

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144484.10 - phase: 0
(589 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
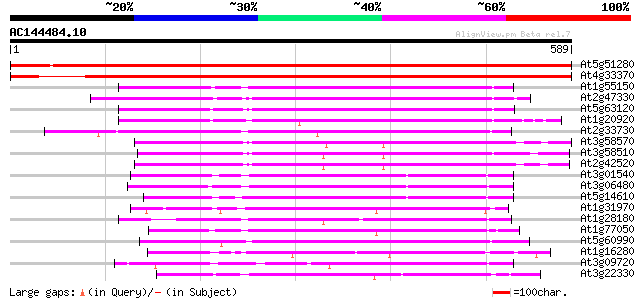
Score E
Sequences producing significant alignments: (bits) Value
At5g51280 DEAD-box protein abstrakt 1025 0.0
At4g33370 putative protein 884 0.0
At1g55150 unknown protein 304 1e-82
At2g47330 putative ATP-dependent RNA helicase 300 1e-81
At5g63120 ATP-dependent RNA helicase-like protein 300 1e-81
At1g20920 putative RNA helicase 294 1e-79
At2g33730 putative U5 small nuclear ribonucleoprotein, an RNA he... 293 1e-79
At3g58570 ATP-dependent RNA helicase-like protein 288 4e-78
At3g58510 ATP-dependent RNA helicase-like protein 288 6e-78
At2g42520 putative ATP-dependent RNA helicase 288 8e-78
At3g01540 AT3g01540/F4P13_9 276 2e-74
At3g06480 putative RNA helicase 271 1e-72
At5g14610 DRH1 DEAD box protein - like 254 9e-68
At1g31970 unknown protein 253 2e-67
At1g28180 hypothetical protein 228 9e-60
At1g77050 224 1e-58
At5g60990 replication protein A1 - like 214 8e-56
At1g16280 putative ATP-dependent RNA helicase 214 1e-55
At3g09720 RNA helicase like protein 209 3e-54
At3g22330 DEAD-Box RNA helicase like protein 207 2e-53
>At5g51280 DEAD-box protein abstrakt
Length = 591
Score = 1025 bits (2649), Expect = 0.0
Identities = 512/589 (86%), Positives = 547/589 (91%), Gaps = 2/589 (0%)
Query: 1 MEEEDDYEEYIPVAKRRAMEAQKILQRKGKATAAIQEDDSEKLKVVETKPSLLVKASQLK 60
MEE D Y EY+ VA+RRA+ AQKILQRKGKA+ +E D EKL E KPSLLV+A+QLK
Sbjct: 5 MEEADSYIEYVSVAERRAIAAQKILQRKGKASELEEEADKEKL--AEAKPSLLVQATQLK 62
Query: 61 KDQPEISVTEQIVQQEKEMIENLSDKKTLMSVRELAKGITYTEPLPTGWKPPLHIRRMSK 120
+D PE+S TEQI+ QEKEM+E+LSDKKTLMSVRELAKGITYTEPL TGWKPPLHIR+MS
Sbjct: 63 RDVPEVSATEQIILQEKEMMEHLSDKKTLMSVRELAKGITYTEPLLTGWKPPLHIRKMSS 122
Query: 121 KDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILS 180
K DLI+KQWHIIVNG++IPPPIKNFKDM+FP P+L LK KGIVQPTPIQVQGLPVIL+
Sbjct: 123 KQRDLIRKQWHIIVNGDDIPPPIKNFKDMKFPRPVLDTLKEKGIVQPTPIQVQGLPVILA 182
Query: 181 GRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELARQTYEVI 240
GRDMIGIAFTGSGKTLVFVLPMIM+A+QEE+MMPI GEGP GLI+CPSRELARQTYEV+
Sbjct: 183 GRDMIGIAFTGSGKTLVFVLPMIMIALQEEMMMPIAAGEGPIGLIVCPSRELARQTYEVV 242
Query: 241 EEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDN 300
E+F+ PL EAGYP LR LLCIGGIDMRSQLE+VK+GVHIVVATPGRLKDMLAKKKM+LD
Sbjct: 243 EQFVAPLVEAGYPPLRSLLCIGGIDMRSQLEVVKRGVHIVVATPGRLKDMLAKKKMSLDA 302
Query: 301 CRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALVKPIIVN 360
CRYLTLDEADRLVDLGFEDDIREVFDHFK+QRQTLLFSATMPTKIQ FARSALVKP+ VN
Sbjct: 303 CRYLTLDEADRLVDLGFEDDIREVFDHFKSQRQTLLFSATMPTKIQIFARSALVKPVTVN 362
Query: 361 VGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTPPPVLIFCENKADVDDIHEYLLLKGV 420
VGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKT PPVLIFCENKADVDDIHEYLLLKGV
