

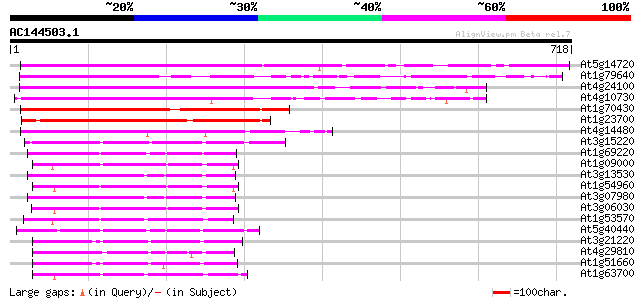
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.1 + phase: 0 /pseudo
(718 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g14720 protein kinase -like protein 550 e-156
At1g79640 hypothetical protein 533 e-151
At4g24100 unknown protein 483 e-136
At4g10730 putative protein kinase 481 e-136
At1g70430 hypothetical protein 462 e-130
At1g23700 putative Ser/Thr protein kinase 328 6e-90
At4g14480 kinase like protein 310 2e-84
At3g15220 putative MAP kinase 217 2e-56
At1g69220 serine/threonine kinase (SIK1) 194 1e-49
At1g09000 NPK1-related protein kinase 1S (ANP1) 152 5e-37
At3g13530 MAP3K epsilon protein kinase 147 2e-35
At1g54960 NPK1-related protein kinase 2 (ANP2) 147 2e-35
At3g07980 putative MAP3K epsilon protein kinase 147 3e-35
At3g06030 NPK1-related protein kinase 3 140 2e-33
At1g53570 MEK kinase MAP3Ka, putative 138 9e-33
At5g40440 MAP kinase kinase 3 (ATMKK3) 137 2e-32
At3g21220 MAP kinase kinase 5 137 3e-32
At4g29810 MAP kinase kinase 2 134 1e-31
At1g51660 unknown protein 133 3e-31
At1g63700 putative protein kinase 131 2e-30
>At5g14720 protein kinase -like protein
Length = 674
Score = 550 bits (1417), Expect = e-156
Identities = 330/713 (46%), Positives = 422/713 (58%), Gaps = 55/713 (7%)
Query: 14 KKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQT 73
+KK+P+ A+ Y+LYEEIG GVSA+VHRALC+ N +VAIK+LD E+ N DL+ I RE QT
Sbjct: 6 EKKFPLNAKDYKLYEEIGDGVSATVHRALCIPLNVVVAIKVLDLEKCNNDLDGIRREVQT 65
Query: 74 MVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLK 133
M L++HPNVL++HCSF + H LWVVMP+M+GGSCLHI+K+++PDGFEE VIAT+LRE LK
Sbjct: 66 MSLINHPNVLQAHCSFTTGHQLWVVMPYMAGGSCLHIIKSSYPDGFEEPVIATLLRETLK 125
Query: 134 GLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMA 193
L YLH HGHIHRDVKAGN+L+DS GAVKL DFGVSAC+FD+GDRQRSRNTFVGTPCWMA
Sbjct: 126 ALVYLHAHGHIHRDVKAGNILLDSNGAVKLADFGVSACMFDTGDRQRSRNTFVGTPCWMA 185
Query: 194 PEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKK 253
PEVM+QLHGY+FKAD+WSFGITALELAHGHAPFSKYPP+KVLLMTLQNAPPGLDYERDK+
Sbjct: 186 PEVMQQLHGYDFKADVWSFGITALELAHGHAPFSKYPPMKVLLMTLQNAPPGLDYERDKR 245
Query: 254 FSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEILK 313
FSK+FK+M+ CLVKDP KRP++ KLLKH FFK AR +DY+ +T+L GLP LGDR +K
Sbjct: 246 FSKAFKEMVGTCLVKDPKKRPTSEKLLKHPFFKHARPADYLVKTILNGLPPLGDRYRQIK 305
Query: 314 RKE-DMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLINDFDDAMSDISHVS 372
KE D+L Q K LSQ EY RGIS WNFNLED+K QA+LI+D D + ++ +
Sbjct: 306 SKEADLLMQNK--SEYEAHLSQQEYIRGISAWNFNLEDLKTQAALISDDDTSHAEEPDFN 363
Query: 373 -SACSLTNLDAQDKQLPSSSHSR----SQTADMSDDDSSITSSSHEP--QTSSCLDDHVD 425
C + A + SSS + + D+ D +SS S +P C D D
Sbjct: 364 QKQCERQDESALSPERASSSATAPSQDDELNDIHDLESSFASFPIKPLQALKGCFDISED 423
Query: 426 HSLGDMENVGRAAEVVVATHPPLHRRGCSSSILPEVTLPPIRAESLRNDREYFKH*CLSN 485
D + V + L + S+ A++ R
Sbjct: 424 E---DNATTPDWKDANVNSGQQLLTKASIGSLAETTKEEDTAAQNTSLPRHVISE---QK 477
Query: 486 FHHSHSICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTGDDVLTE 545
+ S SI + S F+P ++ D E Q S + L D V
Sbjct: 478 KYLSGSIIPE--------STFSPKRITSDADREFQQRRYQTERSYSGSLYRTKRDSV--- 526
Query: 546 LPSRASKTSANSDDTDDKAKVPVVQQRGRFKVTSENVDPEKATPSPVLQKSHSMQVGCLE 605
D+ ++VP V+ +GRFKVTS ++ P+ +T S S
Sbjct: 527 ---------------DETSEVPHVEHKGRFKVTSADLSPKGSTNSTFTPFSGGTSSPSCL 571
Query: 606 VCTPTNILQRVC*VLYGSILSPSANVQD*PSFVHDILTGNHSNFNEANYCG*IC*NIGYT 665
T +IL + SIL +A ++ + + A G N+
Sbjct: 572 NATTASILPSI-----QSILQQNAMQRE-----EILRLIKYLEQTSAKQPGSPETNVD-D 620
Query: 666 LLQLESAHEREKELLHEITDLQWRLICTQEELQKLKTDNAQVLS--NALASNN 716
LLQ A RE+EL ++ LQ EEL+K K N Q+ + NAL N
Sbjct: 621 LLQTPPATSRERELQSQVMLLQQSFSSLTEELKKQKQKNGQLENQLNALTHRN 673
>At1g79640 hypothetical protein
Length = 547
Score = 533 bits (1372), Expect = e-151
Identities = 337/695 (48%), Positives = 397/695 (56%), Gaps = 153/695 (22%)
