+NR P + RRR V S R S QH +L
Sbjct: 816 RSRRKRSSPSSDESSDDSKRKSSSKRKNRSPSPGKSRRRHVSS--RSPHSKHSQHKNTLY 873
Query: 498 FTED 501
+ D
Sbjct: 874 SSHD 877
>At1g68790 putative nuclear matrix constituent protein 1 (NMCP1)
Length = 1085
Score = 31.6 bits (70), Expect = 1.6
Identities = 44/201 (21%), Positives = 88/201 (42%), Gaps = 24/201 (11%)
Query: 5 VYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVHIDSLSISDSDGFSVNG 64
V N + K + +L+ ++S ++ L+ + I + ++ +S+ + D ++
Sbjct: 271 VMENERTIEKKEKILENLQQKISVAKSELTEKEESIK-----IKLNDISLKEKDFEAMKA 325
Query: 65 ALDSEHKEISDLDKWLV-----EFPDLLD---VLLAERRVEEALAALDEGERVVSEAKEM 116
+D + KE+ + ++ L+ E LLD +L RR E + L++ R + E E
Sbjct: 326 KVDIKEKELHEFEENLIEREQMEIGKLLDDQKAVLDSRRREFEM-ELEQMRRSLDEELEG 384
Query: 117 KSLNPSLLLSLQSSITERRQKLADQLAEAACQPSTRGA-----ELRASVSALKKLGDGPH 171
K + LQ I+ + +KLA + EAA + G +L A + +K+
Sbjct: 385 KKAE---IEQLQVEISHKEEKLAKR--EAALEKKEEGVKKKEKDLDARLKTVKEKEKALK 439
Query: 172 AHSLLLNAHLQRYQYNMQSLR 192
A L+ +R + + LR
Sbjct: 440 AEEKKLHMENERLLEDKECLR 460
>At1g15800 hypothetical protein
Length = 251
Score = 31.6 bits (70), Expect = 1.6
Identities = 23/94 (24%), Positives = 40/94 (42%), Gaps = 9/94 (9%)
Query: 434 DELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRH-----PEQREWRRRLVGSVDRLKDSF 488
DE+ P A+ +S+ P+K +++T + E+ R L D LK
Sbjct: 149 DEITPVHAITQTSVTTSPFKRSRKKKTLAQLKEEESVLLKERNGLRNELATMQDLLKQQR 208
Query: 489 CRQHALSLIFTE----DGDSHLTADMYISMERNA 518
R +L + E D S L D+ I+++ N+
Sbjct: 209 ARNESLKKLQAESQKNDDSSFLLPDLNIALDNNS 242
>At1g09770 putative DNA-binding protein, Myb
Length = 844
Score = 31.6 bits (70), Expect = 1.6
Identities = 27/65 (41%), Positives = 32/65 (48%), Gaps = 2/65 (3%)
Query: 377 LEGLFQVFNSYVNMLIKALPESMEEEESFE-DSGNKIVR-MAETEAQQIALLANASLLAD 434
L GL Q N Y + ES E EE E D ++I R AE EA+Q ALL S +
Sbjct: 481 LTGLPQPKNEYQIVAQPPPEESEEPEEKIEEDMSDRIAREKAEEEARQQALLKKRSKVLQ 540
Query: 435 ELLPR 439
LPR
Sbjct: 541 RDLPR 545
>At3g19770 unknown protein
Length = 472
Score = 31.2 bits (69), Expect = 2.2
Identities = 30/94 (31%), Positives = 43/94 (44%), Gaps = 13/94 (13%)
Query: 19 ISDLEGELSSIRNLLSTQATLIHGLADGVHI--DSLSISDSDGFSVNGALDSEHKEISDL 76
I D+EG L+S + L+ L GL DG + S + S GF N + +SE S
Sbjct: 318 IGDVEGLLNSYKQLVFKYVCLTKGLGDGTSLAPSSSPLQASSGF--NTSKESEDHRRSSS 375
Query: 77 DKWLVEFPDLLDVLLAERRVEEALAAL-DEGERV 109
D + + D R V++ + AL EGE V
Sbjct: 376 DVQMTKETD--------RSVDDLIRALHGEGEDV 401
>At4g32420 cyclophilin (AtCYP95)
Length = 837
Score = 30.8 bits (68), Expect = 2.8
Identities = 25/95 (26%), Positives = 42/95 (43%), Gaps = 13/95 (13%)
Query: 398 SMEEEESFEDSGNKIVRMAETEAQQIALLANASLLADELLPRAAMKLSSLNQDPYKDDNR 457