Sbjct: 363 VGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTSPPVLIFCENKADVDDIHEYLLLKGV 422
Query: 421 EAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYV 480
EAVAIHGGKDQE+REYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYV
Sbjct: 423 EAVAIHGGKDQEDREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYV 482
Query: 481 HRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPMEDNEEIT 540
HRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAEL DPME+ E I
Sbjct: 483 HRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELNDPMEEAETIA 542
Query: 541 GISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDYFGSGGYRGEI 589
SGVKGCAYCGGLGHRIRDCPKLEHQKSVAI+N+RKDYFGSGGYRGEI
Sbjct: 543 NASGVKGCAYCGGLGHRIRDCPKLEHQKSVAISNSRKDYFGSGGYRGEI 591
>At4g33370 putative protein
Length = 542
Score = 884 bits (2285), Expect = 0.0
Identities = 444/589 (75%), Positives = 487/589 (82%), Gaps = 47/589 (7%)
Query: 1 MEEEDDYEEYIPVAKRRAMEAQKILQRKGKATAAIQEDDSEKLKVVETKPSLLVKASQLK 60
ME +D Y EY+PV +R A +K+++ GK
Sbjct: 1 MEVDDGYVEYVPVEERLAQMKRKVVEEPGKG----------------------------- 31
Query: 61 KDQPEISVTEQIVQQEKEMIENLSDKKTLMSVRELAKGITYTEPLPTGWKPPLHIRRMSK 120
M+E+LSDKK LMSV ELA+GITYTEPL T WKPPLH+R+MS
Sbjct: 32 ------------------MMEHLSDKKKLMSVGELARGITYTEPLSTWWKPPLHVRKMST 73
Query: 121 KDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILS 180
K DLI+KQWHI VNGE+IPPPIKNF DM+FP P+L+MLK KGI+ PTPIQVQGLPV+LS
Sbjct: 74 KQMDLIRKQWHITVNGEDIPPPIKNFMDMKFPSPLLRMLKDKGIMHPTPIQVQGLPVVLS 133
Query: 181 GRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELARQTYEVI 240
GRDMIGIAFTGSGKTLVFVLPMI++A+QEEIMMPI GEGP L+ICPSRELA+QTY+V+
Sbjct: 134 GRDMIGIAFTGSGKTLVFVLPMIILALQEEIMMPIAAGEGPIALVICPSRELAKQTYDVV 193
Query: 241 EEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDN 300
E+F+ L E GYP LR LLCIGG+DMRSQL++VKKGVHIVVATPGRLKD+LAKKKM+LD
Sbjct: 194 EQFVASLVEDGYPRLRSLLCIGGVDMRSQLDVVKKGVHIVVATPGRLKDILAKKKMSLDA 253
Query: 301 CRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALVKPIIVN 360
CR LTLDEADRLVDLGFEDDIR VFDHFK+QRQTLLFSATMP KIQ FA SALVKP+ VN
Sbjct: 254 CRLLTLDEADRLVDLGFEDDIRHVFDHFKSQRQTLLFSATMPAKIQIFATSALVKPVTVN 313
Query: 361 VGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTPPPVLIFCENKADVDDIHEYLLLKGV 420
VGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKT PPVLIFCENKADVDDIHEYLLLKGV
Sbjct: 314 VGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTTPPVLIFCENKADVDDIHEYLLLKGV 373
Query: 421 EAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYV 480
EAVAIHGGKDQE+R+YAIS FKAGKKDVLVATDVASKGLDFPDIQHVINYDMP EIENYV
Sbjct: 374 EAVAIHGGKDQEDRDYAISLFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPGEIENYV 433
Query: 481 HRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPMEDNEEIT 540