Query: 13 EKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQ 72
EKKKYPIG EHY LYE IGQGVSA VHRALC+ F+E+VAIKILDFERDNCDLNNISREAQ
Sbjct: 2 EKKKYPIGPEHYTLYEFIGQGVSALVHRALCIPFDEVVAIKILDFERDNCDLNNISREAQ 61
Query: 73 TMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVL 132
TM+LVDHPNVLKSHCSFVSDHNLWV+MP+MSGGSCLHILKAA+PDGFEE +IAT+LRE L
Sbjct: 62 TMMLVDHPNVLKSHCSFVSDHNLWVIMPYMSGGSCLHILKAAYPDGFEEAIIATILREAL 121
Query: 133 KGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWM 192
KGL+YLH HGHIHRDVKAGN+L+ +RGAVKLGDFGVSACLFDSGDRQR+RNTFVGTPCW
Sbjct: 122 KGLDYLHQHGHIHRDVKAGNILLGARGAVKLGDFGVSACLFDSGDRQRTRNTFVGTPCW- 180
Query: 193 APEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDK 252
ADIWSFGIT LELAHGHAPFSKYPP+
Sbjct: 181 --------------ADIWSFGITGLELAHGHAPFSKYPPM-------------------- 206
Query: 253 KFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEIL 312
KSFKQMIA CLVKDPSKRPSA KLLKHSFFKQARSSDYI R LL+GLP L +R++ +
Sbjct: 207 ---KSFKQMIASCLVKDPSKRPSAKKLLKHSFFKQARSSDYIARKLLDGLPDLVNRVQAI 263
Query: 313 KRKEDMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLINDFDDAMSDISHVS 372
K NEYKRGISG + + ++A D D S+I +
Sbjct: 264 K---------------------NEYKRGISGLSGSATSLQAL-----DSQDTQSEIQEDT 297
Query: 373 SACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSSSHEPQTSSCLDDHVDHSLGDME 432
+TN Q P S S D SDDDSS+ S S++ S H D SL +
Sbjct: 298 G--QITNKYLQ----PLIHRSLSIARDKSDDDSSLASPSYDSYVYSS-PRHEDLSLNNTH 350
Query: 433 NVGRAAEVVVATHPPLHRRGCSSSILPEVTLPPIRAESLRNDREYFKH*CLSNFHHSHSI 492
V +TH + ++SI I S+ D
Sbjct: 351 --------VGSTHANNGKPTDATSIPTNQPTEIIAGSSVLAD------------------ 384
Query: 493 CSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTGDDVLTELPSRASK 552
N P N +S+K Q N S+ N T GDDV TE+ + K
Sbjct: 385 -----------GNGAP----NKGESDKTQEQLQNGSNCNGTHPTVGGDDVPTEMAVKPPK 429
Query: 553 TSANSDDTDDKAKVPVVQQRGRFKVTSENVDPEKATPSPVLQKSHSMQVGCLEVCTPTNI 612
+++ D++DDK+K PVVQQRGRFKVTSEN+D EK VL + S + +V P
Sbjct: 430 AASSLDESDDKSKPPVVQQRGRFKVTSENLDIEK-----VLCQHSSASLPHSDVTLPNLT 484
Query: 613 LQRVC*VLYGSILSPSANVQD*PSFVHDILTGNHSNFNEANYCG*IC*NIGYTLLQLESA 672
V ++Y P +IL N+ Y +C QL +
Sbjct: 485 SSYVYPLVY-------------PVLQTNILERMDVQLNKEVYNNLLC-------PQLRN- 523
Query: 673 HEREKELLHEITDLQWRLICTQEELQKLKTDNAQV 707
W LIC +EELQK KT++AQV
Sbjct: 524 --------------PW-LICAEEELQKYKTEHAQV 543
>At4g24100 unknown protein
Length = 709
Score = 483 bits (1244), Expect = e-136
Identities = 280/610 (45%), Positives = 366/610 (59%), Gaps = 55/610 (9%)
Query: 13 EKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQ 72
+++ + + + Y+L EEIG G SA V+RA+ + NE+VAIK LD +R N +L++I RE+Q
Sbjct: 22 QQRGFSMNPKDYKLMEEIGHGASAVVYRAIYLPTNEVVAIKCLDLDRCNSNLDDIRRESQ 81
Query: 73 TMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVL 132
TM L+DHPNV+KS CSF DH+LWVVMPFM+ GSCLH++K A+ DGFEE I VL+E L
Sbjct: 82 TMSLIDHPNVIKSFCSFSVDHSLWVVMPFMAQGSCLHLMKTAYSDGFEESAICCVLKETL 141
Query: 133 KGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWM 192
K L+YLH GHIHRDVKAGN+L+D G +KLGDFGVSACLFD+GDRQR+RNTFVGTPCWM
Sbjct: 142 KALDYLHRQGHIHRDVKAGNILLDDNGEIKLGDFGVSACLFDNGDRQRARNTFVGTPCWM 201
Query: 193 APEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDK 252
APEV++ +GYN KADIWSFGITALELAHGHAPFSKYPP+KVLLMT+QNAPPGLDY+RDK
Sbjct: 202 APEVLQPGNGYNSKADIWSFGITALELAHGHAPFSKYPPMKVLLMTIQNAPPGLDYDRDK 261
Query: 253 KFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEIL 312
KFSKSFK+M+A CLVKD +KRP+A KLLKHS FK + + + L LP L R++ L
Sbjct: 262 KFSKSFKEMVAMCLVKDQTKRPTAEKLLKHSCFKHTKPPEQTVKILFSDLPPLWTRVKSL 321
Query: 313 KRKE-DMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLINDFDDAMSDISHV 371
+ K+ LA K+M E +SQ+EY+RG+S WNF++ D+K QASL+ D DD
Sbjct: 322 QDKDAQQLALKRMATADEEAISQSEYQRGVSAWNFDVRDLKTQASLLIDDDDLEESKEDE 381
Query: 372 SSACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSSSHEPQTSSCLDDHVDHSLGDM 431
