

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.3 + phase: 0
(544 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
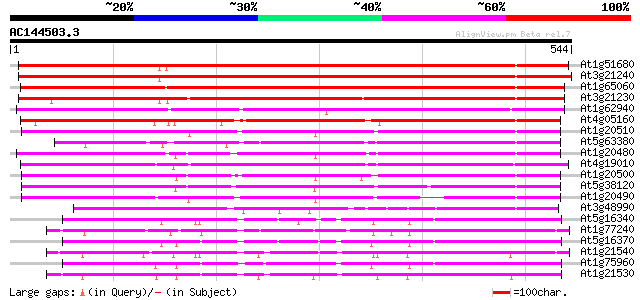
Score E
Sequences producing significant alignments: (bits) Value
At1g51680 4-coumarate--coenzyme A ligase (At4CL1) 785 0.0
At3g21240 putative 4-coumarate:CoA ligase 2 773 0.0
At1g65060 4-coumarate:CoA ligase 3 (4CL3) 683 0.0
At3g21230 putative 4-coumarate:CoA ligase 2 660 0.0
At1g62940 4-coumarate-CoA ligase-like protein 404 e-113
At4g05160 4-coumarate--CoA ligase - like protein 401 e-112
At1g20510 4-coumarate:CoA ligase like protein 356 2e-98
At5g63380 4-coumarate-CoA ligase-like protein 337 8e-93
At1g20480 unknown protein 337 1e-92
At4g19010 4-coumarate-CoA ligase - like 333 2e-91
At1g20500 hypothetical protein 332 2e-91
At5g38120 4-coumarate--CoA ligase -like protein 321 6e-88
At1g20490 unknown protein 290 2e-78
At3g48990 4-coumarate-CoA ligase -like protein 197 1e-50
At5g16340 AMP-binding protein 184 1e-46
At1g77240 unknown protein 177 1e-44
At5g16370 AMP-binding protein 176 3e-44
At1g21540 amp-binding protein, putative 176 3e-44
At1g75960 adenosine monophosphate binding protein 8 AMPBP8 171 1e-42
At1g21530 amp-binding protein, putative 166 4e-41
>At1g51680 4-coumarate--coenzyme A ligase (At4CL1)
Length = 561
Score = 785 bits (2028), Expect = 0.0
Identities = 391/541 (72%), Positives = 455/541 (83%), Gaps = 8/541 (1%)
Query: 9 ELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASG 68
++IF+SKLPDIYIP HL LH Y F+N+S++ ++PCLIN PT +YTY DV + ++++A+
Sbjct: 22 DVIFRSKLPDIYIPNHLSLHDYIFQNISEFATKPCLINGPTGHVYTYSDVHVISRQIAAN 81
Query: 69 LNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLL 128
+KLG+ Q DV+M+LLPNCPEFV +FL ASFRGA TAANPFFT AEIAKQAKASNTKL+
Sbjct: 82 FHKLGVNQNDVVMLLLPNCPEFVLSFLAASFRGATATAANPFFTPAEIAKQAKASNTKLI 141
Query: 129 VTQACYYDKVKDLEN---VKLVFVD---SSPEEDNHMHFSELIQADQNEMEEV-KVNIKP 181
+T+A Y DK+K L+N V +V +D S P + + F+EL Q+ E + V I P
Sbjct: 142 ITEARYVDKIKPLQNDDGVVIVCIDDNESVPIPEGCLRFTELTQSTTEASEVIDSVEISP 201
Query: 182 DDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYS 241
DDVVALPYSSGTTGLPKGVMLTHKGLVTS+AQQVDGENPNLY+HS+DVILCVLPMFHIY+
Sbjct: 202 DDVVALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDGENPNLYFHSDDVILCVLPMFHIYA 261
Query: 242 LNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLS 301
LNS++LCGLR A+IL+MPKF+IN L+ + KVT+AP+VPPIVLAIAKS E +KYDLS
Sbjct: 262 LNSIMLCGLRVGAAILIMPKFEINLLLELIQRCKVTVAPMVPPIVLAIAKSSETEKYDLS 321
Query: 302 SIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGAC 361
SIRV+KSG APLGKELED V AKFP AKLGQGYGMTEAGPVL M L FAKEP VKSGAC
Sbjct: 322 SIRVVKSGAAPLGKELEDAVNAKFPNAKLGQGYGMTEAGPVLAMSLGFAKEPFPVKSGAC 381
Query: 362 GTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGD 421
GTVVRNAEMKIVDP SL RNQPGEICIRG QIMKGYLNNP AT ETIDK+GWLHTGD
Sbjct: 382 GTVVRNAEMKIVDPDTGDSLSRNQPGEICIRGHQIMKGYLNNPAATAETIDKDGWLHTGD 441
Query: 422 IGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPV 481
IG IDDDDELFIVDRLKELIKYKGFQVAPAELEA+++ HP I+DVAVV M +EAAGEVPV
Sbjct: 442 IGLIDDDDELFIVDRLKELIKYKGFQVAPAELEALLIGHPDITDVAVVAMKEEAAGEVPV 501
Query: 482 AFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAG 541
AFVV+S S + +EDD+K+FVSKQVVFYKRIN+VFF ++IPK+PSGKILRKDLRAKLA G
Sbjct: 502 AFVVKSKDS-ELSEDDVKQFVSKQVVFYKRINKVFFTESIPKAPSGKILRKDLRAKLANG 560
Query: 542 V 542
+
Sbjct: 561 L 561
>At3g21240 putative 4-coumarate:CoA ligase 2
Length = 556
Score = 773 bits (1995), Expect = 0.0
Identities = 378/538 (70%), Positives = 456/538 (84%), Gaps = 3/538 (0%)
Query: 9 ELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASG 68
++IF+S+LPDIYIP HLPLH Y FEN+S++ ++PCLIN PT E+YTY DV +T++K+A+G
Sbjct: 20 DVIFRSRLPDIYIPNHLPLHDYIFENISEFAAKPCLINGPTGEVYTYADVHVTSRKLAAG 79
Query: 69 LNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLL 128
L+ LG++Q DV+M+LLPN PE V FL ASF GAI T+ANPFFT AEI+KQAKAS KL+
Sbjct: 80 LHNLGVKQHDVVMILLPNSPEVVLTFLAASFIGAITTSANPFFTPAEISKQAKASAAKLI 139
Query: 129 VTQACYYDKVKDLEN--VKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDDVVA 186
VTQ+ Y DK+K+L+N V +V DS +N + FSEL Q+++ ++ + I P+DVVA
Sbjct: 140 VTQSRYVDKIKNLQNDGVLIVTTDSDAIPENCLRFSELTQSEEPRVDSIPEKISPEDVVA 199
Query: 187 LPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVL 246
LP+SSGTTGLPKGVMLTHKGLVTS+AQQVDGENPNLY++ +DVILCVLPMFHIY+LNS++
Sbjct: 200 LPFSSGTTGLPKGVMLTHKGLVTSVAQQVDGENPNLYFNRDDVILCVLPMFHIYALNSIM 259
Query: 247 LCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVL 306
LC LR A+IL+MPKF+I + + KVT+A VVPPIVLAIAKSPE +KYDLSS+R++
Sbjct: 260 LCSLRVGATILIMPKFEITLLLEQIQRCKVTVAMVVPPIVLAIAKSPETEKYDLSSVRMV 319
Query: 307 KSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVR 366
KSG APLGKELED + AKFP AKLGQGYGMTEAGPVL M L FAKEP VKSGACGTVVR
Sbjct: 320 KSGAAPLGKELEDAISAKFPNAKLGQGYGMTEAGPVLAMSLGFAKEPFPVKSGACGTVVR 379
Query: 367 NAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFID 426
NAEMKI+DP SLPRN+PGEICIRG+QIMKGYLN+P AT TIDK+GWLHTGD+GFID
Sbjct: 380 NAEMKILDPDTGDSLPRNKPGEICIRGNQIMKGYLNDPLATASTIDKDGWLHTGDVGFID 439
Query: 427 DDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVR 486
DDDELFIVDRLKELIKYKGFQVAPAELE++++ HP+I+DVAVV M +E AGEVPVAFVVR
