

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.2 + phase: 0
(322 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
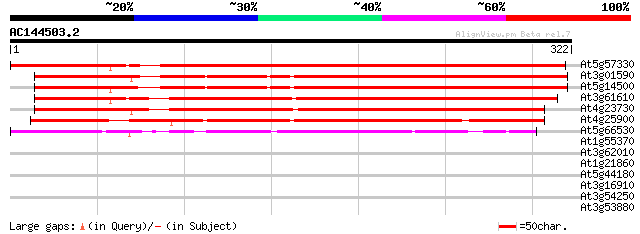
Score E
Sequences producing significant alignments: (bits) Value
At5g57330 apospory-associated protein C 429 e-120
At3g01590 unknown protein 351 3e-97
At5g14500 unknown protein 343 6e-95
At3g61610 unknown protein 341 3e-94
At4g23730 unknown protein 311 3e-85
At4g25900 possible apospory-associated like protein 287 5e-78
At5g66530 apospory-associated protein C-like 104 6e-23
At1g55370 unknown protein 37 0.019
At3g62010 putative protein 30 2.3
At1g21860 pectinesterase, putative 29 3.9
At5g44180 unknown protein 28 6.7
At3g16910 AMP-binding protein, putative 28 6.7
At3g54250 DIPHOSPHOMEVALONATE DECARBOXYLASE - like protein 28 8.7
At3g53880 putative alcohol dehydrogenase 28 8.7
>At5g57330 apospory-associated protein C
Length = 312
Score = 429 bits (1102), Expect = e-120
Identities = 214/323 (66%), Positives = 252/323 (77%), Gaps = 16/323 (4%)
Query: 1 MSRETMYVKHCMGVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK---- 56
M+ E + + G+NGL+KI+LRE+RG SAEVYLYG VTSWKNE GEELL +SSK
Sbjct: 1 MATERHHFELAKGINGLDKIVLRESRGRSAEVYLYGSHVTSWKNENGEELLHLSSKAIFK 60
Query: 57 PEKEVKKTNQYNCFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNS 116
P K ++ CF +N + + F +N++W ++ NPPP P NS S
Sbjct: 61 PPKPIRGGIPL-CFPQFSN-----------FGTLESHGFARNRIWEVEANPPPLPLNSCS 108
Query: 117 RAFVDLVLKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHT 176
AFVDL+L+ +EDD K WP+ +EFRLRIALG G+L LTSRIRNTN DGK FTFTFAYHT
Sbjct: 109 SAFVDLILRPTEDDLKIWPNNFEFRLRIALGTEGELTLTSRIRNTNSDGKPFTFTFAYHT 168
Query: 177 YLYVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHE 236
Y V+DISEVR+EGLETLDYLD+LK+RERFTEQGDAITFE EVDK+YLSTPTKIAI+DHE
Sbjct: 169 YFSVSDISEVRVEGLETLDYLDNLKDRERFTEQGDAITFESEVDKIYLSTPTKIAILDHE 228
Query: 237 RKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWK 296
+KRTF IRKDGL DAVVWNPWDKKSK ISDLGD++YKHMLCV+ A +E+ ITLKPGEEWK
Sbjct: 229 KKRTFVIRKDGLADAVVWNPWDKKSKTISDLGDEDYKHMLCVEAAAIERPITLKPGEEWK 288
Query: 297 GRQEISAVPSSYCSGQLDPRKVL 319
GR E+SAVPSSY SGQLDP+KVL
Sbjct: 289 GRLELSAVPSSYSSGQLDPKKVL 311
>At3g01590 unknown protein
Length = 306
Score = 351 bits (901), Expect = 3e-97
Identities = 177/309 (57%), Positives = 225/309 (72%), Gaps = 18/309 (5%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYN---CFD 71
+G +IIL E RG +AEV L+GGQV SWKNER EELL++SSK + + K + CF
Sbjct: 10 DGSSRIILTEPRGSTAEVLLFGGQVISWKNERREELLYMSSKAQYKPPKAIRGGIPVCFP 69
Query: 72 MNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDDT 131
N + ++ F +NK W D +P P P +N ++ VDL+LK++EDD
Sbjct: 70 QFGN-----------FGGLERHGFARNKFWSHDEDPSPLPP-ANKQSSVDLILKSTEDDL 117