C+ N +Q+ S Q + + ++++ E T
Sbjct: 382 EILCAQFNKVNDREQVFDS----LQLYENMNGKEKVSNTEVEEPTC-------------- 423
Query: 432 ENVGRAAEVVVATHPPLHRRGCSSS-ILPEVTLPPIRAESLRNDREYFKH*CLSNFHHSH 490
+ V T L R +S +PE + P+R +S + HS
Sbjct: 424 ----KEKFTFVTTTSSLERMSPNSEHDIPEAKVKPLRRQSQSGPLT-----SRTVLSHSA 474
Query: 491 SICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTGDDVLTELPSRA 550
S S F ++ P + + S L NLST SS L +
Sbjct: 475 SEKSHIFERSESEPQTAPTVRRAPSFSGPL-NLSTRASS--------------NSLSAPI 519
Query: 551 SKTSANSDDTDDKAKVPVVQQRGRFKVTSENVD----------PEKATPSPVLQKSHSMQ 600
+ D DDK+K +V Q+GRF VTS NVD P ++ + L+KS S+
Sbjct: 520 KYSGGFRDSLDDKSKANLV-QKGRFSVTSGNVDLAKDVPLSIVPRRSPQATPLRKSASVG 578
Query: 601 VGCLEVCTPT 610
LE PT
Sbjct: 579 NWILEPKMPT 588
>At4g10730 putative protein kinase
Length = 693
Score = 481 bits (1238), Expect = e-136
Identities = 280/620 (45%), Positives = 371/620 (59%), Gaps = 92/620 (14%)
Query: 7 DKEKETEKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNN 66
DK+K KK + + + Y+L EE+G G SA VHRA+ + NE+VAIK LD +R N +L++
Sbjct: 13 DKKK---KKGFSVNPKDYKLMEEVGYGASAVVHRAIYLPTNEVVAIKSLDLDRCNSNLDD 69
Query: 67 ISREAQTMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIAT 126
I REAQTM L+DHPNV+KS CSF DH+LWVVMPFM+ GSCLH++KAA+PDGFEE I +
Sbjct: 70 IRREAQTMTLIDHPNVIKSFCSFAVDHHLWVVMPFMAQGSCLHLMKAAYPDGFEEAAICS 129
Query: 127 VLREVLKGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFV 186
+L+E LK L+YLH GHIHRDVKAGN+L+D G +KLGDFGVSACLFD+GDRQR+RNTFV
Sbjct: 130 MLKETLKALDYLHRQGHIHRDVKAGNILLDDTGEIKLGDFGVSACLFDNGDRQRARNTFV 189
Query: 187 GTPCWMAPEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGL 246
GTPCWMAPEV++ GYN KADIWSFGITALELAHGHAPFSKYPP+KVLLMT+QNAPPGL
Sbjct: 190 GTPCWMAPEVLQPGSGYNSKADIWSFGITALELAHGHAPFSKYPPMKVLLMTIQNAPPGL 249
Query: 247 DYERDKKFSK-------SFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLL 299
DY+RDKKFSK SFK+++A CLVKD +KRP+A KLLKHSFFK + + + L
Sbjct: 250 DYDRDKKFSKKMSSCAQSFKELVALCLVKDQTKRPTAEKLLKHSFFKNVKPPEICVKKLF 309
Query: 300 EGLPALGDRMEILKRKEDMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLIN 359
LP L R++ L Q+EY+RG+S WNFN+ED+K QASL++
Sbjct: 310 VDLPPLWTRVKAL---------------------QSEYQRGVSAWNFNIEDLKEQASLLD 348
Query: 360 DFDDAMSDISHVSSACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSSSHEPQTSSC 419
D DD +++ S ++ +QL + + R Q + S + S + + +
Sbjct: 349 D-DDILTE--------SREEEESFGEQLHNKVNDRGQVS-----GSQLLSENMNGKEKAS 394
Query: 420 LDDHVDHSLGDMENVGRAAEVVVATHPPLHRRGCSSSILPEVTLPPIRAESLRNDREYFK 479
+ V+ + + V S +P+ P+R ++
Sbjct: 395 DTEVVEPICEEKSTLNSTTSSVE------QPASSSEQDVPQAKGKPVRLQT--------- 439
Query: 480 H*CLSNFHHSHSICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTG 539
HS + S N + SE + L ++V A + +G
Sbjct: 440 --------HSGPLSSGVVLINSDSEKVH-----GYERSESERQLKSSVRRAPSF----SG 482
Query: 540 DDVLTELPSRASKTSANS---------DDTDDKAKVPVVQQRGRFKVTSENVDPEKATPS 590
LP+RAS S ++ D DDK+K VVQ +GRF VTSEN+D +A+P
Sbjct: 483 P---LNLPNRASANSLSAPIKSSGGFRDSIDDKSKANVVQIKGRFSVTSENLDLARASP- 538
Query: 591 PVLQKSHSMQVGCLEVCTPT 610
L+KS S+ L+ PT
Sbjct: 539 --LRKSASVGNWILDSKMPT 556
>At1g70430 hypothetical protein
Length = 594
Score = 462 bits (1188), Expect = e-130
Identities = 218/344 (63%), Positives = 276/344 (79%), Gaps = 13/344 (3%)
Query: 15 KKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTM 74
K++P+ A+ Y+L+EE+G+GVSA+V+RA C++ NEIVA+KILD E+ DL I +E M
Sbjct: 7 KRFPLYAKDYELFEEVGEGVSATVYRARCIALNEIVAVKILDLEKCRNDLETIRKEVHIM 66
Query: 75 VLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKG 134
L+DHPN+LK+HCSF+ +LW+VMP+MSGGSC H++K+ +P+G E+ +IAT+LREVLK
Sbjct: 67 SLIDHPNLLKAHCSFIDSSSLWIVMPYMSGGSCFHLMKSVYPEGLEQPIIATLLREVLKA 126
Query: 135 LEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAP 194
L YLH GHIHRDVKAGN+LI S+G VKLGDFGVSAC+FDSG+R ++RNTFVGTPCWMAP