HRIGRTGRCGKTGIATTFINKNQSE TLLDLKHLLQEAKQRIPPVLAEL PME+ E I
Sbjct: 434 HRIGRTGRCGKTGIATTFINKNQSEITLLDLKHLLQEAKQRIPPVLAELNGPMEETETIA 493
Query: 541 GISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDYFGSGGYRGEI 589
SGVKGCAYCGGLGHRI CPK EHQKSVAI+++RKD+FGS GYRGE+
Sbjct: 494 NASGVKGCAYCGGLGHRILQCPKFEHQKSVAISSSRKDHFGSDGYRGEV 542
>At1g55150 unknown protein
Length = 501
Score = 304 bits (778), Expect = 1e-82
Identities = 163/418 (38%), Positives = 251/418 (59%), Gaps = 13/418 (3%)
Query: 115 IRRMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQG 174
+ M+ + + +K I V G++IP P+K+F+D+ FPD +L+ +K G +PTPIQ QG
Sbjct: 70 VAAMTDTEVEEYRKLREITVEGKDIPKPVKSFRDVGFPDYVLEEVKKAGFTEPTPIQSQG 129
Query: 175 LPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELAR 234
P+ + GRD+IGIA TGSGKTL ++LP I+ + ++ G+GP L++ P+RELA
Sbjct: 130 WPMAMKGRDLIGIAETGSGKTLSYLLPAIVHVNAQPML---AHGDGPIVLVLAPTRELAV 186
Query: 235 QTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKK 294
Q + +F +++ GG+ Q+ ++KGV IV+ATPGRL DM+
Sbjct: 187 QIQQEASKF------GSSSKIKTTCIYGGVPKGPQVRDLQKGVEIVIATPGRLIDMMESN 240
Query: 295 KMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALV 354
NL YL LDEADR++D+GF+ IR++ H + RQTL +SAT P +++ ++ L
Sbjct: 241 NTNLRRVTYLVLDEADRMLDMGFDPQIRKIVSHIRPDRQTLYWSATWPKEVEQLSKKFLY 300
Query: 355 KPIIVNVGRAGA-ANLDVIQEVEYVKQEAKIVYLLECLQKTPPP--VLIFCENKADVDDI 411
P V +G + AN + Q V+ + + K L++ L+ +L+F + K D I
Sbjct: 301 NPYKVIIGSSDLKANRAIRQIVDVISESQKYNKLVKLLEDIMDGSRILVFLDTKKGCDQI 360
Query: 412 HEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYD 471
L + G A++IHG K Q ER++ +S F++GK ++ ATDVA++GLD D+++VINYD
Sbjct: 361 TRQLRMDGWPALSIHGDKSQAERDWVLSEFRSGKSPIMTATDVAARGLDVKDVKYVINYD 420
Query: 472 MPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAEL 529
P +E+YVHRIGRTGR G G A TF + +L ++LQEA Q++ P LA +
Sbjct: 421 FPGSLEDYVHRIGRTGRAGAKGTAYTFFTVANARFA-KELTNILQEAGQKVSPELASM 477
>At2g47330 putative ATP-dependent RNA helicase
Length = 760
Score = 300 bits (769), Expect = 1e-81
Identities = 187/468 (39%), Positives = 264/468 (55%), Gaps = 17/468 (3%)
Query: 85 DKKTLMSVRELAKGITYTEPLPTGWKPPLH-IRRMSKKDCDLIQKQWHIIVNGEEIPPPI 143
DK+ + + L EP+ + L I M++++ +++ I V+G ++ P+
Sbjct: 168 DKRKIEPITALDHSSIDYEPINKDFYEELESISGMTEQETTDYRQRLGIRVSGFDVHRPV 227
Query: 144 KNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKTLVFVLPMI 203
K F+D F I+ +K + +PT IQ Q LP++LSGRD+IGIA TGSGKT FVLPMI
Sbjct: 228 KTFEDCGFSSQIMSAIKKQAYEKPTAIQCQALPIVLSGRDVIGIAKTGSGKTAAFVLPMI 287
Query: 204 MMAMQEEIMMPIVPGEGPFGLIICPSRELARQTYEVIEEFLLPLKEAGYPELRPLLCIGG 263
+ M + + EGP G+I P+RELA Q + ++F K G LR GG
Sbjct: 288 VHIMDQPELQR---DEGPIGVICAPTRELAHQIFLEAKKFS---KAYG---LRVSAVYGG 338
Query: 264 IDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDNCRYLTLDEADRLVDLGFEDDIRE 323
+ Q + +K G IVVATPGRL DML K + + YL LDEADR+ DLGFE +R