Query: 132 KNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEGL 191
K WPH +E R+RI++ P G L L R+RN D K+F+F FA YLYV+DISEVR+EGL
Sbjct: 118 KTWPHSFELRIRISISP-GKLTLIPRVRNI--DSKAFSFMFALRNYLYVSDISEVRVEGL 174
Query: 192 ETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDA 251
ETLDYLD+L +ERFTEQ DAITF+GEVD+VYL+TPTKIA+IDHERKRT E+RK+G+P+A
Sbjct: 175 ETLDYLDNLIGKERFTEQADAITFDGEVDRVYLNTPTKIAVIDHERKRTIELRKEGMPNA 234
Query: 252 VVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCSG 311
VVWNPWDKK+K I+D+GD++YK MLCV +E + LKP EEWKGRQE+S V SSYCSG
Sbjct: 235 VVWNPWDKKAKTIADMGDEDYKTMLCVDSGVIEPLVLLKPREEWKGRQELSIVSSSYCSG 294
Query: 312 QLDPRKVLF 320
QLDPRKVL+
Sbjct: 295 QLDPRKVLY 303
>At5g14500 unknown protein
Length = 306
Score = 343 bits (881), Expect = 6e-95
Identities = 174/310 (56%), Positives = 223/310 (71%), Gaps = 20/310 (6%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEKEVKKTNQYNCF 70
+G +I+L + G +AEV LYGGQV SWKNER E+LL++S+K P K ++ +
Sbjct: 10 DGSSRILLTDPAGSTAEVLLYGGQVVSWKNERREKLLYMSTKAQLKPPKAIRGGLPISFP 69
Query: 71 DMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDD 130
N + ++ F +N+ W LD +P P P +N ++ VDLVLK++EDD
Sbjct: 70 QFGN------------FGALERHGFARNRFWSLDNDPSPLPP-ANQQSTVDLVLKSTEDD 116
Query: 131 TKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEG 190
K WPH +E R+RI++ P G L + R+RNT D K+F+F F+ YLYV+DISEVR+EG
Sbjct: 117 LKIWPHSFELRVRISISP-GKLTIIPRVRNT--DTKAFSFMFSLRNYLYVSDISEVRVEG 173
Query: 191 LETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPD 250
LETLDYLD+L RERFTEQ DAITF+GEVDKVYL+TPTKIAIIDHERKRT E+RK+G+P+
Sbjct: 174 LETLDYLDNLMRRERFTEQADAITFDGEVDKVYLNTPTKIAIIDHERKRTIELRKEGMPN 233
Query: 251 AVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCS 310
A VWNPWDKK+K I+D+GD++Y MLCV +E I LKP EEWKGRQE+S V SSYCS
Sbjct: 234 AAVWNPWDKKAKSIADMGDEDYTTMLCVDSGAIESPIVLKPHEEWKGRQELSIVSSSYCS 293
Query: 311 GQLDPRKVLF 320
GQLDPRKVL+
Sbjct: 294 GQLDPRKVLY 303
>At3g61610 unknown protein
Length = 317
Score = 341 bits (875), Expect = 3e-94
Identities = 172/305 (56%), Positives = 221/305 (72%), Gaps = 19/305 (6%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEKEVKKTNQYNCF 70
NG+++++LR G SA++ L+GGQV SW+NE GEELLF S+K P K ++ Q C+
Sbjct: 21 NGVDQVLLRNPHGASAKISLHGGQVISWRNELGEELLFTSNKAIFKPPKSMRGGIQI-CY 79
Query: 71 DMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSN-SRAFVDLVLKNSED 129
+ L Q+ F +NK+W++D NPPP +N + ++FVDL+LK SED
Sbjct: 80 PQFGDCGSLD-----------QHGFARNKIWVIDENPPPLNSNESLGKSFVDLLLKPSED 128
Query: 130 DTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIE 189
D K WPH +EFRLR++L GDL LTSRIRN N GK F+F+FAYHTYL V+DISEVRIE
Sbjct: 129 DLKQWPHSFEFRLRVSLAVDGDLTLTSRIRNIN--GKPFSFSFAYHTYLSVSDISEVRIE 186