Sbjct: 440 DDDELFIVDRLKELIKYKGFQVAPAELESLLIGHPEINDVAVVAMKEEDAGEVPVAFVVR 499
Query: 487 SNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAGVPN 544
S S + +ED+IK+FVSKQVVFYKRIN+VFF D+IPK+PSGKILRKDLRA+LA G+ N
Sbjct: 500 SKDS-NISEDEIKQFVSKQVVFYKRINKVFFTDSIPKAPSGKILRKDLRARLANGLMN 556
>At1g65060 4-coumarate:CoA ligase 3 (4CL3)
Length = 561
Score = 683 bits (1762), Expect = 0.0
Identities = 339/529 (64%), Positives = 426/529 (80%), Gaps = 3/529 (0%)
Query: 11 IFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASGLN 70
IF+SKLPDI IP HLPLH+YCFE LS +PCLI T + YTY + L ++VASGL
Sbjct: 34 IFRSKLPDIDIPNHLPLHTYCFEKLSSVSDKPCLIVGSTGKSYTYGETHLICRRVASGLY 93
Query: 71 KLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVT 130
KLGI++GDVIM+LL N EFVF+F+GAS GA+ T ANPF+TS E+ KQ K+S KL++T
Sbjct: 94 KLGIRKGDVIMILLQNSAEFVFSFMGASMIGAVSTTANPFYTSQELYKQLKSSGAKLIIT 153
Query: 131 QACYYDKVKDL-ENVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDDVVALPY 189
+ Y DK+K+L EN+ L+ D P +N + FS LI D+ + V+I DD ALP+
Sbjct: 154 HSQYVDKLKNLGENLTLITTDE-PTPENCLPFSTLITDDETNPFQETVDIGGDDAAALPF 212
Query: 190 SSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCG 249
SSGTTGLPKGV+LTHK L+TS+AQQVDG+NPNLY S DVILCVLP+FHIYSLNSVLL
Sbjct: 213 SSGTTGLPKGVVLTHKSLITSVAQQVDGDNPNLYLKSNDVILCVLPLFHIYSLNSVLLNS 272
Query: 250 LRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSG 309
LR+ A++LLM KF+I A L+ +++VTIA +VPP+V+A+AK+P ++ YDLSS+R + SG
Sbjct: 273 LRSGATVLLMHKFEIGALLDLIQRHRVTIAALVPPLVIALAKNPTVNSYDLSSVRFVLSG 332
Query: 310 GAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAE 369
APLGKEL+D++R + P+A LGQGYGMTEAGPVL+M L FAKEPI KSG+CGTVVRNAE
Sbjct: 333 AAPLGKELQDSLRRRLPQAILGQGYGMTEAGPVLSMSLGFAKEPIPTKSGSCGTVVRNAE 392
Query: 370 MKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDD 429
+K+V + SL NQPGEICIRG QIMK YLN+PEAT TID+EGWLHTGDIG++D+DD
Sbjct: 393 LKVVHLETRLSLGYNQPGEICIRGQQIMKEYLNDPEATSATIDEEGWLHTGDIGYVDEDD 452
Query: 430 ELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNG 489
E+FIVDRLKE+IK+KGFQV PAELE+++++H I+D AVVP DE AGEVPVAFVVRSNG
Sbjct: 453 EIFIVDRLKEVIKFKGFQVPPAELESLLINHHSIADAAVVPQNDEVAGEVPVAFVVRSNG 512
Query: 490 SIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKL 538
+ D TE+D+K++V+KQVVFYKR+++VFF+ +IPKSPSGKILRKDL+AKL
Sbjct: 513 N-DITEEDVKEYVAKQVVFYKRLHKVFFVASIPKSPSGKILRKDLKAKL 560
>At3g21230 putative 4-coumarate:CoA ligase 2
Length = 570
Score = 660 bits (1702), Expect = 0.0
Identities = 329/543 (60%), Positives = 415/543 (75%), Gaps = 17/543 (3%)
Query: 9 ELIFKSKLPDIYIPKHLPLHSYCFENLSKYG----SRPCLINAPTAEIYTYYDVQLTAQK 64
+ IF+SKLPDI+IP HLPL Y F+ S G S C+I+ T I TY DVQ ++
Sbjct: 26 DFIFRSKLPDIFIPNHLPLTDYVFQRFSGDGDGDSSTTCIIDGATGRILTYADVQTNMRR 85
Query: 65 VASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASN 124
+A+G+++LGI+ GDV+M+LLPN PEF +FL ++ GA+ T ANPF+T EIAKQAKAS
Sbjct: 86 IAAGIHRLGIRHGDVVMLLLPNSPEFALSFLAVAYLGAVSTTANPFYTQPEIAKQAKASA 145
Query: 125 TKLLVTQACYYDKVKDLEN--VKLVFVDS-------SPEEDNHMHFSELIQADQNEMEEV 175
K+++T+ C DK+ +L+N V +V +D S +D + F+EL QAD+ E+ +
Sbjct: 146 AKMIITKKCLVDKLTNLKNDGVLIVCLDDDGDNGVVSSSDDGCVSFTELTQADETEL--L 203
Query: 176 KVNIKPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLP 235
K I P+D VA+PYSSGTTGLPKGVM+THKGLVTSIAQ+VDGENPNL + + DVILC LP
Sbjct: 204 KPKISPEDTVAMPYSSGTTGLPKGVMITHKGLVTSIAQKVDGENPNLNFTANDVILCFLP 263
Query: 236 MFHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPEL 295
MFHIY+L++++L +R A++L++P+F++N L+ +YKVT+ PV PP+VLA KSPE
Sbjct: 264 MFHIYALDALMLSAMRTGAALLIVPRFELNLVMELIQRYKVTVVPVAPPVVLAFIKSPET 323
Query: 296 DKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPID 355
++YDLSS+R++ SG A L KELED VR KFP A GQGYGMTE+G V L+FAK P
Sbjct: 324 ERYDLSSVRIMLSGAATLKKELEDAVRLKFPNAIFGQGYGMTESGTV-AKSLAFAKNPFK 382
Query: 356 VKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEG 415
KSGACGTV+RNAEMK+VD + SLPRN+ GEIC+RG Q+MKGYLN+PEAT TIDK+G
Sbjct: 383 TKSGACGTVIRNAEMKVVDTETGISLPRNKSGEICVRGHQLMKGYLNDPEATARTIDKDG 442
Query: 416 WLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEA 475
WLHTGDIGF+DDDDE+FIVDRLKELIK+KG+QVAPAELEA+++SHP I D AVV M DE
Sbjct: 443 WLHTGDIGFVDDDDEIFIVDRLKELIKFKGYQVAPAELEALLISHPSIDDAAVVAMKDEV 502
Query: 476 AGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLR 535
A EVPVAFV RS GS TEDD+K +V+KQVV YKRI VFFI+ IPK+ SGKILRKDLR
Sbjct: 503 ADEVPVAFVARSQGS-QLTEDDVKSYVNKQVVHYKRIKMVFFIEVIPKAVSGKILRKDLR 561
Query: 536 AKL 538
AKL
Sbjct: 562 AKL 564
>At1g62940 4-coumarate-CoA ligase-like protein
Length = 542
Score = 404 bits (1038), Expect = e-113
Identities = 213/534 (39%), Positives = 321/534 (59%), Gaps = 7/534 (1%)
Query: 7 EQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVA 66
+ E IF+S P + IP L L + + + +Y + A T + TY DV +++A
Sbjct: 8 DNEYIFRSLYPSVPIPDKLTLPEFVLQGVEEYTENVAFVEAVTGKAVTYGDVVRDTKRLA 67
Query: 67 SGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTK 126
L LG+++G V++V+LPN E+ LG G + + ANP +EI KQ +AS +
Sbjct: 68 KALTSLGLRKGQVMVVVLPNVAEYGIIALGIMSAGGVFSGANPTALVSEIKKQVEASGAR 127
Query: 127 LLVTQACYYDKVKDLENVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDDVVA 186
++T A Y+KVK L +V + E +++ +L++A + I D+ A
Sbjct: 128 GIITDATNYEKVKSLGLPVIVLGEEKIE--GAVNWKDLLEAGDKCGDTDNEEILQTDLCA 185
