

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144502.7 - phase: 2
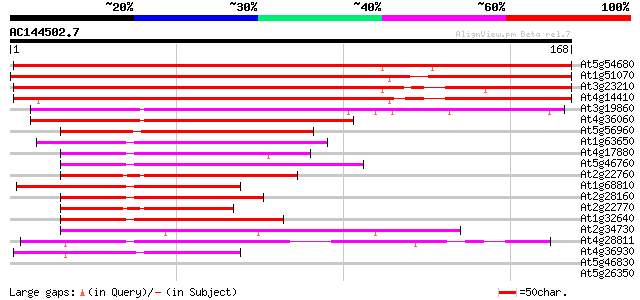
(168 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g54680 putative bHLH transcription factor (bHLH105) 278 1e-75
At1g51070 bHLH transcription factor (bHLH115) like protein 234 2e-62
At3g23210 putative bHLH transcription factor (bHLH034) 170 4e-43
At4g14410 putative bHLH transcription factor (bHLH104) 166 5e-42
At3g19860 putative bHLH transcription factor (bHLH121) 102 1e-22
At4g36060 transcription factor BHLH11 like protein 84 4e-17
At5g56960 putative protein 50 4e-07
At1g63650 putative transcription factor (BHLH2) 45 2e-05
At4g17880 putative transcription factor BHLH4 44 4e-05
At5g46760 putative transcription factor BHLH5 43 7e-05
At2g22760 putative bHLH transcription factor (bHLH019) 42 2e-04
At1g68810 putative bHLH transcription factor (bHLH030) 42 2e-04
At2g28160 putative bHLH transcription factor (bHLH029) 41 3e-04
At2g22770 putative bHLH transcription factor (bHLH020) 41 3e-04
At1g32640 transcription factor like protein, BHLH6 41 3e-04
At2g34730 putative myosin heavy chain 40 5e-04
At4g28811 AtbHLH119 40 6e-04
At4g36930 putative bHLH transcription factor (AtbHLH024) / SPATU... 40 8e-04
At5g46830 putative transcription factor bHLH28 39 0.001
At5g26350 putative protein 39 0.002
>At5g54680 putative bHLH transcription factor (bHLH105)
Length = 234
Score = 278 bits (711), Expect = 1e-75
Identities = 142/169 (84%), Positives = 152/169 (89%), Gaps = 2/169 (1%)
Query: 2 RSESCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGE 61
R ES +ATSSKACREK RRDRLNDKF+ELG+ILEPG P KTDKAAIL+DAVRMVTQLRGE
Sbjct: 66 RCESSSATSSKACREKQRRDRLNDKFMELGAILEPGNPPKTDKAAILVDAVRMVTQLRGE 125
Query: 62 AQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAP-PSFLPTPTAL 120
AQKLKD+NS LQ+KIKELK EKNELRDEKQRLK EKEKLEQQLK+MNAP PSF P P +
Sbjct: 126 AQKLKDSNSSLQDKIKELKTEKNELRDEKQRLKTEKEKLEQQLKAMNAPQPSFFPAPPMM 185
Query: 121 PAAFA-AQGQAHGNKLVPFISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 168
P AFA AQGQA GNK+VP ISYPGVAMWQFMPPA+VDTSQDHVLRPPVA
Sbjct: 186 PTAFASAQGQAPGNKMVPIISYPGVAMWQFMPPASVDTSQDHVLRPPVA 234
>At1g51070 bHLH transcription factor (bHLH115) like protein
Length = 226
Score = 234 bits (596), Expect = 2e-62
Identities = 119/172 (69%), Positives = 136/172 (78%), Gaps = 9/172 (5%)
Query: 1 VRSESCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRG 60
+++ESC ++SKACREK RRDRLNDKF EL S+LEPGR KTDK AI+ DA+RMV Q R
Sbjct: 60 IKTESCTGSNSKACREKQRRDRLNDKFTELSSVLEPGRTPKTDKVAIINDAIRMVNQARD 119
Query: 61 EAQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAPPS----FLPT 116
EAQKLKD NS LQEKIKELK EKNELRDEKQ+LK EKE+++QQLK++ P FLP
Sbjct: 120 EAQKLKDLNSSLQEKIKELKDEKNELRDEKQKLKVEKERIDQQLKAIKTQPQPQPCFLPN 179