Query: 190 GLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLP 249
GLETLDYLD+L R+ TEQGDAITFE E+D+ YL +P +A++DHERKRT+ I K+GLP
Sbjct: 187 GLETLDYLDNLSQRQLLTEQGDAITFESEMDRTYLRSPKVVAVLDHERKRTYVIGKEGLP 246
Query: 250 DAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYC 309
D VVWNPW+KKSK ++D GD+EYK MLCV A VE+ ITLKPGEEW GR ++AV SS+C
Sbjct: 247 DTVVWNPWEKKSKTMADFGDEEYKSMLCVDGAAVERPITLKPGEEWTGRLILTAVKSSFC 306
Query: 310 SGQLD 314
QL+
Sbjct: 307 FDQLE 311
>At4g23730 unknown protein
Length = 306
Score = 311 bits (797), Expect = 3e-85
Identities = 154/298 (51%), Positives = 213/298 (70%), Gaps = 18/298 (6%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYN---CFD 71
NG ++I+L+ RG S ++ L+GGQV SWK ++G+ELLF S+K + + CF
Sbjct: 22 NGTDQILLQNPRGASVKISLHGGQVLSWKTDKGDELLFNSTKANLKPPHPVRGGIPICFP 81
Query: 72 MNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPT-NSNSRAFVDLVLKNSEDD 130
+ L Q+ F +NK+W+++ NPP P+ +S +A+VDLVLK+S++D
Sbjct: 82 QFGTRGSLE-----------QHGFARNKMWLVENNPPALPSFDSTGKAYVDLVLKSSDED 130
Query: 131 TKN-WPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIE 189
T WP+ +EF LR++L G+L L SR+RN N K F+F+ AYHTY ++DISEVR+E
Sbjct: 131 TMRIWPYSFEFHLRVSLALDGNLTLISRVRNINS--KPFSFSIAYHTYFSISDISEVRLE 188
Query: 190 GLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLP 249
GLETLDYLD++ +RERFTEQGDA+TFE E+D+VYL++ +AI DHERKRTF I+K+GLP
Sbjct: 189 GLETLDYLDNMHDRERFTEQGDALTFESEIDRVYLNSKDVVAIFDHERKRTFLIKKEGLP 248
Query: 250 DAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSS 307
D VVWNPW+KK++ ++DLGDDEY+HMLCV A +EK ITLKPGEEW G+ +S V S+
Sbjct: 249 DVVVWNPWEKKARALTDLGDDEYRHMLCVDGAAIEKPITLKPGEEWTGKLHLSLVLST 306
>At4g25900 possible apospory-associated like protein
Length = 318
Score = 287 bits (735), Expect = 5e-78
Identities = 149/298 (50%), Positives = 199/298 (66%), Gaps = 21/298 (7%)
Query: 13 GVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYNCFDM 72
GVNGL+KII+R+ RG SAEVYLYGGQV+SWKNE GEELL +SSK F
Sbjct: 36 GVNGLDKIIIRDRRGRSAEVYLYGGQVSSWKNENGEELLVMSSKA-----------IFQP 84
Query: 73 NNNKCRLILLLQDLYAEAF---QYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSED 129
I +L Y+ + F++ + W ++ PPP P S S A VDL++++S +
Sbjct: 85 PTPIRGGIPVLFPQYSNTGPLPSHGFVRQRFWEVETKPPPLP--SLSTAHVDLIVRSSNE 142
Query: 130 DTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIE 189
D K WPH++E+RLR+ALG GDL LTSR++NT D K F FTFA H Y V++ISE+ +E
Sbjct: 143 DLKIWPHKFEYRLRVALGHDGDLTLTSRVKNT--DTKPFNFTFALHPYFAVSNISEIHVE 200
Query: 190 GLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLP 249
GL LDYLD KNR RFT+ ITF ++D++YLSTP ++ I+DH++K+T + K+G
Sbjct: 201 GLHNLDYLDQQKNRTRFTDHEKVITFNAQLDRLYLSTPDQLRIVDHKKKKTIVVHKEGQV 260
Query: 250 DAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSS 307