Query: 187 LPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVL 246
LP+SSGTTGL KGVMLTH+ L+ ++ + G + + V L ++P FHIY + +
Sbjct: 186 LPFSSGTTGLQKGVMLTHRNLIANLCSTLFGVRSEMI--GQIVTLGLIPFFHIYGIVGIC 243
Query: 247 LCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRV- 305
++ K ++ M ++D+ F + ++V+ AP+VPPI+L + K+P +D++DLS +++
Sbjct: 244 CATMKNKGKVVAMSRYDLRIFLNALIAHEVSFAPIVPPIILNLVKNPIVDEFDLSKLKLQ 303
Query: 306 -LKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTV 364
+ + APL EL AKFP ++ + YG+TE + K K + G +
Sbjct: 304 SVMTAAAPLAPELLTAFEAKFPNVQVQEAYGLTEHSCITLTHGDPEKGQGIAKRNSVGFI 363
Query: 365 VRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGF 424
+ N E+K +DP SLP+N GE+C+R +M+GY N E T +TID++GWLHTGDIG+
Sbjct: 364 LPNLEVKFIDPDTGRSLPKNTSGELCVRSQCVMQGYFMNKEETDKTIDEQGWLHTGDIGY 423
Query: 425 IDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFV 484
IDDD ++FIVDR+KELIKYKGFQVAPAELEAI+L+HP + DVAVVP+ DE AGE+P A V
Sbjct: 424 IDDDGDIFIVDRIKELIKYKGFQVAPAELEAILLTHPSVEDVAVVPLPDEEAGEIPAACV 483
Query: 485 VRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKL 538
V N E+DI FV+ V YK++ V F+D+IPKS SGKI+R+ LR K+
Sbjct: 484 V-INPKATEKEEDILNFVAANVAHYKKVRAVHFVDSIPKSLSGKIMRRLLRDKI 536
>At4g05160 4-coumarate--CoA ligase - like protein
Length = 544
Score = 401 bits (1030), Expect = e-112
Identities = 226/539 (41%), Positives = 330/539 (60%), Gaps = 28/539 (5%)
Query: 11 IFKSKLPDIYIPK--HLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASG 68
I++S P + +PK + L S+ F N S Y S+ + ++ T + T+ ++ ++A G
Sbjct: 11 IYRSLRPTLVLPKDPNTSLVSFLFRNSSSYPSKLAIADSDTGDSLTFSQLKSAVARLAHG 70
Query: 69 LNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLL 128
++LGI++ DV+++ PN +F FL + G + T ANP +T E++KQ K SN K++
Sbjct: 71 FHRLGIRKNDVVLIFAPNSYQFPLCFLAVTAIGGVFTTANPLYTVNEVSKQIKDSNPKII 130
Query: 129 VTQACYYDKVK--DLENVKLVFVDSS--PEEDNH--MHFSELIQADQNEMEEVKVNIKPD 182
++ +DK+K DL V L D+ P N + F +++ + E V IK
Sbjct: 131 ISVNQLFDKIKGFDLPVVLLGSKDTVEIPPGSNSKILSFDNVMELSEPVSEYPFVEIKQS 190
Query: 183 DVVALPYSSGTTGLPKGVMLTH-----KGLVTSIAQQVDGENPNLYYHSEDVILCVLPMF 237
D AL YSSGTTG KGV LTH L+ ++ Q + GE YH V LC LPMF
Sbjct: 191 DTAALLYSSGTTGTSKGVELTHGNFIAASLMVTMDQDLMGE-----YHG--VFLCFLPMF 243
Query: 238 HIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDK 297
H++ L + L+ +++ M +F++ + K++VT VVPP+ LA++K + K
Sbjct: 244 HVFGLAVITYSQLQRGNALVSMARFELELVLKNIEKFRVTHLWVVPPVFLALSKQSIVKK 303
Query: 298 YDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVK 357
+DLSS++ + SG APLGK+L + P L QGYGMTE ++++ ++P K
Sbjct: 304 FDLSSLKYIGSGAAPLGKDLMEECGRNIPNVLLMQGYGMTETCGIVSV-----EDPRLGK 358
Query: 358 --SGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEG 415
SG+ G + E +IV + S P NQ GEI +RG +MKGYLNNP+AT+ETIDK+
Sbjct: 359 RNSGSAGMLAPGVEAQIVSVETGKSQPPNQQGEIWVRGPNMMKGYLNNPQATKETIDKKS 418
Query: 416 WLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEA 475
W+HTGD+G+ ++D L++VDR+KELIKYKGFQVAPAELE +++SHP I D V+P DE
Sbjct: 419 WVHTGDLGYFNEDGNLYVVDRIKELIKYKGFQVAPAELEGLLVSHPDILDAVVIPFPDEE 478
Query: 476 AGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDL 534
AGEVP+AFVVRS S TE DI+KF++KQV YKR+ RV FI +PKS +GKILR++L
Sbjct: 479 AGEVPIAFVVRSPNS-SITEQDIQKFIAKQVAPYKRLRRVSFISLVPKSAAGKILRREL 536
>At1g20510 4-coumarate:CoA ligase like protein
Length = 546
Score = 356 bits (914), Expect = 2e-98
Identities = 206/527 (39%), Positives = 303/527 (57%), Gaps = 11/527 (2%)
Query: 12 FKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASGLNK 71
F SK I +P + L F + + R I+A T + T+ ++ + VA L++
Sbjct: 17 FYSKRTPIPLPPNPSLDVTTFISSQAHRGRIAFIDASTGQNLTFTELWRAVESVADCLSE 76
Query: 72 LGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVTQ 131
+GI++G V+++L PN F L GAI+T NP TS EIAKQ K SN L T
Sbjct: 77 IGIRKGHVVLLLSPNSILFPVVCLSVMSLGAIITTTNPLNTSNEIAKQIKDSNPVLAFTT 136
Query: 132 ACYYDKVKDLENVKLVFVDSSPEEDNHMHFSELIQADQNEME--EVKVNIKPDDVVALPY 189
+ K+ + + D+ L++ + E VK + DD L Y
Sbjct: 137 SQLLPKISAAAKKLPIVLMDEERVDSVGDVRRLVEMMKKEPSGNRVKERVDQDDTATLLY 196
Query: 190 SSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCG 249
SSGTTG+ KGV+ +H+ L+ + V+ + E +C +PMFHIY L +
Sbjct: 197 SSGTTGMSKGVISSHRNLIAMVQTIVNRFGSD---DGEQRFICTVPMFHIYGLAAFATGL 253
Query: 250 LRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPEL--DKYDLSSIRVLK 307
L ++I+++ KF+++ + KY+ T P+VPPI++A+ + KYDLSS+ +
Sbjct: 254 LAYGSTIIVLSKFEMHEMMSAIGKYQATSLPLVPPILVAMVNGADQIKAKYDLSSMHTVL 313
Query: 308 SGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRN 367
GGAPL KE+ + K+P K+ QGYG+TE+ + + + + G G + +
Sbjct: 314 CGGAPLSKEVTEGFAEKYPTVKILQGYGLTESTGIGASTDTVEESR---RYGTAGKLSAS 370
Query: 368 AEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDD 427
E +IVDP L Q GE+ ++G IMKGY +N EAT T+D EGWL TGD+ +ID+
Sbjct: 371 MEGRIVDPVTGQILGPKQTGELWLKGPSIMKGYFSNEEATSSTLDSEGWLRTGDLCYIDE 430
Query: 428 DDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRS 487
D +F+VDRLKELIKYKG+QVAPAELEA++L+HP+I+D AV+P D+ G+ P+A+VVR
Sbjct: 431 DGFIFVVDRLKELIKYKGYQVAPAELEALLLTHPEITDAAVIPFPDKEVGQFPMAYVVRK 490
Query: 488 NGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDL 534
GS +E I +FV+KQV YKRI +V F+ +IPK+PSGKILRKDL
Sbjct: 491 TGS-SLSEKTIMEFVAKQVAPYKRIRKVAFVSSIPKNPSGKILRKDL 536