Sbjct: 127 LVYLHRQGHIHRDVKAGNILIHSKGVVKLGDFGVSACMFDSGERMQTRNTFVGTPCWMAP 186
Query: 195 EVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKF 254
EVM+QL GY+FK LAHGHAPFSKYPP+KVLLMTLQNAPP LDY+RDKKF
Sbjct: 187 EVMQQLDGYDFK-----------YLAHGHAPFSKYPPMKVLLMTLQNAPPRLDYDRDKKF 235
Query: 255 SKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEILKR 314
SKSF+++IA CLVKDP KRP+A+KLLKH FFK ARS+DY++R +L GL LG+R + LK
Sbjct: 236 SKSFRELIAACLVKDPKKRPTAAKLLKHPFFKHARSTDYLSRKILHGLSPLGERFKKLKE 295
Query: 315 KEDMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLI 358
E L K +G E+LSQ+EY RGIS WNF+LE ++ QASL+
Sbjct: 296 AEAELF--KGINGDKEQLSQHEYMRGISAWNFDLEALRRQASLV 337
>At1g23700 putative Ser/Thr protein kinase
Length = 473
Score = 328 bits (841), Expect = 6e-90
Identities = 172/322 (53%), Positives = 231/322 (71%), Gaps = 16/322 (4%)
Query: 16 KYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTMV 75
++P+ A+ Y++ EEIG GV +RA C+ +EIVAIKI + E+ DL I +E +
Sbjct: 8 RFPLVAKDYEILEEIGDGV----YRARCILLDEIVAIKIWNLEKCTNDLETIRKEVHRLS 63
Query: 76 LVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGL 135
L+DHPN+L+ HCSF+ +LW+VMPFMS GS L+I+K+ +P+G EE VIA +LRE+LK L
Sbjct: 64 LIDHPNLLRVHCSFIDSSSLWIVMPFMSCGSSLNIMKSVYPNGLEEPVIAILLREILKAL 123
Query: 136 EYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQR--SRNTFVGTPCWMA 193
YLH GHIHR+VKAGNVL+DS G VKLGDF VSA +FDS +R R S NTFVG P MA
Sbjct: 124 VYLHGLGHIHRNVKAGNVLVDSEGTVKLGDFEVSASMFDSVERMRTSSENTFVGNPRRMA 183
Query: 194 PEV-MEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDK 252
PE M+Q+ GY+FK DIWSFG+TALELAHGH+P + VL + LQN+ P +YE D
Sbjct: 184 PEKDMQQVDGYDFKVDIWSFGMTALELAHGHSPTT------VLPLNLQNS-PFPNYEEDT 236
Query: 253 KFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEIL 312
KFSKSF++++A CL++DP KRP+AS+LL++ F +Q S++Y+ T L+GL LG+R L
Sbjct: 237 KFSKSFRELVAACLIEDPEKRPTASQLLEYPFLQQTLSTEYLASTFLDGLSPLGERYRKL 296
Query: 313 KRKEDMLAQKKMPDGQMEELSQ 334
K ++ L K DG E++SQ
Sbjct: 297 KEEKAKLV--KGVDGNKEKVSQ 316
>At4g14480 kinase like protein
Length = 487
Score = 310 bits (794), Expect = 2e-84
Identities = 175/419 (41%), Positives = 242/419 (56%), Gaps = 46/419 (10%)
Query: 14 KKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNE-IVAIKILDFERDNCDLNNISREAQ 72
K ++P+ AE Y++ +IG GVSASV++A+C+ N +VAIK +D ++ D +++ RE +
Sbjct: 5 KLEFPLDAEAYEIICKIGVGVSASVYKAICIPMNSMVVAIKAIDLDQSRADFDSLRRETK 64
Query: 73 TMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVL 132
TM L+ HPN+L ++CSF D LWVVMPFMS GS I+ ++ P G E I+ L+E L
Sbjct: 65 TMSLLSHPNILNAYCSFTVDRCLWVVMPFMSCGSLHSIVSSSFPSGLPENCISVFLKETL 124
Query: 133 KGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFD-----SGDRQRS--RNTF 185
+ YLH GH+HRD+KAGN+L+DS G+VKL DFGVSA +++ SG S
Sbjct: 125 NAISYLHDQGHLHRDIKAGNILVDSDGSVKLADFGVSASIYEPVTSSSGTTSSSLRLTDI 184
Query: 186 VGTPCWMAPEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPG 245
GTP WMAPEV+ GY FKADIWSFGITALELAHG P S PPLK LLM +
Sbjct: 185 AGTPYWMAPEVVHSHTGYGFKADIWSFGITALELAHGRPPLSHLPPLKSLLMKITKRFHF 244
Query: 246 LDYE---------RDKKFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITR 296
DYE +KKFSK+F++M+ CL +DP+KRPSA KLLKH FFK + D++ +
Sbjct: 245 SDYEINTSGSSKKGNKKFSKAFREMVGLCLEQDPTKRPSAEKLLKHPFFKNCKGLDFVVK 304
Query: 297 TLLEGLPALGDRMEILKRKEDMLAQKKMPDGQMEELSQNEY--KRGISGWNFNLEDMKAQ 354
+L L E + + +L + D + EE E R ISGWNF +D++
Sbjct: 305 NVLHSL----SNAEQMFMESQILIKSVGDDDEEEEEEDEEIVKNRRISGWNFREDDLQ-- 358
Query: 355 ASLINDFDDAMSDISHVSSACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSSSHE 413
+S T D+ + SS Q+ D +DD ++T + +E
Sbjct: 359 ----------------LSPVFPATESDSSE----SSPREEDQSKDKKEDD-NVTITGYE 396
>At3g15220 putative MAP kinase
Length = 690
Score = 217 bits (552), Expect = 2e-56
Identities = 122/335 (36%), Positives = 191/335 (56%), Gaps = 17/335 (5%)
Query: 20 GAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTMVLVDH 79