Sbjct: 339 MSKHEQFKELKAGCEIVVATPGRLIDMLKMKALTMMRASYLVLDEADRMFDLGFEPQVRS 398
Query: 324 VFDHFKAQRQTLLFSATMPTKIQNFARSALVKPIIVNVGRAGAANLDVIQEVEYVKQEA- 382
+ + RQTLLFSATMP K++ AR L PI V VG G AN D+ Q V + +A
Sbjct: 399 IVGQIRPDRQTLLFSATMPWKVEKLAREILSDPIRVTVGEVGMANEDITQVVNVIPSDAE 458
Query: 383 KIVYLLECL--QKTPPPVLIFCENKADVDDIHEYLLLKGVEAVAIHGGKDQEEREYAISS 440
K+ +LLE L VL+F KA VD+I L L + A+HG KDQ R +
Sbjct: 459 KLPWLLEKLPGMIDEGDVLVFASKKATVDEIEAQLTLNSFKVAALHGDKDQASRMETLQK 518
Query: 441 FKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYVHRIGRTGRCG-KTGIATTFI 499
FK+G VL+ATDVA++GLD ++ V+NYD+ +++ +VHRIGRTGR G + G+A T +
Sbjct: 519 FKSGVHHVLIATDVAARGLDIKSLKTVVNYDIAKDMDMHVHRIGRTGRAGDRDGVAYTLV 578
Query: 500 NKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPMEDNEEITGISGVKG 547
+ ++ +L + L A Q +PP L +L M+D + G KG
Sbjct: 579 TQREARFA-GELVNSLVAAGQNVPPELTDLA--MKDGRFKSKRDGRKG 623
>At5g63120 ATP-dependent RNA helicase-like protein
Length = 591
Score = 300 bits (768), Expect = 1e-81
Identities = 172/419 (41%), Positives = 248/419 (59%), Gaps = 13/419 (3%)
Query: 115 IRRMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQG 174
++ M+++D + + + I V G ++P P+K F+D FPD IL+ + G +PTPIQ QG
Sbjct: 136 VQAMTEQDVAMYRTERDISVEGRDVPKPMKMFQDANFPDNILEAIAKLGFTEPTPIQAQG 195
Query: 175 LPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELAR 234
P+ L GRD+IGIA TGSGKTL ++LP ++ + + +GP LI+ P+RELA
Sbjct: 196 WPMALKGRDLIGIAETGSGKTLAYLLPALVHVSAQP---RLGQDDGPIVLILAPTRELAV 252
Query: 235 QTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKK 294
Q E +F L +G +R GG Q+ +++GV IV+ATPGRL DML +
Sbjct: 253 QIQEESRKFGL---RSG---VRSTCIYGGAPKGPQIRDLRRGVEIVIATPGRLIDMLECQ 306
Query: 295 KMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALV 354
NL YL LDEADR++D+GFE IR++ + RQTLL+SAT P +++ AR L
Sbjct: 307 HTNLKRVTYLVLDEADRMLDMGFEPQIRKIVSQIRPDRQTLLWSATWPREVETLARQFLR 366
Query: 355 KPIIVNVGRAGA-ANLDVIQEVEYVKQEAKIVYLLECLQKTPP--PVLIFCENKADVDDI 411
P +G AN + Q +E V K LL L++ +LIF E K D +
Sbjct: 367 DPYKAIIGSTDLKANQSINQVIEIVPTPEKYNRLLTLLKQLMDGSKILIFVETKRGCDQV 426
Query: 412 HEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYD 471
L + G A+AIHG K Q ER+ ++ FK+G+ ++ ATDVA++GLD DI+ V+NYD
Sbjct: 427 TRQLRMDGWPALAIHGDKTQSERDRVLAEFKSGRSPIMTATDVAARGLDVKDIKCVVNYD 486
Query: 472 MPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELV 530
P +E+Y+HRIGRTGR G G+A TF + ++ +L +LQEA Q +PP L+ LV
Sbjct: 487 FPNTLEDYIHRIGRTGRAGAKGMAFTFFTHDNAKFA-RELVKILQEAGQVVPPTLSALV 544
>At1g20920 putative RNA helicase
Length = 1166
Score = 294 bits (752), Expect = 1e-79
Identities = 170/470 (36%), Positives = 267/470 (56%), Gaps = 19/470 (4%)
Query: 115 IRRMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQG 174
I RM++++ + +K+ + V+G+++P PIK + IL +K +P PIQ Q