>At5g63380 4-coumarate-CoA ligase-like protein
Length = 562
Score = 337 bits (865), Expect = 8e-93
Identities = 198/500 (39%), Positives = 302/500 (59%), Gaps = 27/500 (5%)
Query: 44 LINAPTAEIYTYYDVQLTAQKVASGLNKL--GIQQGDVIMVLLPNCPEFVFAFLGASFRG 101
L+N+ + + TY ++ + +A L + + +V +L P+ + +L G
Sbjct: 69 LVNSSSGDNLTYGELLRRVRSLAVSLRERFPSLASRNVAFILSPSSLDIPVLYLALMSIG 128
Query: 102 AIMTAANPFFTSAEIAKQAKASNTKLLVTQACYYDKVKDLENVKL----VFVDSSPEEDN 157
+++ ANP + +E++ Q + S + + VK L++ L V +DS+
Sbjct: 129 VVVSPANPIGSESEVSHQVEVSEPVIAFATS---QTVKKLQSSSLPLGTVLMDSTE---- 181
Query: 158 HMHFSELIQADQNEMEEVKVNIKPDDVVALPYSSGTTGLPKGVMLTHKGLV--TSIAQQV 215
S L ++D + + +V + D A+ +SSGTTG KGV+LTH+ L+ T+++ Q
Sbjct: 182 --FLSWLNRSDSSSVNPFQVQVNQSDPAAILFSSGTTGRVKGVLLTHRNLIASTAVSHQR 239
Query: 216 DGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYK 275
++P Y + V L LP+FH++ +++ + +++L+ +F++ A F V KYK
Sbjct: 240 TLQDPVNY---DRVGLFSLPLFHVFGF-MMMIRAISLGETLVLLGRFELEAMFKAVEKYK 295
Query: 276 VTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYG 335
VT PV PP+++A+ KS KYDL S+R L GGAPLGK++ + + KFP + QGYG
Sbjct: 296 VTGMPVSPPLIVALVKSELTKKYDLRSLRSLGCGGAPLGKDIAERFKQKFPDVDIVQGYG 355
Query: 336 MTEA-GPVLTMCLSFAKEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGD 394
+TE+ GP + +F E + VK G+ G + N E KIVDP SLP + GE+ +RG
Sbjct: 356 LTESSGPAAS---TFGPEEM-VKYGSVGRISENMEAKIVDPSTGESLPPGKTGELWLRGP 411
Query: 395 QIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELE 454
IMKGY+ N +A+ ET+DKEGWL TGD+ + D +D L+IVDRLKELIKYK +QV P ELE
Sbjct: 412 VIMKGYVGNEKASAETVDKEGWLKTGDLCYFDSEDFLYIVDRLKELIKYKAYQVPPVELE 471
Query: 455 AIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINR 514
I+ S+P + D AVVP DE AGE+P+AF+VR GS + E I FV+KQV YK++ R
Sbjct: 472 QILHSNPDVIDAAVVPFPDEDAGEIPMAFIVRKPGS-NLNEAQIIDFVAKQVTPYKKVRR 530
Query: 515 VFFIDAIPKSPSGKILRKDL 534
V FI+AIPK+P+GKILR++L
Sbjct: 531 VAFINAIPKNPAGKILRREL 550
>At1g20480 unknown protein
Length = 565
Score = 337 bits (863), Expect = 1e-92
Identities = 207/543 (38%), Positives = 299/543 (54%), Gaps = 29/543 (5%)
Query: 7 EQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVA 66
E IF SK + +P + L F + + ++A T ++ ++ L ++VA
Sbjct: 29 ESTSIFYSKREPMALPPNQFLDVTSFIASQPHRGKTVFVDAVTGRRLSFPELWLGVERVA 88
Query: 67 SGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTK 126
L LG+++G+V+++L PN F L GAI+T ANP TS EI+KQ S
Sbjct: 89 GCLYALGVRKGNVVIILSPNSILFPIVSLSVMSLGAIITTANPINTSDEISKQIGDSRPV 148
Query: 127 LLVTQACYYDKVKDLENVKLVFVDSSPEEDNHM-------------HFSELIQADQNEME 173
L T K+ N L V +D H+ +I+ + +E
Sbjct: 149 LAFTTCKLVSKLAAASNFNLPVVLM---DDYHVPSQSYGDRVKLVGRLETMIETEPSE-S 204
Query: 174 EVKVNIKPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCV 233
VK + DD AL YSSGTTG KGVML+H+ L+ + + E +C
Sbjct: 205 RVKQRVNQDDTAALLYSSGTTGTSKGVMLSHRNLIALVQAY------RARFGLEQRTICT 258
Query: 234 LPMFHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSP 293
+PM HI+ + +I+++PKFD+ V ++ + +VPPIV+A+
Sbjct: 259 IPMCHIFGFGGFATGLIALGWTIVVLPKFDMAKLLSAVETHRSSYLSLVPPIVVAMVNGA 318
Query: 294 EL--DKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAK 351
KYDLSS+ + +GGAPL +E+ + +PK K+ QGYG+TE+ + F K
Sbjct: 319 NEINSKYDLSSLHTVVAGGAPLSREVTEKFVENYPKVKILQGYGLTESTAIAASM--FNK 376
Query: 352 EPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETI 411
E + GA G + N E KIVDP L NQ GE+ IR +MKGY N EAT TI
Sbjct: 377 EETK-RYGASGLLAPNVEGKIVDPDTGRVLGVNQTGELWIRSPTVMKGYFKNKEATASTI 435
Query: 412 DKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPM 471
D EGWL TGD+ +ID D +F+VDRLKELIK G+QVAPAELEA++L+HP+I+D AV+P+
Sbjct: 436 DSEGWLKTGDLCYIDGDGFVFVVDRLKELIKCNGYQVAPAELEALLLAHPEIADAAVIPI 495
Query: 472 LDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILR 531
D AG+ P+A++VR GS + +E +I FV+KQV YK+I +V F+ +IPK+PSGKILR
Sbjct: 496 PDMKAGQYPMAYIVRKVGS-NLSESEIMGFVAKQVSPYKKIRKVTFLASIPKNPSGKILR 554
Query: 532 KDL 534
++L
Sbjct: 555 REL 557
>At4g19010 4-coumarate-CoA ligase - like
Length = 566
Score = 333 bits (853), Expect = 2e-91
Identities = 202/539 (37%), Positives = 313/539 (57%), Gaps = 14/539 (2%)
Query: 11 IFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASGL- 69
I+ SK P +++P L + K+ LI++ T ++ ++Q+ Q +A+G+
Sbjct: 31 IYTSKFPSLHLPVDPNLDAVSALFSHKHHGDTALIDSLTGFSISHTELQIMVQSMAAGIY 90
Query: 70 NKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLV 129
+ LG++QGDV+ ++LPN F FL GAI+T NP + EI KQ + L
Sbjct: 91 HVLGVRQGDVVSLVLPNSVYFPMIFLSLISLGAIVTTMNPSSSLGEIKKQVSECSVGLAF 150
Query: 130 TQACYYDKVKDLENVKLVFVDSSPEEDN----HMHFSELIQADQNEMEEVKVNIKPDDVV 185
T +K+ L V ++ V S + D+ + F +++ + K IK DDV
Sbjct: 151 TSTENVEKLSSL-GVSVISVSESYDFDSIRIENPKFYSIMKESFGFVP--KPLIKQDDVA 207
Query: 186 ALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYH-SEDVILCVLPMFHIYSLNS 244
A+ YSSGTTG KGV+LTH+ L+ S+ V E Y S +V L LP+ HIY L+
Sbjct: 208 AIMYSSGTTGASKGVLLTHRNLIASMELFVRFEASQYEYPGSSNVYLAALPLCHIYGLSL 267
Query: 245 VLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPE-LDKYDLSSI 303
++ L ++I++M +FD + ++ ++K+T PVVPP+++A+ K + + S+
Sbjct: 268 FVMGLLSLGSTIVVMKRFDASDVVNVIERFKITHFPVVPPMLMALTKKAKGVCGEVFKSL 327
Query: 304 RVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGT 363
+ + SG APL ++ + P L QGYGMTE+ V T F E + + + G