GA Q+ E IG+G V++A N+ VAIK++D E ++ +I +E +
Sbjct: 12 GARFSQI-ELIGRGSFGDVYKAFDKDLNKEVAIKVIDLEESEDEIEDIQKEISVLSQCRC 70
Query: 80 PNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLH 139
P + + + S++ LW++M +M+GGS +L++ +P +E IA + R++L +EYLH
Sbjct: 71 PYITEYYGSYLHQTKLWIIMEYMAGGSVADLLQSNNP--LDETSIACITRDLLHAVEYLH 128
Query: 140 HHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQ 199
+ G IHRD+KA N+L+ G VK+ DFGVSA L + R R TFVGTP WMAPEV++
Sbjct: 129 NEGKIHRDIKAANILLSENGDVKVADFGVSAQLTRTISR---RKTFVGTPFWMAPEVIQN 185
Query: 200 LHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKFSKSFK 259
GYN KADIWS GIT +E+A G P + P++VL + + PP L D+ FS+ K
Sbjct: 186 SEGYNEKADIWSLGITVIEMAKGEPPLADLHPMRVLFIIPRETPPQL----DEHFSRQVK 241
Query: 260 QMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEILKRKEDML 319
+ ++ CL K P++RPSA +L+KH F K AR S + + E + ++ + +E
Sbjct: 242 EFVSLCLKKAPAERPSAKELIKHRFIKNARKSPKLLERIRE-----RPKYQVKEDEETPR 296
Query: 320 AQKKMP--DGQMEELSQNEYKRGISGWNFNLEDMK 352
K P ++++E +G G++F +K
Sbjct: 297 NGAKAPVESSGTVRIARDERSQGAPGYSFQGNTVK 331
>At1g69220 serine/threonine kinase (SIK1)
Length = 836
Score = 194 bits (493), Expect = 1e-49
Identities = 103/267 (38%), Positives = 159/267 (58%), Gaps = 7/267 (2%)
Query: 24 YQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTMVLVDHPNVL 83
Y+ E+G+G SV++A + +EIVA+K++ I E + + +HPNV+
Sbjct: 249 YEFLNELGKGSYGSVYKARDLKTSEIVAVKVISLTEGEEGYEEIRGEIEMLQQCNHPNVV 308
Query: 84 KSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGH 143
+ S+ + LW+VM + GGS ++ + EE IA + RE LKGL YLH
Sbjct: 309 RYLGSYQGEDYLWIVMEYCGGGSVADLMNVTE-EALEEYQIAYICREALKGLAYLHSIYK 367
Query: 144 IHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHGY 203
+HRD+K GN+L+ +G VKLGDFGV+A L + RNTF+GTP WMAPEV+++ + Y
Sbjct: 368 VHRDIKGGNILLTEQGEVKLGDFGVAAQLTRT---MSKRNTFIGTPHWMAPEVIQE-NRY 423
Query: 204 NFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKFSKSFKQMIA 263
+ K D+W+ G++A+E+A G P S P++VL M P L E +K+S F +A
Sbjct: 424 DGKVDVWALGVSAIEMAEGLPPRSSVHPMRVLFMISIEPAPML--EDKEKWSLVFHDFVA 481
Query: 264 CCLVKDPSKRPSASKLLKHSFFKQARS 290
CL K+P RP+A+++LKH F ++ ++
Sbjct: 482 KCLTKEPRLRPTAAEMLKHKFVERCKT 508
>At1g09000 NPK1-related protein kinase 1S (ANP1)
Length = 666
Score = 152 bits (385), Expect = 5e-37
Identities = 93/277 (33%), Positives = 152/277 (54%), Gaps = 23/277 (8%)
Query: 30 IGQGVSASVHRALCVSFNEIVAIK-------ILDFERDNCDLNNISREAQTMVLVDHPNV 82
IG+G +V+ + + E++A+K E+ + + E + + + HPN+
Sbjct: 75 IGRGAFGTVYMGMNLDSGELLAVKQVLIAANFASKEKTQAHIQELEEEVKLLKNLSHPNI 134
Query: 83 LKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHG 142
++ + D L +++ F+ GGS +L+ P F E V+ T R++L GLEYLH+H
Sbjct: 135 VRYLGTVREDDTLNILLEFVPGGSISSLLEKFGP--FPESVVRTYTRQLLLGLEYLHNHA 192
Query: 143 HIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHG 202
+HRD+K N+L+D++G +KL DFG S + + ++ + GTP WMAPEV+ Q G
Sbjct: 193 IMHRDIKGANILVDNKGCIKLADFGASKQVAELATMTGAK-SMKGTPYWMAPEVILQT-G 250
Query: 203 YNFKADIWSFGITALELAHGHAPFS-KYPPLKVLLM--TLQNAPPGLDYERDKKFSKSFK 259
++F ADIWS G T +E+ G AP+S +Y + + T ++ PP D S K
Sbjct: 251 HSFSADIWSVGCTVIEMVTGKAPWSQQYKEVAAIFFIGTTKSHPPIPD-----TLSSDAK 305
Query: 260 QMIACCLVKDPSKRPSASKLLKHSF----FKQARSSD 292
+ CL + P+ RP+AS+LLKH F K++ S+D
Sbjct: 306 DFLLKCLQEVPNLRPTASELLKHPFVMGKHKESASTD 342
>At3g13530 MAP3K epsilon protein kinase
Length = 1368
Score = 147 bits (372), Expect = 2e-35
Identities = 90/267 (33%), Positives = 138/267 (50%), Gaps = 9/267 (3%)
Query: 24 YQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERD-NCDLNNISREAQTMVLVDHPNV 82
Y L +EIG+G V++ L + + VAIK + E DLN I +E + ++H N+
Sbjct: 20 YMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIVQEDLNTIMQEIDLLKNLNHKNI 79
Query: 83 LKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHG 142
+K S + +L +++ ++ GS +I+K F E ++A + +VL+GL YLH G