Query: 117 PTALPAAFAAQGQAHGNKLVPFISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 168
P L +Q QA G+KLVPF +YPG AMWQFMPPAAVDTSQDHVLRPPVA
Sbjct: 180 PQTL-----SQAQAPGSKLVPFTTYPGFAMWQFMPPAAVDTSQDHVLRPPVA 226
>At3g23210 putative bHLH transcription factor (bHLH034)
Length = 291
Score = 170 bits (430), Expect = 4e-43
Identities = 95/172 (55%), Positives = 120/172 (69%), Gaps = 13/172 (7%)
Query: 2 RSESCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGE 61
R+ SC+ +KACREKLRR++LNDKF++L S+LEPGR KTDK+AIL DA+R+V QLRGE
Sbjct: 128 RTGSCSKPGTKACREKLRREKLNDKFMDLSSVLEPGRTPKTDKSAILDDAIRVVNQLRGE 187
Query: 62 AQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAP-PSFLPTPTAL 120
A +L++ N L E+IK LK +KNELR+EK LKAEKEK+EQQLKSM P P F+P+
Sbjct: 188 AHELQETNQKLLEEIKSLKADKNELREEKLVLKAEKEKMEQQLKSMVVPSPGFMPSQH-- 245
Query: 121 PAAFAAQGQAHGNKLVPFISY----PGVAMWQFMPPAAVDTSQDHVLRPPVA 168
PAAF H +K+ Y P + MW +PPA DTS+D PPVA
Sbjct: 246 PAAF------HSHKMAVAYPYGYYPPNMPMWSPLPPADRDTSRDLKNLPPVA 291
>At4g14410 putative bHLH transcription factor (bHLH104)
Length = 283
Score = 166 bits (420), Expect = 5e-42
Identities = 92/169 (54%), Positives = 120/169 (70%), Gaps = 11/169 (6%)
Query: 2 RSESCA-ATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRG 60
R+ SC+ +KACRE+LRR++LN++F++L S+LEPGR KTDK AIL DA+R++ QLR
Sbjct: 124 RTGSCSRGGGTKACRERLRREKLNERFMDLSSVLEPGRTPKTDKPAILDDAIRILNQLRD 183
Query: 61 EAQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAPPS-FLPTPTA 119
EA KL++ N L E+IK LK EKNELR+EK LKA+KEK EQQLKSM AP S F+P
Sbjct: 184 EALKLEETNQKLLEEIKSLKAEKNELREEKLVLKADKEKTEQQLKSMTAPSSGFIP---H 240
Query: 120 LPAAFAAQGQAHGNKLVPFISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 168
+PAAF + NK+ + SY + MW +MP + DTS+D LRPP A
Sbjct: 241 IPAAF------NHNKMAVYPSYGYMPMWHYMPQSVRDTSRDQELRPPAA 283
>At3g19860 putative bHLH transcription factor (bHLH121)
Length = 284
Score = 102 bits (253), Expect = 1e-22
Identities = 70/185 (37%), Positives = 99/185 (52%), Gaps = 26/185 (14%)
Query: 7 AATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLK 66
A S KA REKLRR++LN+ F+ELG++L+P RP K DKA IL D V+++ +L E KLK
Sbjct: 5 ARKSQKAGREKLRREKLNEHFVELGNVLDPERP-KNDKATILTDTVQLLKELTSEVNKLK 63
Query: 67 DANSGLQEKIKELKVEKNELRDEKQRLKAEKEKL----EQQLKSMN-------------A 109
+ L ++ +EL EKN+LR+EK LK++ E L +Q+L+SM+
Sbjct: 64 SEYTALTDESRELTQEKNDLREEKTSLKSDIENLNLQYQQRLRSMSPWGAAMDHTVMMAP 123
Query: 110 PPSF-LPTPTALPAAFAAQGQA------HGNKLVPFISYPGVAMWQFMPPAAVDTSQD-H 161
PPSF P P A+P + GN+ I P +MPP V Q H
Sbjct: 124 PPSFPYPMPIAMPPGSIPMHPSMPSYTYFGNQNPSMIPAPCPTYMPYMPPNTVVEQQSVH 183
Query: 162 VLRPP 166
+ + P
Sbjct: 184 IPQNP 188
>At4g36060 transcription factor BHLH11 like protein
Length = 286
Score = 84.0 bits (206), Expect = 4e-17