Sbjct: 328 KQVSSGAAPLSRKFIEDFLQTLPHVDLIQGYGMTESTAVGTR--GFNSEKLS-RYSSVGL 384
Query: 364 VVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIG 423
+ N + K+VD + S LP GE+ I+G +MKGYLNNP+AT+ +I ++ WL TGDI
Sbjct: 385 LAPNMQAKVVDWSSGSFLPPGNRGELWIQGPGVMKGYLNNPKATQMSIVEDSWLRTGDIA 444
Query: 424 FIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAF 483
+ D+D LFIVDR+KE+IKYKGFQ+APA+LEA+++SHP I D AV +E GE+PVAF
Sbjct: 445 YFDEDGYLFIVDRIKEIIKYKGFQIAPADLEAVLVSHPLIIDAAVTAAPNEECGEIPVAF 504
Query: 484 VVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAGV 542
VVR +E+D+ +V+ QV Y+++ +V +++IPKSP+GKILRK+L+ L V
Sbjct: 505 VVRRQ-ETTLSEEDVISYVASQVAPYRKVRKVVMVNSIPKSPTGKILRKELKRILTNSV 562
>At1g20500 hypothetical protein
Length = 550
Score = 332 bits (852), Expect = 2e-91
Identities = 195/532 (36%), Positives = 307/532 (57%), Gaps = 21/532 (3%)
Query: 12 FKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASGL-N 70
F SK + +P +L F + + + I+A T + T+ D+ +VA L +
Sbjct: 23 FYSKRQPLSLPPNLSRDVTTFISSQPHRGKTAFIDAATGQCLTFSDLWRAVDRVADCLYH 82
Query: 71 KLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVT 130
++GI++GDV+++L PN L GA+ T AN TS EI+KQ SN L+ T
Sbjct: 83 EVGIRRGDVVLILSPNSIFIPVVCLSVMSLGAVFTTANTLNTSGEISKQIADSNPTLVFT 142
Query: 131 QACYYDKVKDLENVKLVFVDSSPEEDNHMH----FSELIQADQNEMEEVKVNIKPDDVVA 186
K+ +V L + E + + SE+++ + + + V+ + DD
Sbjct: 143 TRQLAPKLPVAISVVLTDDEVYQELTSAIRVVGILSEMVKKEPSG-QRVRDRVNQDDTAM 201
Query: 187 LPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVL 246
+ YSSGTTG KGV+ +H+ L +A+ + + NL +D+ +C +PMFH Y L +
Sbjct: 202 MLYSSGTTGPSKGVISSHRNLTAHVARFI---SDNL--KRDDIFICTVPMFHTYGLLTFA 256
Query: 247 LCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPEL--DKYDLSSIR 304
+ + ++++++ +F ++ V K++ T + PP+++A+ +L KYDLSS++
Sbjct: 257 MGTVALGSTVVILRRFQLHDMMDAVEKHRATALALAPPVLVAMINDADLIKAKYDLSSLK 316
Query: 305 VLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEA--GPVLTMCLSFAKEPIDVKSGACG 362
++ GGAPL KE+ + K+P + QGY +TE+ G T ++ + G G
Sbjct: 317 TVRCGGAPLSKEVTEGFLEKYPTVDILQGYALTESNGGGAFTNSAEESR-----RYGTAG 371
Query: 363 TVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDI 422
T+ + E +IVDP + NQ GE+ ++G I KGY N EAT ETI+ EGWL TGD+
Sbjct: 372 TLTSDVEARIVDPNTGRFMGINQTGELWLKGPSISKGYFKNQEATNETINLEGWLKTGDL 431
Query: 423 GFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVA 482
+ID+D LF+VDRLKELIKYKG+QV PAELEA++++HP I D AV+P D+ AG+ P+A
Sbjct: 432 CYIDEDGFLFVVDRLKELIKYKGYQVPPAELEALLITHPDILDAAVIPFPDKEAGQYPMA 491
Query: 483 FVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDL 534
+VVR + S + +E + F+SKQV YK+I +V FI++IPK+ SGK LRKDL
Sbjct: 492 YVVRKHES-NLSEKQVIDFISKQVAPYKKIRKVSFINSIPKTASGKTLRKDL 542
>At5g38120 4-coumarate--CoA ligase -like protein
Length = 550
Score = 321 bits (823), Expect = 6e-88
Identities = 192/531 (36%), Positives = 293/531 (55%), Gaps = 20/531 (3%)
Query: 12 FKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASGL-N 70
F SK + +P L F + Y + I+A T ++ D+ + +VA L +
Sbjct: 24 FYSKRKPLALPSKESLDITTFISSQTYRGKTAFIDAATDHRISFSDLWMAVDRVADCLLH 83
Query: 71 KLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVT 130
+GI++GDV++VL PN L GA++T ANP T++EI +Q SN KL T
Sbjct: 84 DVGIRRGDVVLVLSPNTISIPIVCLSVMSLGAVLTTANPLNTASEILRQIADSNPKLAFT 143
Query: 131 QACYYDKVKDLE-NVKLVFVDSSPEEDNHM----HFSELIQADQNEMEEVKVNIKPDDVV 185
K+ ++ L V+ + + + +E+++ + + + V+ + DD
Sbjct: 144 TPELAPKIASSGISIVLERVEDTLRVPRGLKVVGNLTEMMKKEPSG-QAVRNQVHKDDTA 202
Query: 186 ALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSV 245
L YSSGTTG KGV +H L+ +A+ + + + +C +P+FH + L +
Sbjct: 203 MLLYSSGTTGRSKGVNSSHGNLIAHVARYI----AEPFEQPQQTFICTVPLFHTFGLLNF 258
Query: 246 LLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPE--LDKYDLSSI 303
+L L +++++P+FD+ V KY+ T +VPP+++ + + + KYD+S +
Sbjct: 259 VLATLALGTTVVILPRFDLGEMMAAVEKYRATTLILVPPVLVTMINKADQIMKKYDVSFL 318
Query: 304 RVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGT 363
R ++ GGAPL KE+ K+P + QGY +TE+ S + + GA G
Sbjct: 319 RTVRCGGAPLSKEVTQGFMKKYPTVDVYQGYALTESNGAGASIESVEESR---RYGAVGL 375
Query: 364 VVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIG 423
+ E +IVDP + NQ GE+ ++G I KGY N E E I EGWL TGD+
Sbjct: 376 LSCGVEARIVDPNTGQVMGLNQTGELWLKGPSIAKGYFRNEE---EIITSEGWLKTGDLC 432
Query: 424 FIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAF 483
+ID+D LFIVDRLKELIKYKG+QV PAELEA++L+HP I D AV+P D+ AG+ P+A+
Sbjct: 433 YIDNDGFLFIVDRLKELIKYKGYQVPPAELEALLLNHPDILDAAVIPFPDKEAGQFPMAY 492
Query: 484 VVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDL 534
V R S + E + F+SKQV YK+I +V FID+IPK+PSGK LRKDL
Sbjct: 493 VARKPES-NLCEKKVIDFISKQVAPYKKIRKVAFIDSIPKTPSGKTLRKDL 542
>At1g20490 unknown protein
Length = 530
Score = 290 bits (741), Expect = 2e-78
Identities = 179/533 (33%), Positives = 284/533 (52%), Gaps = 43/533 (8%)
Query: 12 FKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASGL-N 70
F SK + +P + L F + I+A T T+ D+ +VA L +
Sbjct: 23 FYSKRNPLCLPPNPSLDVTTFISSQPQRGTTAFIDASTGHRLTFSDLWRVVDRVADCLYH 82
Query: 71 KLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVT 130
++GI++GDV+++L PN L GA++T AN TS EI+KQ SN L+ T
Sbjct: 83 EVGIRRGDVVLILSPNSIYIPVVCLSVMSLGAVVTTANTLNTSGEISKQIAQSNPTLVFT 142