Sbjct: 80 VKYLGSSKTKTHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQG 139
Query: 143 HIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHG 202
IHRD+K N+L G VKL DFGV+ L + + ++ VGTP WMAPEV+E + G
Sbjct: 140 VIHRDIKGANILTTKEGLVKLADFGVATKL---NEADVNTHSVVGTPYWMAPEVIE-MSG 195
Query: 203 YNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKFSKSFKQMI 262
+DIWS G T +EL P+ P+ L +Q+ P + S +
Sbjct: 196 VCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDDNPPI----PDSLSPDITDFL 251
Query: 263 ACCLVKDPSKRPSASKLLKHSFFKQAR 289
C KD +RP A LL H + + +R
Sbjct: 252 RQCFKKDSRQRPDAKTLLSHPWIRNSR 278
>At1g54960 NPK1-related protein kinase 2 (ANP2)
Length = 651
Score = 147 bits (372), Expect = 2e-35
Identities = 90/277 (32%), Positives = 151/277 (54%), Gaps = 23/277 (8%)
Query: 30 IGQGVSASVHRALCVSFNEIVAIKIL-------DFERDNCDLNNISREAQTMVLVDHPNV 82
IG+G +V+ + + E++A+K + E+ + + E + + + HPN+
Sbjct: 74 IGRGAFGTVYMGMNLDSGELLAVKQVLITSNCASKEKTQAHIQELEEEVKLLKNLSHPNI 133
Query: 83 LKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHG 142
++ + D L +++ F+ GGS +L+ F E V+ T ++L GLEYLH+H
Sbjct: 134 VRYLGTVREDETLNILLEFVPGGSISSLLEKF--GAFPESVVRTYTNQLLLGLEYLHNHA 191
Query: 143 HIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHG 202
+HRD+K N+L+D++G +KL DFG S + + ++ + GTP WMAPEV+ Q G
Sbjct: 192 IMHRDIKGANILVDNQGCIKLADFGASKQVAELATISGAK-SMKGTPYWMAPEVILQT-G 249
Query: 203 YNFKADIWSFGITALELAHGHAPFS-KYPPLKVL--LMTLQNAPPGLDYERDKKFSKSFK 259
++F ADIWS G T +E+ G AP+S +Y + + + T ++ PP D S
Sbjct: 250 HSFSADIWSVGCTVIEMVTGKAPWSQQYKEIAAIFHIGTTKSHPPIPD-----NISSDAN 304
Query: 260 QMIACCLVKDPSKRPSASKLLKHSFF----KQARSSD 292
+ CL ++P+ RP+AS+LLKH F K++ S D
Sbjct: 305 DFLLKCLQQEPNLRPTASELLKHPFVTGKQKESASKD 341
>At3g07980 putative MAP3K epsilon protein kinase
Length = 1367
Score = 147 bits (370), Expect = 3e-35
Identities = 90/267 (33%), Positives = 136/267 (50%), Gaps = 9/267 (3%)
Query: 24 YQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFER-DNCDLNNISREAQTMVLVDHPNV 82
Y L +EIG+G V+ L + + VAIK + E DLN I +E + ++H N+
Sbjct: 20 YMLGDEIGKGAYGRVYIGLDLENGDFVAIKQVSLENIGQEDLNTIMQEIDLLKNLNHKNI 79
Query: 83 LKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHG 142
+K S + +L +++ ++ GS +I+K F E ++ + +VL+GL YLH G
Sbjct: 80 VKYLGSLKTKTHLHIILEYVENGSLANIIKPNKFGPFPESLVTVYIAQVLEGLVYLHEQG 139
Query: 143 HIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHG 202
IHRD+K N+L G VKL DFGV+ L + + ++ VGTP WMAPEV+E L G
Sbjct: 140 VIHRDIKGANILTTKEGLVKLADFGVATKL---NEADFNTHSVVGTPYWMAPEVIE-LSG 195
Query: 203 YNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKFSKSFKQMI 262
+DIWS G T +EL P+ P+ L +Q+ P + S +
Sbjct: 196 VCAASDIWSVGCTIIELLTCVPPYYDLQPMPALYRIVQDDTPPI----PDSLSPDITDFL 251
Query: 263 ACCLVKDPSKRPSASKLLKHSFFKQAR 289
C KD +RP A LL H + + +R
Sbjct: 252 RLCFKKDSRQRPDAKTLLSHPWIRNSR 278
>At3g06030 NPK1-related protein kinase 3
Length = 651
Score = 140 bits (353), Expect = 2e-33
Identities = 91/275 (33%), Positives = 145/275 (52%), Gaps = 17/275 (6%)
Query: 28 EEIGQGVSASVHRALCVSFNEIVAIKIL-------DFERDNCDLNNISREAQTMVLVDHP 80
E IG G V+ + + E++AIK + E+ + + E Q + + HP
Sbjct: 72 ELIGCGAFGRVYMGMNLDSGELLAIKQVLIAPSSASKEKTQGHIRELEEEVQLLKNLSHP 131
Query: 81 NVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHH 140
N+++ + +L ++M F+ GGS +L+ F E VI +++L GLEYLH+
Sbjct: 132 NIVRYLGTVRESDSLNILMEFVPGGSISSLLEKF--GSFPEPVIIMYTKQLLLGLEYLHN 189
Query: 141 HGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQL 200
+G +HRD+K N+L+D++G ++L DFG S + + ++ + GTP WMAPEV+ Q
Sbjct: 190 NGIMHRDIKGANILVDNKGCIRLADFGASKKVVELATVNGAK-SMKGTPYWMAPEVILQT 248
Query: 201 HGYNFKADIWSFGITALELAHGHAPFSKYPP--LKVLLMTLQNAPPGLDYERDKKFSKSF 258
G++F ADIWS G T +E+A G P+S+ VL + A P + + S
Sbjct: 249 -GHSFSADIWSVGCTVIEMATGKPPWSEQYQQFAAVLHIGRTKAHPPI----PEDLSPEA 303
Query: 259 KQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDY 293