Identities = 44/97 (45%), Positives = 66/97 (67%), Gaps = 1/97 (1%)
Query: 7 AATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLK 66
A S KA REKLRRD+L ++F+ELG+ L+P RP K+DKA++L D ++M+ + + +LK
Sbjct: 44 AVCSQKAEREKLRRDKLKEQFLELGNALDPNRP-KSDKASVLTDTIQMLKDVMNQVDRLK 102
Query: 67 DANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQ 103
L ++ +EL EK+ELR+EK LK++ E L Q
Sbjct: 103 AEYETLSQESRELIQEKSELREEKATLKSDIEILNAQ 139
>At5g56960 putative protein
Length = 466
Score = 50.4 bits (119), Expect = 4e-07
Identities = 28/76 (36%), Positives = 47/76 (61%), Gaps = 2/76 (2%)
Query: 16 EKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDANSGLQEK 75
E+ RR++LN+ F L S+L PG K DKA++L A ++ L+GE KL + N ++ K
Sbjct: 294 ERKRREKLNESFQALRSLLPPG--TKKDKASVLSIAREQLSSLQGEISKLLERNREVEAK 351
Query: 76 IKELKVEKNELRDEKQ 91
+ + +N+LR E++
Sbjct: 352 LAGEREIENDLRPEER 367
>At1g63650 putative transcription factor (BHLH2)
Length = 596
Score = 44.7 bits (104), Expect = 2e-05
Identities = 25/87 (28%), Positives = 47/87 (53%), Gaps = 2/87 (2%)
Query: 9 TSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDA 68
T + A EK RR++LN++F+ L SI+ +K DK +IL D + + L+ Q+L+
Sbjct: 403 TGNHALSEKKRREKLNERFMTLRSIIP--SISKIDKVSILDDTIEYLQDLQKRVQELESC 460
Query: 69 NSGLQEKIKELKVEKNELRDEKQRLKA 95
+ + +++ + DE++R A
Sbjct: 461 RESADTETRITMMKRKKPDDEEERASA 487
>At4g17880 putative transcription factor BHLH4
Length = 589
Score = 43.9 bits (102), Expect = 4e-05
Identities = 25/79 (31%), Positives = 47/79 (58%), Gaps = 6/79 (7%)
Query: 16 EKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDANSGLQEK 75
E+ RR++LN +F L +++ +K DKA++L DA+ +++L+ + QK + LQ++
Sbjct: 421 ERQRREKLNQRFYSLRAVVP--NVSKMDKASLLGDAISYISELKSKLQKAESDKEELQKQ 478
Query: 76 I----KELKVEKNELRDEK 90
I KE K+ ++D K
Sbjct: 479 IDVMNKEAGNAKSSVKDRK 497
>At5g46760 putative transcription factor BHLH5
Length = 592
Score = 43.1 bits (100), Expect = 7e-05
Identities = 24/91 (26%), Positives = 47/91 (51%), Gaps = 2/91 (2%)
Query: 16 EKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDANSGLQEK 75
E+ RR++LN +F L +++ +K DKA++L DA+ + +L+ + Q+ + +Q+K
Sbjct: 420 ERQRREKLNQRFYSLRAVVP--NVSKMDKASLLGDAISYINELKSKLQQAESDKEEIQKK 477
Query: 76 IKELKVEKNELRDEKQRLKAEKEKLEQQLKS 106
+ + E N + R K K + S
Sbjct: 478 LDGMSKEGNNGKGCGSRAKERKSSNQDSTAS 508
>At2g22760 putative bHLH transcription factor (bHLH019)
Length = 295
Score = 42.0 bits (97), Expect = 2e-04
Identities = 24/71 (33%), Positives = 44/71 (61%), Gaps = 2/71 (2%)
Query: 16 EKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDANSGLQEK 75
E+ RR++L++KFI L ++L PG K DK IL DA+ + QL+ + + LK+ ++
Sbjct: 124 ERKRREKLSEKFIALSALL-PGLK-KADKVTILDDAISRMKQLQEQLRTLKEEKEATRQM 181
Query: 76 IKELKVEKNEL 86
+ V+K+++
Sbjct: 182 ESMILVKKSKV 192
>At1g68810 putative bHLH transcription factor (bHLH030)