S E ES DS ET++ + +++ L + L ++ + + K D
Sbjct: 205 SSSESESSSDS--------ETDSSESDSESDSDLSSPSFLSSSSHERQKKRKRSSKKDKH 256
Query: 458 RRTTERQNRHPEQREWR-----RRLVGSVDRLKDS 487
RR+ +R RH ++R R R+ S D L+DS
Sbjct: 257 RRSKQRDKRHEKKRSMRDKRPKRKSRRSPDSLEDS 291
>At3g04160 hypothetical protein
Length = 712
Score = 30.8 bits (68), Expect = 2.8
Identities = 20/65 (30%), Positives = 32/65 (48%), Gaps = 2/65 (3%)
Query: 8 NYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVHID--SLSISDSDGFSVNGA 65
N + + + I +L G LSS+++LLS + L+ + +D SL D +G V
Sbjct: 43 NNYSIVPSPPPIRELSGTLSSLKSLLSECQRTLDSLSQNLALDHSSLLQKDENGCFVRCP 102
Query: 66 LDSEH 70
DS H
Sbjct: 103 FDSNH 107
>At1g04600 putative myosin heavy chain
Length = 1730
Score = 30.8 bits (68), Expect = 2.8
Identities = 32/133 (24%), Positives = 63/133 (47%), Gaps = 13/133 (9%)
Query: 16 SKEISDLEGELSSI----RNLLSTQATLIHGLADGVHIDSLSISD-SDGFSVNGALDSEH 70
SKEISDL+ L+ I R+ T++ I L + L I + S G + L +E+
Sbjct: 944 SKEISDLQSVLTDIKLQLRDTQETKSKEISDLQSALQDMQLEIEELSKGLEMTNDLAAEN 1003
Query: 71 KE----ISDLDKWLVEFPDLLDVL--LAERRVEEALAALDEGERVVSEAKEMKSLNPSLL 124
++ +S L + E + + ++E R+++ + +D+ + E + K +L+
Sbjct: 1004 EQLKESVSSLQNKIDESERKYEEISKISEERIKDEVPVIDQSAIIKLETENQKL--KALV 1061
Query: 125 LSLQSSITERRQK 137
S++ I E +K
Sbjct: 1062 SSMEEKIDELDRK 1074
>At3g55760 unknown protein
Length = 578
Score = 30.4 bits (67), Expect = 3.7
Identities = 17/58 (29%), Positives = 31/58 (53%)
Query: 65 ALDSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPS 122
A+D E ++ DL+K EF +L+V + ER + + +D +S A+ + + N S
Sbjct: 91 AVDGEKEKGGDLEKKSDEFQKILEVSVEERDRIQRMQVVDRAAAAISAARAILASNNS 148
>At1g58300 hypothetical protein
Length = 283
Score = 30.4 bits (67), Expect = 3.7
Identities = 19/74 (25%), Positives = 36/74 (47%), Gaps = 2/74 (2%)
Query: 614 DDIEEGPRPLGPLGLQQFYLDMKFVVCFASNGRYLSRNLQRIVNEIIRKAMSAFSATGMD 673
D ++EG L + + ++ + F + + + L+RI+NE +A + TG++
Sbjct: 94 DQVKEGKSDSNDL-VSTWNFTIEGYLKFLVDSKLVFETLERIINESAIQAYAGLKNTGLE 152
Query: 674 PYSDLPED-EWFNE 686
+L D EWF E
Sbjct: 153 RAENLSRDLEWFKE 166
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.132 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,704,382
Number of Sequences: 26719
Number of extensions: 591212
Number of successful extensions: 1948
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 1920
Number of HSP's gapped (non-prelim): 40
length of query: 729
length of database: 11,318,596
effective HSP length: 107
effective length of query: 622
effective length of database: 8,459,663
effective search space: 5261910386
effective search space used: 5261910386
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144503.8