K + CL K+PS R SA++LL+H F R Y
Sbjct: 304 KDFLMKCLHKEPSLRLSATELLQHPFVTGKRQEPY 338
>At1g53570 MEK kinase MAP3Ka, putative
Length = 609
Score = 138 bits (348), Expect = 9e-33
Identities = 87/275 (31%), Positives = 142/275 (51%), Gaps = 17/275 (6%)
Query: 18 PIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIK----ILDFERDNCDLNNISREAQT 73
P G ++ + +G G V+ ++ AIK I D + L +++E
Sbjct: 208 PSGFSTWKKGKFLGSGTFGQVYLGFNSEKGKMCAIKEVKVISDDQTSKECLKQLNQEINL 267
Query: 74 MVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLK 133
+ + HPN+++ + S +S+ L V + ++SGGS +LK F E VI R++L
Sbjct: 268 LNQLCHPNIVQYYGSELSEETLSVYLEYVSGGSIHKLLKDY--GSFTEPVIQNYTRQILA 325
Query: 134 GLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMA 193
GL YLH +HRD+K N+L+D G +KL DFG++ + + +F G+P WMA
Sbjct: 326 GLAYLHGRNTVHRDIKGANILVDPNGEIKLADFGMAKHV----TAFSTMLSFKGSPYWMA 381
Query: 194 PEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVL--LMTLQNAPPGLDYERD 251
PEV+ +GY DIWS G T LE+A P+S++ + + + ++ P D+
Sbjct: 382 PEVVMSQNGYTHAVDIWSLGCTILEMATSKPPWSQFEGVAAIFKIGNSKDTPEIPDH--- 438
Query: 252 KKFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFK 286
S K I CL ++P+ RP+AS+LL+H F +
Sbjct: 439 --LSNDAKNFIRLCLQRNPTVRPTASQLLEHPFLR 471
>At5g40440 MAP kinase kinase 3 (ATMKK3)
Length = 520
Score = 137 bits (346), Expect = 2e-32
Identities = 99/317 (31%), Positives = 163/317 (51%), Gaps = 20/317 (6%)
Query: 9 EKETEKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILD-FERDNCDLNNI 67
E E+ + Y + +++ IG G S+ V RA+ + + I+A+K ++ FER+ +
Sbjct: 68 ESESSETTYQCASHEMRVFGAIGSGASSVVQRAIHIPNHRILALKKINIFEREK--RQQL 125
Query: 68 SREAQTMVLVD-HPNVLKSHCSFVSDHN--LWVVMPFMSGGSCLHILKAAHPDGFEEVVI 124
E +T+ H ++ H +F S + + + + +M+GGS ILK E V+
Sbjct: 126 LTEIRTLCEAPCHEGLVDFHGAFYSPDSGQISIALEYMNGGSLADILKVTKK--IPEPVL 183
Query: 125 ATVLREVLKGLEYLHHHGH-IHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRN 183
+++ ++L+GL YLH H +HRD+K N+LI+ +G K+ DFG+SA L +S
Sbjct: 184 SSLFHKLLQGLSYLHGVRHLVHRDIKPANLLINLKGEPKITDFGISAGLENS---MAMCA 240
Query: 184 TFVGTPCWMAPEVMEQLHGYNFKADIWSFGITALELAHGHAPF-SKYPPLKVLLMTLQNA 242
TFVGT +M+PE + Y++ ADIWS G+ E G P+ + P+ ++L L +
Sbjct: 241 TFVGTVTYMSPERIRN-DSYSYPADIWSLGLALFECGTGEFPYIANEGPVNLMLQILDDP 299
Query: 243 PPGLDYERDKKFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGL 302
P ++FS F I CL KDP RP+A +LL H F + T ++
Sbjct: 300 SP---TPPKQEFSPEFCSFIDACLQKDPDARPTADQLLSHPFITKHEKERVDLATFVQ-- 354
Query: 303 PALGDRMEILKRKEDML 319
++ D + LK DML
Sbjct: 355 -SIFDPTQRLKDLADML 370
>At3g21220 MAP kinase kinase 5
Length = 348
Score = 137 bits (344), Expect = 3e-32
Identities = 92/276 (33%), Positives = 144/276 (51%), Gaps = 20/276 (7%)
Query: 30 IGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTMVLVDHPNVLKSHCSF 89
IG G +V++ + + A+K++ ++ I RE + + VDHPNV+K H F
Sbjct: 76 IGSGAGGTVYKVIHTPTSRPFALKVIYGNHEDTVRRQICREIEILRSVDHPNVVKCHDMF 135
Query: 90 VSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGHIHRDVK 149
+ + V++ FM GS L+ AH ++E +A + R++L GL YLH +HRD+K
Sbjct: 136 DHNGEIQVLLEFMDQGS----LEGAHI--WQEQELADLSRQILSGLAYLHRRHIVHRDIK 189
Query: 150 AGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVM--EQLHGY--NF 205
N+LI+S VK+ DFGVS L + D N+ VGT +M+PE + + HG +
Sbjct: 190 PSNLLINSAKNVKIADFGVSRILAQTMD---PCNSSVGTIAYMSPERINTDLNHGRYDGY 246
Query: 206 KADIWSFGITALELAHGHAPF--SKYPPLKVLLMTL-QNAPPGLDYERDKKFSKSFKQMI 262
D+WS G++ LE G PF S+ L+ + + PP E S+ F+ +
Sbjct: 247 AGDVWSLGVSILEFYLGRFPFAVSRQGDWASLMCAICMSQPP----EAPATASQEFRHFV 302
Query: 263 ACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTL 298
+CCL DP KR SA +LL+H F +A + + L
Sbjct: 303 SCCLQSDPPKRWSAQQLLQHPFILKATGGPNLRQML 338
>At4g29810 MAP kinase kinase 2
Length = 363
Score = 134 bits (338), Expect = 1e-31
Identities = 91/270 (33%), Positives = 140/270 (51%), Gaps = 25/270 (9%)