DAVVWNPWDKK +SDLG ++YK + V+ A V K IT+ PG+EWKG +S VPS+
Sbjct: 261 DAVVWNPWDKK---VSDLGVEDYKRFVTVESAAVAKPITVNPGKEWKGILHVSVVPSN 315
>At5g66530 apospory-associated protein C-like
Length = 307
Score = 104 bits (260), Expect = 6e-23
Identities = 89/310 (28%), Positives = 142/310 (45%), Gaps = 41/310 (13%)
Query: 1 MSRETMYVKHCMGVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKE 60
M+ T V+ G L K++L + AE+YL+GG +TSWK G++LLFV +P+
Sbjct: 29 MASSTTGVRVAEGEGNLPKLVLTSPQNSEAEIYLFGGCITSWKVASGKDLLFV--RPDAV 86
Query: 61 VKKTNQY-----NCFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLW-ILDPNPPPFPTNS 114
K +CF L+Q Q+ F +N W ++D N+
Sbjct: 87 FNKIKPISGGIPHCFPQFGPG-----LIQ-------QHGFGRNMDWSVVDSQ------NA 128
Query: 115 NSRAFVDLVLKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAY 174
+ A V L LK+ W ++ ++ +G L++ ++ TN D K F+F+ A
Sbjct: 129 DDNAAVTLELKDGPYSRAMWDFAFQALYKVIVGADS---LSTELKITNTDDKPFSFSTAL 185
Query: 175 HTYLYVTDI-SEVR-IEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAI 232
HTY + + VR ++G +TL+ KN E DA+TF G VD VYL P ++
Sbjct: 186 HTYFRASSAGASVRGLKGCKTLNKDPDPKNPIEGKEDRDAVTFPGFVDTVYLDAPNELQ- 244
Query: 233 IDHERKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPG 292
D+ I+ DAV+WNP + Y+ +CV+ A + + L+PG
Sbjct: 245 FDNGLGDKIIIKNTNWSDAVLWNPHTQMEAC--------YRDFVCVENAKLGD-VKLEPG 295
Query: 293 EEWKGRQEIS 302
+ W Q +S
Sbjct: 296 QSWTATQLLS 305
>At1g55370 unknown protein
Length = 354
Score = 36.6 bits (83), Expect = 0.019
Identities = 25/138 (18%), Positives = 54/138 (39%), Gaps = 23/138 (16%)
Query: 176 TYLYVTDISEVRIEGLETLDYLDSLKNRERF----------------TEQGDAITFEGEV 219
+YL V+ GLE D++++ RF E+ + E+
Sbjct: 215 SYLTVSTPEATYAVGLEGSDFVETTPFLPRFGVVQGEKEEEKPGFGGEEESNYKQLNREM 274
Query: 220 DKVYLSTPTKIAIIDHERKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQ 279
++Y P +ID R+ + + ++G + +++P + +C+
Sbjct: 275 SRIYTCAPKSFTVIDRGRRNSVVVGREGFEEVYMYSPGSRLESYTKSA-------YVCIG 327
Query: 280 PACVEKAITLKPGEEWKG 297
P+ + I+L+ G W+G
Sbjct: 328 PSSLLSPISLESGCVWRG 345
>At3g62010 putative protein
Length = 1254
Score = 29.6 bits (65), Expect = 2.3
Identities = 17/64 (26%), Positives = 30/64 (46%), Gaps = 6/64 (9%)
Query: 36 GGQVTSWKNERGEELLFVSSKP------EKEVKKTNQYNCFDMNNNKCRLILLLQDLYAE 89
GG+ W N R E +++++ KP E+ K +Y D + + L +D+ E
Sbjct: 501 GGRPVFWHNMREEPVIYINGKPFVLREVERPYKNMLEYTGIDRDRVEGMEARLKEDILRE 560
Query: 90 AFQY 93
A +Y
Sbjct: 561 AKRY 564
>At1g21860 pectinesterase, putative
Length = 538
Score = 28.9 bits (63), Expect = 3.9
Identities = 13/29 (44%), Positives = 20/29 (68%), Gaps = 4/29 (13%)
Query: 110 FPTNS----NSRAFVDLVLKNSEDDTKNW 134
FPT S + RAFV+++ +NSED ++W
Sbjct: 406 FPTTSVMQADFRAFVEVIFENSEDIVQSW 434
>At5g44180 unknown protein