Query: 131 QACYYDKVKDLENVKLVFVDSSPEEDNHMHFSELIQADQNEM-------EEVKVNIKPDD 183
+ K+ + +V D E+ + + +EM + V+ + DD
Sbjct: 143 TSQLAPKLAAA--ISVVLTDEEDEKRVELTSGVRVVGILSEMMKKETSGQRVRDRVNQDD 200
Query: 184 VVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLN 243
+ YSSGTTG KGV+ +H+ L +A+ +D + + +++ +C +PMFH + L
Sbjct: 201 TAMMLYSSGTTGTSKGVISSHRNLTAYVAKYIDDK-----WKRDEIFVCTVPMFHSFGLL 255
Query: 244 SVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPEL--DKYDLS 301
+ + + + ++++++ +F ++ V KYK TI + PP+++A+ + KYDL+
Sbjct: 256 AFAMGSVASGSTVVILRRFGLDDMMQAVEKYKATILSLAPPVLVAMINGADQLKAKYDLT 315
Query: 302 SIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGAC 361
S+R ++ GGAPL KE+ D+ K+P + QGY +TE+ S + +K GA
Sbjct: 316 SLRKVRCGGAPLSKEVMDSFLEKYPTVNIFQGYALTESHGSGASTESVEES---LKYGAV 372
Query: 362 GTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGD 421
G + E +IVDP + NQPGE+ ++G I K D
Sbjct: 373 GLLSSGIEARIVDPDTGRVMGVNQPGELWLKGPSISK----------------------D 410
Query: 422 IGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPV 481
+ +ID+D LF+VDRLKELIKYKG+QV PAELEA++++HP I D AV+P D AG+ P+
Sbjct: 411 LCYIDEDGFLFVVDRLKELIKYKGYQVPPAELEALLIAHPHILDAAVIPFPDREAGQYPM 470
Query: 482 AFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDL 534
A+V R S + +E ++ F+S QV YK+I +V FI +IPK+ SGK LRKDL
Sbjct: 471 AYVARKPES-NLSEKEVIDFISNQVAPYKKIRKVAFISSIPKTASGKTLRKDL 522
>At3g48990 4-coumarate-CoA ligase -like protein
Length = 514
Score = 197 bits (501), Expect = 1e-50
Identities = 157/481 (32%), Positives = 235/481 (48%), Gaps = 28/481 (5%)
Query: 63 QKVASGL-NKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAK 121
++ AS L + GI+ GDV+ + PN EFV FL A N +T+ E
Sbjct: 41 ERAASRLVSDAGIKPGDVVALTFPNTVEFVIMFLAVIRARATAAPLNAAYTAEEFEFYLS 100
Query: 122 ASNTKLLVTQACYYDKVKDLEN-VKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIK 180
S++KLL+T ++ + +K+ V ++ + + +D ++
Sbjct: 101 DSDSKLLLTSKEGNAPAQEAASKLKISHVTATLLDAGSDLVLSVADSDSVVDSATELVNH 160
Query: 181 PDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYH--SEDVILCVLPMFH 238
PDD ++SGTT PKGV LT L +S+ +N Y D + VLP+FH
Sbjct: 161 PDDGALFLHTSGTTSRPKGVPLTQLNLASSV------KNIKAVYKLTESDSTVIVLPLFH 214
Query: 239 IYSLNSVLLCGLRAKASILLMP--KFDINAFFGLVTKYKVTIAPVVPPIVLAI----AKS 292
++ L + LL L A A++ L +F F+ + KY T VP I I A
Sbjct: 215 VHGLLAGLLSSLGAGAAVTLPAAGRFSATTFWPDMKKYNATWYTAVPTIHQIILDRHASH 274
Query: 293 PELDKYDLSSIRVLKSGGAP-LGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAK 351
PE + L IR + AP + LE+ A +A Y MTEA +++ +
Sbjct: 275 PETEYPKLRFIRSCSASLAPVILSRLEEAFGAPVLEA-----YAMTEATHLMSS--NPLP 327
Query: 352 EPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETI 411
E K G+ G V EM I++ + + P N+ GE+CIRG + KGY NNPEA +
Sbjct: 328 EEGPHKPGSVGKPV-GQEMAILNEKGEIQEPNNK-GEVCIRGPNVTKGYKNNPEANKAGF 385
Query: 412 DKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPM 471
+ GW HTGDIG+ D D L +V R+KELI G +++P E++A++L+HP +S +
Sbjct: 386 EF-GWFHTGDIGYFDTDGYLHLVGRIKELINRGGEKISPIEVDAVLLTHPDVSQGVAFGV 444
Query: 472 LDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILR 531
DE GE V+ G+ TE+DIK F K + +K RVF D +PK+ SGKI R
Sbjct: 445 PDEKYGEEINCAVIPREGT-TVTEEDIKAFCKKNLAAFKVPKRVFITDNLPKTASGKIQR 503
Query: 532 K 532
+
Sbjct: 504 R 504
>At5g16340 AMP-binding protein
Length = 550
Score = 184 bits (467), Expect = 1e-46
Identities = 152/513 (29%), Positives = 240/513 (46%), Gaps = 43/513 (8%)
Query: 52 IYTYYDVQLTAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFF 111
+YT+ + L +VAS L+ +GI + DV+ VL PN P GAI+ N
Sbjct: 40 VYTWRETNLRCLRVASSLSSIGIGRSDVVSVLSPNTPAMYELQFAVPMSGAILNNINTRL 99
Query: 112 TSAEIAKQAKASNTKLLVTQACYYDKVKDLENVK------LVFVDSSPEEDNHMHFSELI 165
+ ++ + +KLL D + ++ LV + EE ++L
Sbjct: 100 DARTVSVLLRHCESKLLFVDVFSVDLAVEAVSMMTTDPPILVVIADKEEEGGVADVADLS 159
Query: 166 QADQNEMEEVKVN------IKPD---DVVALPYSSGTTGLPKGVMLTHKGL-VTSIAQQV 215
+ + ++ I+P+ D V L Y+SGTT PKGV+ H+G+ V S+ +
Sbjct: 160 KFSYTYDDLIERGDPGFKWIRPESEWDPVVLNYTSGTTSAPKGVVHCHRGIFVMSVDSLI 219
Query: 216 DGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYK 275
D P V L LP+FH + ++ L KFD + L+ +
Sbjct: 220 DWAVPK-----NPVYLWTLPIFHSNGWTNPWGIAAVGGTNVCLR-KFDAPLIYRLIRDHG 273
Query: 276 VTI---APVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQ 332
VT APVV ++ A +S L+ + +L +G P L +RA+ +
Sbjct: 274 VTHMCGAPVVLNMLSATQESQPLNH----PVNILTAGSPPPATVL---LRAESIGFVISH 326
Query: 333 GYGMTEAGPVLTMCLSFAK-------EPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQ 385
GYG+TE V+ C K + +K+ V E+ +VDP++ S+ RN
Sbjct: 327 GYGLTETAGVIVSCAWKPKWNHLPASDRARLKARQGVRTVGFTEIDVVDPESGLSVERNG 386
Query: 386 P--GEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKY 443
GEI +RG +M GYL +P T + + K GW +TGD+G I D L I DR K++I
Sbjct: 387 ETVGEIVMRGSSVMLGYLKDPVGTEKAL-KNGWFYTGDVGVIHSDGYLEIKDRSKDIIIT 445
Query: 444 KGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNG-SIDTTEDDIKKFV 502
G V+ E+E ++ + P +++VAVV DE GE P AFV NG S TE+++ ++
Sbjct: 446 GGENVSSVEVETVLYTIPAVNEVAVVARPDEFWGETPCAFVSLKNGFSGKPTEEELMEYC 505
Query: 503 SKQVVFYKRINRVFFIDAIPKSPSGKILRKDLR 535
K++ Y V F+D +PKS +GK+ + LR
Sbjct: 506 RKKMPKYMVPKTVSFMDELPKSSTGKVTKFVLR 538
>At1g77240 unknown protein
Length = 545
Score = 177 bits (450), Expect = 1e-44