Length = 368
Score = 41.6 bits (96), Expect = 2e-04
Identities = 22/67 (32%), Positives = 41/67 (60%), Gaps = 2/67 (2%)
Query: 3 SESCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEA 62
+++ AA+ S + E+ RR+R+N+ +L SIL KTDKA++L + ++ V +L+ E
Sbjct: 169 AKALAASKSHSEAERRRRERINNHLAKLRSILP--NTTKTDKASLLAEVIQHVKELKRET 226
Query: 63 QKLKDAN 69
+ + N
Sbjct: 227 SVISETN 233
>At2g28160 putative bHLH transcription factor (bHLH029)
Length = 318
Score = 40.8 bits (94), Expect = 3e-04
Identities = 24/61 (39%), Positives = 37/61 (60%), Gaps = 2/61 (3%)
Query: 16 EKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDANSGLQEK 75
E+ RR R+ DK L S++ K DKA+I+ DAV V +L+ +A+KLK +GL+
Sbjct: 136 ERRRRGRMKDKLYALRSLVP--NITKMDKASIVGDAVLYVQELQSQAKKLKSDIAGLEAS 193
Query: 76 I 76
+
Sbjct: 194 L 194
>At2g22770 putative bHLH transcription factor (bHLH020)
Length = 320
Score = 40.8 bits (94), Expect = 3e-04
Identities = 22/52 (42%), Positives = 36/52 (68%), Gaps = 2/52 (3%)
Query: 16 EKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKD 67
E+ RR +LN++ I L ++L PG KTDKA +L DA++ + QL+ +KL++
Sbjct: 137 ERKRRQKLNERLIALSALL-PGLK-KTDKATVLEDAIKHLKQLQERVKKLEE 186
>At1g32640 transcription factor like protein, BHLH6
Length = 623
Score = 40.8 bits (94), Expect = 3e-04
Identities = 19/67 (28%), Positives = 42/67 (62%), Gaps = 2/67 (2%)
Query: 16 EKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDANSGLQEK 75
E+ RR++LN +F L +++ +K DKA++L DA+ + +L+ + K + ++ +
Sbjct: 457 ERQRREKLNQRFYALRAVVP--NVSKMDKASLLGDAIAYINELKSKVVKTESEKLQIKNQ 514
Query: 76 IKELKVE 82
++E+K+E
Sbjct: 515 LEEVKLE 521
>At2g34730 putative myosin heavy chain
Length = 829
Score = 40.4 bits (93), Expect = 5e-04
Identities = 32/126 (25%), Positives = 62/126 (48%), Gaps = 6/126 (4%)
Query: 16 EKLRRDRLNDKFIELGSILEPGRPAKTDKA-AILIDAVRMVTQLRGEAQKLKDANSGLQ- 73
E+ + + ++ + +L S +E DK A+ + + R + +++G K+ L+
Sbjct: 604 ERKKIEVVSQQINDLQSQVERQETEIQDKIEALSVVSARELEKVKGYETKISSLREELEL 663
Query: 74 --EKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMN--APPSFLPTPTALPAAFAAQGQ 129
E +KE+K EK + ++ KAEKE L++QL S++ PP + L A + Q
Sbjct: 664 ARESLKEMKDEKRKTEEKLSETKAEKETLKKQLVSLDLVVPPQLIKGFDILEGLIAEKTQ 723
Query: 130 AHGNKL 135
++L
Sbjct: 724 KTNSRL 729
>At4g28811 AtbHLH119
Length = 470
Score = 40.0 bits (92), Expect = 6e-04
Identities = 43/170 (25%), Positives = 69/170 (40%), Gaps = 36/170 (21%)
Query: 4 ESCAATSSKACR--------EKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMV 55
E+ +TS K R E+ RR+R+N++ L +L R KTDK ++L D + V
Sbjct: 269 EAHGSTSRKRSRAADMHNLSERRRRERINERMKTLQELLP--RCRKTDKVSMLEDVIEYV 326
Query: 56 TQLRGEAQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAPPSFLP 115
L+ + Q + + + + E ++ + +K MN PP F+P