Query: 30 IGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTMVLVDHPNVLKSHCSF 89
IG+G S V + A+K++ D I++E + PN++ S+ SF
Sbjct: 76 IGKGSSGVVQLVQHKWTGQFFALKVIQLNIDEAIRKAIAQELKINQSSQCPNLVTSYQSF 135
Query: 90 VSDHNLWVVMPFMSGGSCLHILKA--AHPDGFEEVVIATVLREVLKGLEYLHHHGHI-HR 146
+ + +++ +M GGS LK+ A PD + ++ + R+VL+GL YLHH HI HR
Sbjct: 136 YDNGAISLILEYMDGGSLADFLKSVKAIPDSY----LSAIFRQVLQGLIYLHHDRHIIHR 191
Query: 147 DVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHGYNFK 206
D+K N+LI+ RG VK+ DFGVS + ++ NTFVGT +M+PE + + Y K
Sbjct: 192 DLKPSNLLINHRGEVKITDFGVSTVMTNTAG---LANTFVGTYNYMSPERIVG-NKYGNK 247
Query: 207 ADIWSFGITALELAHGHAPFSKYPP---------LKVLLMTLQNAPPGLDYERDKKFSKS 257
+DIWS G+ LE A G P++ PP +++ + PP L FS
Sbjct: 248 SDIWSLGLVVLECATGKFPYA--PPNQEETWTSVFELMEAIVDQPPPALP---SGNFSPE 302
Query: 258 FKQMIACCLVKDPSKRPSASKLLKHSFFKQ 287
I+ CL KDP+ R SA +L++H F +
Sbjct: 303 LSSFISTCLQKDPNSRSSAKELMEHPFLNK 332
>At1g51660 unknown protein
Length = 366
Score = 133 bits (335), Expect = 3e-31
Identities = 91/269 (33%), Positives = 140/269 (51%), Gaps = 20/269 (7%)
Query: 30 IGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTMVLVDHPNVLKSHCSF 89
IG G +V++ + + + A+K++ + I RE + + V+HPNV+K H F
Sbjct: 85 IGSGAGGTVYKVIHRPSSRLYALKVIYGNHEETVRRQICREIEILRDVNHPNVVKCHEMF 144
Query: 90 VSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGHIHRDVK 149
+ + V++ FM GS L+ AH ++E +A + R++L GL YLH +HRD+K
Sbjct: 145 DQNGEIQVLLEFMDKGS----LEGAHV--WKEQQLADLSRQILSGLAYLHSRHIVHRDIK 198
Query: 150 AGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPE----VMEQLHGYNF 205
N+LI+S VK+ DFGVS L + D N+ VGT +M+PE + Q +
Sbjct: 199 PSNLLINSAKNVKIADFGVSRILAQTMD---PCNSSVGTIAYMSPERINTDLNQGKYDGY 255
Query: 206 KADIWSFGITALELAHGHAPF--SKYPPLKVLLMTL-QNAPPGLDYERDKKFSKSFKQMI 262
DIWS G++ LE G PF S+ L+ + + PP E S F+ I
Sbjct: 256 AGDIWSLGVSILEFYLGRFPFPVSRQGDWASLMCAICMSQPP----EAPATASPEFRHFI 311
Query: 263 ACCLVKDPSKRPSASKLLKHSFFKQARSS 291
+CCL ++P KR SA +LL+H F +A S
Sbjct: 312 SCCLQREPGKRRSAMQLLQHPFILRASPS 340
>At1g63700 putative protein kinase
Length = 883
Score = 131 bits (329), Expect = 2e-30
Identities = 83/281 (29%), Positives = 139/281 (48%), Gaps = 20/281 (7%)
Query: 30 IGQGVSASVHRALCVSFNEIVAIKIL----DFERDNCDLNNISREAQTMVLVDHPNVLKS 85
+G G V+ E+ A+K + D + + +E + + H N+++
Sbjct: 406 LGMGSFGHVYLGFNSESGEMCAMKEVTLCSDDPKSRESAQQLGQEISVLSRLRHQNIVQY 465
Query: 86 HCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGHIH 145
+ S D L++ + ++SGGS +L+ F E I +++L GL YLH +H
Sbjct: 466 YGSETVDDKLYIYLEYVSGGSIYKLLQEYGQ--FGENAIRNYTQQILSGLAYLHAKNTVH 523
Query: 146 RDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHGYNF 205
RD+K N+L+D G VK+ DFG++ + Q +F G+P WMAPEV++ +G N
Sbjct: 524 RDIKGANILVDPHGRVKVADFGMAKHI----TAQSGPLSFKGSPYWMAPEVIKNSNGSNL 579
Query: 206 KADIWSFGITALELAHGHAPFSKYP--PLKVLLMTLQNAPPGLDYERDKKFSKSFKQMIA 263
DIWS G T LE+A P+S+Y P + + P D+ S+ K +
Sbjct: 580 AVDIWSLGCTVLEMATTKPPWSQYEGVPAMFKIGNSKELPDIPDH-----LSEEGKDFVR 634
Query: 264 CCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPA 304
CL ++P+ RP+A++LL H+F + + R ++ G PA
Sbjct: 635 KCLQRNPANRPTAAQLLDHAFVRNVMPME---RPIVSGEPA 672
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,520,407
Number of Sequences: 26719
Number of extensions: 714595
Number of successful extensions: 5064
Number of sequences better than 10.0: 1001
Number of HSP's better than 10.0 without gapping: 784
Number of HSP's successfully gapped in prelim test: 217
Number of HSP's that attempted gapping in prelim test: 2545
Number of HSP's gapped (non-prelim): 1222
length of query: 718
length of database: 11,318,596
effective HSP length: 106
effective length of query: 612
effective length of database: 8,486,382
effective search space: 5193665784
effective search space used: 5193665784
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144503.1