Identities = 147/538 (27%), Positives = 248/538 (45%), Gaps = 47/538 (8%)
Query: 36 SKYGSRPCLINAPTAEIYTYYDVQLTAQKVASGLNK--LGIQQGDVIMVLLPNCPEFVFA 93
S +G P L++ T ++T+ + ++AS L+ LGI +G V+ V+ PN P
Sbjct: 25 SVFGDSPSLLHTTT--VHTWSETHSRCLRIASTLSSASLGINRGQVVSVIGPNVPSVYEL 82
Query: 94 FLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVTQACYYDKVKDLENVKLVFVDSSP 153
GA++ NP + ++ + S +KL+ ++ LE V + D P
Sbjct: 83 QFAVPMSGAVLNNINPRLDAHALSVLLRHSESKLVFVD--HHSSSLVLEAVSFLPKDERP 140
Query: 154 E-----EDNHMHFSELIQADQNEMEEVKVNIKPDDV-------------VALPYSSGTTG 195
+ N M S AD + ++ + ++ D+ + L Y+SGTT
Sbjct: 141 RLVILNDGNDMPSSS--SADMDFLDTYEGFMERGDLRFKWVRPKSEWTPMVLNYTSGTTS 198
Query: 196 LPKGVMLTHKGL-VTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCGLRAKA 254
PKGV+ +H+ + +++I +D PN V L LPMFH S A
Sbjct: 199 SPKGVVHSHRSVFMSTINSLLDWSLPN-----RPVYLWTLPMFHANGW-SYTWATAAVGA 252
Query: 255 SILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLG 314
+ + + D+ F L+ KY+VT P ++ + P K S ++V+ +G P
Sbjct: 253 RNICVTRVDVPTIFNLIDKYQVTHMCAAPMVLNMLTNHPA-QKPLQSPVKVMTAGAPPPA 311
Query: 315 KELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAK----EPIDVKSGACGTVVRNA-- 368
+ F + GYGMTE G ++ C + EP + +R A
Sbjct: 312 TVISKAEALGFD---VSHGYGMTETGGLVVSCALKPEWDRLEPDERAKQKSRQGIRTAVF 368
Query: 369 -EMKIVDPQNDSSLPRNQP--GEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFI 425
E+ + DP + S+ + GEI RG +M GY +PE T ++ ++GW +TGDIG +
Sbjct: 369 AEVDVRDPISGKSVKHDGATVGEIVFRGGSVMLGYYKDPEGTAASMREDGWFYTGDIGVM 428
Query: 426 DDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVV 485
D L + DR K+++ G ++ ELEA++ ++P I + AVV D+ GE P AFV
Sbjct: 429 HPDGYLEVKDRSKDVVICGGENISSTELEAVLYTNPAIKEAAVVAKPDKMWGETPCAFVS 488
Query: 486 RSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAGVP 543
TE +I++F ++ Y V F++ +PK+ +GKI +K L ++A +P
Sbjct: 489 LKYHDGSVTEREIREFCKTKLPKYMVPRNVVFLEELPKTSTGKI-QKFLLRQMAKSLP 545
>At5g16370 AMP-binding protein
Length = 552
Score = 176 bits (446), Expect = 3e-44
Identities = 145/511 (28%), Positives = 235/511 (45%), Gaps = 39/511 (7%)
Query: 52 IYTYYDVQLTAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFF 111
+YT+ + L +VAS L+ +GI + DV+ VL PN P GAI+ N
Sbjct: 40 VYTWRETNLRCLRVASSLSSIGIGRSDVVSVLSPNTPAMYELQFAVPMSGAILNNINTRL 99
Query: 112 TSAEIAKQAKASNTKLLVTQACYYDKVKDLENVK------LVFVDSSPEEDNHMH----- 160
+ ++ + +KLL D + ++ LVF+ EE
Sbjct: 100 DARTVSVLLRHCGSKLLFVDVFSVDLAVEAISMMTTDPPILVFIADKEEEGGDADVADRT 159
Query: 161 -----FSELIQADQNEMEEVKVNIKPDDVVALPYSSGTTGLPKGVMLTHKGL-VTSIAQQ 214
+ +LI + + ++ + D VV L Y+SGTT PKGV+ H+G+ V SI
Sbjct: 160 KFSYTYDDLIHRGDLDFKWIRPESEWDPVV-LNYTSGTTSAPKGVVHCHRGIFVMSIDSL 218
Query: 215 VDGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKY 274
+D P V L LP+FH + ++ L KFD + L+ +
Sbjct: 219 IDWTVPK-----NPVYLWTLPIFHANGWSYPWGIAAVGGTNVCLR-KFDAPLIYRLIRDH 272
Query: 275 KVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGY 334
VT P ++ ++ + E + + +L +G P L +RA+ + GY
Sbjct: 273 GVTHMCGAPVVLNMLSATNEFQPLN-RPVNILTAGAPPPAAVL---LRAESIGFVISHGY 328
Query: 335 GMTEAGPVLTMCLSF-------AKEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQP- 386
G+TE + C A + +K+ V E+ +VDP++ S+ RN
Sbjct: 329 GLTETAGLNVSCAWKPQWNRLPASDRARLKARQGVRTVGFTEIDVVDPESGRSVERNGET 388
Query: 387 -GEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKG 445
GEI +RG IM GYL +P T + + K GW +TGD+G I D L I DR K++I G
Sbjct: 389 VGEIVMRGSSIMLGYLKDPVGTEKAL-KNGWFYTGDVGVIHSDGYLEIKDRSKDIIITGG 447
Query: 446 FQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSID-TTEDDIKKFVSK 504
V+ E+E ++ ++P +++VAVV D GE P AFV +G TE ++ ++ K
Sbjct: 448 ENVSSVEVETVLYTNPAVNEVAVVARPDVFWGETPCAFVSLKSGLTQRPTEVEMIEYCRK 507
Query: 505 QVVFYKRINRVFFIDAIPKSPSGKILRKDLR 535
++ Y V F+D +PK+ +GK+++ LR
Sbjct: 508 KMPKYMVPKTVSFVDELPKTSTGKVMKFVLR 538
>At1g21540 amp-binding protein, putative
Length = 550
Score = 176 bits (446), Expect = 3e-44
Identities = 148/544 (27%), Positives = 261/544 (47%), Gaps = 54/544 (9%)
Query: 36 SKYGSRPCLINAPTAEIYTYYDVQLTAQKVASGL--NKLGIQQGDVIMVLLPNCPEFVFA 93
S YG P +++ T ++T+ + ++AS L + LGI +G V+ V+ PN P
Sbjct: 25 SVYGDCPSILHT-TNTVHTWSETHNRCLRIASALTSSSLGINRGQVVSVVGPNVPSVYEL 83
Query: 94 FLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLL---------VTQACYYDKVKDLENV 144
GAI+ NP + ++ + S +KL+ V +A + ++ E
Sbjct: 84 QFAVPMSGAILNNINPRLDAHALSVLLRHSESKLVFVDPNSISVVLEAVSF--MRQNEKP 141
Query: 145 KLVFVDSSPEEDN--HMHFSELIQADQNEMEEVKVN---IKPD---DVVALPYSSGTTGL 196
LV +D E+ + S+ + Q ME I+P + L Y+SGTT
Sbjct: 142 HLVLLDDDQEDGSLSPSAASDFLDTYQGVMERGDSRFKWIRPQTEWQPMILNYTSGTTSS 201
Query: 197 PKGVMLTHKGL-VTSIAQQVDGENPNLYYHSEDVILCVLPMFHI----YSLNSVLLCGLR 251
PKGV+L+H+ + + +++ +D PN V L LPMFH Y+ + +
Sbjct: 202 PKGVVLSHRAIFMLTVSSLLDWHFPN-----RPVYLWTLPMFHANGWGYTWGTAAV---- 252
Query: 252 AKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGA 311
A+ + + D + L+ K+ VT P ++ + P + ++V+ +G
Sbjct: 253 -GATNVCTRRVDAPTIYDLIDKHHVTHMCAAPMVLNMLTNYPSRKPLK-NPVQVMTAGAP 310
Query: 312 PLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAK----EPID---VKSGACGTV 364
P + RA+ +G GYG+TE G + C A+ +P++ +KS
Sbjct: 311 PPAAIIS---RAETLGFNVGHGYGLTETGGPVVSCAWKAEWDHLDPLERARLKSRQGVRT 367
Query: 365 VRNAEMKIVDPQNDSSLPRN--QPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDI 422