Sbjct: 327 KSLQLQIQMMSMGHGMMPPMMHEGNTQQF------------MPHMAMGMKGMNRPPPFVP 374
Query: 116 TPTAL---PAAFAAQGQAHGNKLVPFISYPGVAMWQFMPPAAVDTSQDHV 162
P P A G ++ P + YP F A D S+ HV
Sbjct: 375 FPGKTFPRPGHMAGVGPSY-----PALRYP------FPDTQASDLSRVHV 413
>At4g36930 putative bHLH transcription factor (AtbHLH024) / SPATULA
(SPT)
Length = 373
Score = 39.7 bits (91), Expect = 8e-04
Identities = 27/76 (35%), Positives = 41/76 (53%), Gaps = 10/76 (13%)
Query: 2 RSESCAATSSKACR--------EKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVR 53
+S + +SSK CR EK RR R+N+K L S++ KTDKA++L +A+
Sbjct: 184 KSGPSSRSSSKRCRAAEVHNLSEKRRRSRINEKMKALQSLIPNSN--KTDKASMLDEAIE 241
Query: 54 MVTQLRGEAQKLKDAN 69
+ QL+ + Q L N
Sbjct: 242 YLKQLQLQVQMLTMRN 257
>At5g46830 putative transcription factor bHLH28
Length = 511
Score = 38.9 bits (89), Expect = 0.001
Identities = 19/65 (29%), Positives = 40/65 (61%), Gaps = 2/65 (3%)
Query: 16 EKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDANSGLQEK 75
E++RR++LN +F L +++ +K DK ++L DAV + +L+ +A+ ++ ++ +
Sbjct: 348 ERMRREKLNHRFYALRAVVP--NVSKMDKTSLLEDAVCYINELKSKAENVELEKHAIEIQ 405
Query: 76 IKELK 80
ELK
Sbjct: 406 FNELK 410
>At5g26350 putative protein
Length = 126
Score = 38.5 bits (88), Expect = 0.002
Identities = 18/46 (39%), Positives = 31/46 (67%)
Query: 62 AQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSM 107
A + K S E +K+ K EKN+L +EK++L+ EK++LE++ K +
Sbjct: 36 ADEEKPFGSATNEDMKKHKEEKNKLEEEKKKLEKEKKQLEEEKKQL 81
Score = 37.7 bits (86), Expect = 0.003
Identities = 17/48 (35%), Positives = 34/48 (70%)
Query: 61 EAQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMN 108
+ +K K+ + L+E+ K+L+ EK +L +EK++L+ EK++LE ++ N
Sbjct: 49 DMKKHKEEKNKLEEEKKKLEKEKKQLEEEKKQLEEEKKQLEFEVMGAN 96
Score = 27.3 bits (59), Expect = 4.0
Identities = 17/48 (35%), Positives = 28/48 (57%), Gaps = 1/48 (2%)
Query: 57 QLRGEAQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQL 104
+L E +KL+ L+E+ K+L+ EK +L E E+EK+ +QL
Sbjct: 59 KLEEEKKKLEKEKKQLEEEKKQLEEEKKQLEFEVMGAN-EREKVLRQL 105
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.130 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,604,205
Number of Sequences: 26719
Number of extensions: 144333
Number of successful extensions: 1644
Number of sequences better than 10.0: 321
Number of HSP's better than 10.0 without gapping: 165
Number of HSP's successfully gapped in prelim test: 157
Number of HSP's that attempted gapping in prelim test: 1110
Number of HSP's gapped (non-prelim): 621
length of query: 168
length of database: 11,318,596
effective HSP length: 92
effective length of query: 76
effective length of database: 8,860,448
effective search space: 673394048
effective search space used: 673394048
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC144502.7