Length = 1694
Score = 28.1 bits (61), Expect = 6.7
Identities = 20/77 (25%), Positives = 39/77 (49%), Gaps = 5/77 (6%)
Query: 14 VNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYNCF--- 70
V G+E + ++ +G++A+ Q+ ++ + EEL S P + ++ N+Y F
Sbjct: 1032 VPGVENLQYQQQQGYTADRERLRAQLKAYVGYKAEELYVYRSLPLGQDRRRNRYWRFSAS 1091
Query: 71 -DMNNNKC-RLILLLQD 85
N+ C R+ + LQD
Sbjct: 1092 ASRNDPGCGRIFVELQD 1108
>At3g16910 AMP-binding protein, putative
Length = 569
Score = 28.1 bits (61), Expect = 6.7
Identities = 32/124 (25%), Positives = 51/124 (40%), Gaps = 20/124 (16%)
Query: 186 VRIEGLETLDYLDSLKNRERFTEQGDA--ITFEGE-VDKVYLSTPTKIAIIDHERKRTFE 242
VR G+E LD +D+ + + A I F G V K YL P K TF
Sbjct: 378 VRYTGMEQLDVIDTQTGKPVPADGKTAGEIVFRGNMVMKGYLKNP-------EANKETFA 430
Query: 243 IRKDGLPDAVVWNP------WDKKSKIISDLGDD----EYKHMLCVQPACVEKAITLKPG 292
D V +P D+ +I G++ E ++++ PA +E ++ +P
Sbjct: 431 GGWFHSGDIAVKHPDNYIEIKDRSKDVIISGGENISSVEVENVVYHHPAVLEASVVARPD 490
Query: 293 EEWK 296
E W+
Sbjct: 491 ERWQ 494
>At3g54250 DIPHOSPHOMEVALONATE DECARBOXYLASE - like protein
Length = 419
Score = 27.7 bits (60), Expect = 8.7
Identities = 35/152 (23%), Positives = 62/152 (40%), Gaps = 11/152 (7%)
Query: 103 LDPNPPPFPTNSNSRAFVDLVLK-NSEDDTKNWPHRYEFRLRIALGPTGDLMLTSR-IRN 160
LD +PP F N S + LV K N + T + ++ GP L+ +R +
Sbjct: 273 LDTSPPIFYMNDTSHRIISLVEKWNRSEGTPQVAYTFD------AGPNAVLIARNRKVAV 326
Query: 161 TNKDGKSFTFTFAYHTYL---YVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEG 217
G + F T + V D S ++ GL+ +++L+ + + +G
Sbjct: 327 QLLQGLLYYFPPKSDTDMKSYVVGDNSILKEAGLDGASGVENLQPPPEIKDNIGSQDQKG 386
Query: 218 EVDKVYLSTPTKIAIIDHERKRTFEIRKDGLP 249
EV + P K I+ H++ + + GLP
Sbjct: 387 EVSYFICTRPGKGPIVLHDQTQALLDPETGLP 418
>At3g53880 putative alcohol dehydrogenase
Length = 315
Score = 27.7 bits (60), Expect = 8.7
Identities = 18/73 (24%), Positives = 34/73 (45%), Gaps = 4/73 (5%)
Query: 68 NCFDMNNNKCRLILLLQDLYAEAF---QYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVL 124
+C N+ + +L+ L+ + + +F+ +K+W+ D +PP N R DL L
Sbjct: 47 DCASRYGNEIEIGKVLKKLFDDGVVKREKLFITSKIWLTDLDPPDVQDALN-RTLQDLQL 105
Query: 125 KNSEDDTKNWPHR 137
+ +WP R
Sbjct: 106 DYVDLYLMHWPVR 118
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,036,994
Number of Sequences: 26719
Number of extensions: 365670
Number of successful extensions: 974
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 941
Number of HSP's gapped (non-prelim): 16
length of query: 322
length of database: 11,318,596
effective HSP length: 99
effective length of query: 223
effective length of database: 8,673,415
effective search space: 1934171545
effective search space used: 1934171545
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144503.2