+ AE+ + DP+ S+ + GEI ++G +M GY +PE T + ++GW ++GD+
Sbjct: 368 IGFAEVDVRDPRTGKSVEHDGVSVGEIVLKGGSVMLGYYKDPEGTAACMREDGWFYSGDV 427
Query: 423 GFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVA 482
G I +D L + DR K++I G ++ AE+E ++ ++P + + AVV D+ GE P A
Sbjct: 428 GVIHEDGYLEVKDRSKDVIICGGENISSAEVETVLYTNPVVKEAAVVAKPDKMWGETPCA 487
Query: 483 FV---VRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLA 539
FV SNG+ TE +I++F ++ Y +V F + +PK+ +GKI +K L ++A
Sbjct: 488 FVSLKYDSNGNGLVTEREIREFCKTRLPKYMVPRKVIFQEELPKTSTGKI-QKFLLRQMA 546
Query: 540 AGVP 543
+P
Sbjct: 547 KSLP 550
>At1g75960 adenosine monophosphate binding protein 8 AMPBP8
Length = 544
Score = 171 bits (433), Expect = 1e-42
Identities = 145/511 (28%), Positives = 237/511 (46%), Gaps = 39/511 (7%)
Query: 52 IYTYYDVQLTAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFF 111
+YT+ + VAS L+ +GI + DV+ VL N PE GAI+ N
Sbjct: 40 VYTWRETNHRCLCVASALSSIGIGRSDVVSVLSANTPEMYELQFSVPMSGAILNNINTRL 99
Query: 112 TSAEIAKQAKASNTKLLVTQACYYDKVKD-----LENVKLVFVDSSPEEDNHMH------ 160
+ ++ + +KLL Y D + L LV + + EE+
Sbjct: 100 DARTVSVLLRHCESKLLFVDFFYSDLAVEAITMLLNPPILVLIANEEEEEGGAEVTERSK 159
Query: 161 ----FSELIQADQNEMEEVKVNIKPDDVVALPYSSGTTGLPKGVMLTHKGL-VTSIAQQV 215
+S+LI + + ++ + D +V + Y+SGTT PKGV+ H+G+ V ++
Sbjct: 160 FCYLYSDLITRGNPDFKWIRPGSEWDPIV-VNYTSGTTSSPKGVVHCHRGIFVMTLDSLT 218
Query: 216 DGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCGLRAKASI-LLMPKFDINAFFGLVTKY 274
D P V L LP+FH G+ A + + K + + L+ +
Sbjct: 219 DWAVPKT-----PVYLWTLPIFHANGWTYPW--GIAAVGGTNVCVRKLHAPSIYHLIRDH 271
Query: 275 KVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGY 334
VT P ++ ++ S E D+ S + L +G +P L +RA+ + GY
Sbjct: 272 GVTHMYGAPIVLQILSASQESDQPLKSPVNFLTAGSSPPATVL---LRAESLGFIVSHGY 328
Query: 335 GMTEAGPVLTMCLSF-------AKEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQP- 386
G+TE V+ C A + +KS V +E+ +VDP++ S+ R+
Sbjct: 329 GLTETAGVIVSCAWKPNWNRLPASDQAQLKSRQGVRTVGFSEIDVVDPESGRSVERDGET 388
Query: 387 -GEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKG 445
GEI +RG IM GYL NP T+ + K GW TGD+G I D L I DR K++I G
Sbjct: 389 VGEIVLRGSSIMLGYLKNPIGTQNSF-KNGWFFTGDLGVIHGDGYLEIKDRSKDVIISGG 447
Query: 446 FQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNG-SIDTTEDDIKKFVSK 504
V+ E+EA++ ++P +++ AVV DE GE P AFV G + T+ +I ++
Sbjct: 448 ENVSSVEVEAVLYTNPAVNEAAVVARPDEFWGETPCAFVSLKPGLTRKPTDKEIIEYCKY 507
Query: 505 QVVFYKRINRVFFIDAIPKSPSGKILRKDLR 535
++ Y V F++ +PK+ +GKI++ L+
Sbjct: 508 KMPRYMAPKTVSFLEELPKTSTGKIIKSLLK 538
>At1g21530 amp-binding protein, putative
Length = 776
Score = 166 bits (419), Expect = 4e-41
Identities = 139/543 (25%), Positives = 250/543 (45%), Gaps = 58/543 (10%)
Query: 36 SKYGSRPCLINAPTAEIYTYYDVQLTAQKVASGL--NKLGIQQGDVIMVLLPNCPEFVFA 93
S YG P +++ ++T+ + ++AS L + +GI+QG V+ V+ PN P
Sbjct: 25 SVYGDCPSILHTANT-VHTWSETHNRCLRIASALTSSSIGIKQGQVVSVVGPNVPSVYEL 83
Query: 94 FLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVTQACYYDKVKDL-------ENVKL 146
GAI+ NP + ++ + S ++L+ V + E L
Sbjct: 84 QFAVPMSGAILNNINPRLDAHALSVLLRHSESRLVFVDHRSISLVLEAVSLFTQHEKPHL 143
Query: 147 VFVDSSPEEDNHMH------FSELIQADQNEMEEVKVNIKPDDVVALPYSSGTTGLPKGV 200
V +D E D+ + E+++ + + ++ + +V L Y+SGTT PKGV
Sbjct: 144 VLLDDDQENDSSSASDFLDTYEEIMERGNSRFKWIRPQTEWQPMV-LNYTSGTTSSPKGV 202
Query: 201 MLTHKGL-VTSIAQQVDGENPNLYYHSEDVILCVLPMFHI----YSLNSVLLCGLRAKAS 255
+L+H+ + + +++ +D PN V L LPMFH Y+ + + A+
Sbjct: 203 VLSHRAIFMLTVSSLLDWSVPN-----RPVYLWTLPMFHANGWGYTWGTAAV-----GAT 252
Query: 256 ILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSP---------ELDKYDLSSIRVL 306
+ + D + L+ K+ VT P ++ + P ++ + S+ V+
Sbjct: 253 NICTRRVDAPTIYNLIDKHNVTHMCAAPMVLNMLINYPLSTPLKNPVQVHEEKTGSLPVM 312
Query: 307 KSGGAPLGKELEDTVRAKFPKAKLGQGYGMTE-AGPVLTMCLSFAKEPID------VKSG 359
SG P + RA+ + YG+TE +GPV++ + +D +KS
Sbjct: 313 TSGAPPPATIIS---RAESLGFNVSHSYGLTETSGPVVSCAWKPKWDHLDPLERARLKSR 369
Query: 360 ACGTVVRNAEMKIVDPQNDSSLPRN--QPGEICIRGDQIMKGYLNNPEATRETIDKEGWL 417
+ E+ + D + S+ + GEI RG +M GY +P+ T + ++GW
Sbjct: 370 QGVRTLGFTEVDVRDRKTGKSVKHDGVSVGEIVFRGSSVMLGYYKDPQGTAACMREDGWF 429
Query: 418 HTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAG 477
++GDIG I D L I DR K++I G ++ AE+E ++ ++P + + AVV D+ G
Sbjct: 430 YSGDIGVIHKDGYLEIKDRSKDVIICGGENISSAEIETVLYTNPVVKEAAVVAKPDKMWG 489
Query: 478 EVPVAFVV-----RSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRK 532
E P AFV +GS+ TE +I++F ++ Y +V F + +PK+ +GKI +
Sbjct: 490 ETPCAFVSLKCDNNGDGSVPVTEREIREFCKTKLPKYMVPRKVIFQEELPKTSTGKIQKF 549
Query: 533 DLR 535
LR
Sbjct: 550 LLR 552
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,964,563
Number of Sequences: 26719
Number of extensions: 517353
Number of successful extensions: 1440
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 45
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1222
Number of HSP's gapped (non-prelim): 74
length of query: 544
length of database: 11,318,596
effective HSP length: 104
effective length of query: 440
effective length of database: 8,539,820
effective search space: 3757520800
effective search space used: 3757520800
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144503.3