Sbjct: 500 ISRMTQEEVNTYRKELELKVHGKDVPRPIKFWHQTGLTSKILDTMKKLNYEKPMPIQTQA 559
Query: 175 LPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELAR 234
LP+I+SGRD IG+A TGSGKTL FVLPM+ + P+ G+GP GL++ P+REL +
Sbjct: 560 LPIIMSGRDCIGVAKTGSGKTLGFVLPMLRHIKDQP---PVEAGDGPIGLVMAPTRELVQ 616
Query: 235 QTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKK 294
Q + I +F PL +R + GG + Q+ +K+G IVV TPGR+ D+L
Sbjct: 617 QIHSDIRKFSKPLG------IRCVPVYGGSGVAQQISELKRGTEIVVCTPGRMIDILCTS 670
Query: 295 KMNLDNCR---YLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARS 351
+ N R +L +DEADR+ D+GFE I + + + +RQT+LFSAT P +++ AR
Sbjct: 671 SGKITNLRRVTFLVMDEADRMFDMGFEPQITRIIQNIRPERQTVLFSATFPRQVETLARK 730
Query: 352 ALVKPIIVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTPPP--VLIFCENKADVD 409
L KP+ + VG N D+ Q VE + + + LLE L + +L+F +++ D
Sbjct: 731 VLNKPVEIQVGGRSVVNKDITQLVEVRPESDRFLRLLELLGEWSEKGKILVFVQSQEKCD 790
Query: 410 DIHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVIN 469
++ ++ +++HGGKDQ +RE IS FK ++L+AT VA++GLD +++ V+N
Sbjct: 791 ALYRDMIKSSYPCLSLHGGKDQTDRESTISDFKNDVCNLLIATSVAARGLDVKELELVVN 850
Query: 470 YDMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAEL 529
+D P E+YVHR+GRTGR G+ G A TFI+++ ++ DL L+ ++Q +P L L
Sbjct: 851 FDAPNHYEDYVHRVGRTGRAGRKGCAVTFISEDDAKYA-PDLVKALELSEQPVPDDLKAL 909
Query: 530 VDPMEDNEEITGISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDY 579
D + GI G Y GG G + + + E + A K+Y
Sbjct: 910 ADGFMVKVK-QGIEQAHGTGY-GGSGFKFNE--EEEEVRKAAKKAQAKEY 955
>At2g33730 putative U5 small nuclear ribonucleoprotein, an RNA
helicase
Length = 733
Score = 293 bits (751), Expect = 1e-79
Identities = 178/522 (34%), Positives = 274/522 (52%), Gaps = 39/522 (7%)
Query: 37 EDDSEKLKVVETKP--SLLVKASQLKKDQPEISVTEQIVQQEKEMIENLSDKKTLMS--- 91
ED S + V+ P + L+ + +Q + EKEM + + K ++
Sbjct: 194 EDTSRDMNVLYQNPHEAQLLFGRGFRAGMDRREQKKQAAKHEKEMRDEIRKKDGIVEKPE 253
Query: 92 ------VRELAKGI--TYTEPLPTGWKPPLHIRRMSKKDCDLIQKQWHIIVNGEEIPPPI 143
VRE A ++ + W + M+++D + ++ ++I G IP P+
Sbjct: 254 EAAAQRVREEAADTYDSFDMRVDRHWSDK-RLEEMTERDWRIFREDFNISYKGSRIPRPM 312
Query: 144 KNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKTLVFVLPMI 203
+++++ + +LK ++ G +P+PIQ+ +P+ L RD+IGIA TGSGKT FVLPM+
Sbjct: 313 RSWEESKLTSELLKAVERAGYKKPSPIQMAAIPLGLQQRDVIGIAETGSGKTAAFVLPML 372
Query: 204 MMAMQEEIMMPIVPGEGPFGLIICPSRELARQTYEVIEEFLLPLKEAGYPELRPLLCIGG 263
+ M EGP+ +++ P+RELA+Q E +F A Y R +GG
Sbjct: 373 AYISRLPPMSEENETEGPYAVVMAPTRELAQQIEEETVKF------AHYLGFRVTSIVGG 426
Query: 264 IDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDNCRYLTLDEADRLVDLGFEDDI-- 321
+ Q + +G IV+ATPGRL D L ++ L+ C Y+ LDEADR++D+GFE +
Sbjct: 427 QSIEEQGLKITQGCEIVIATPGRLIDCLERRYAVLNQCNYVVLDEADRMIDMGFEPQVAG 486
Query: 322 ---------------REVFDHFKAQRQTLLFSATMPTKIQNFARSALVKPIIVNVGRAGA 366
E D K R T +FSATMP ++ AR L P++V +G AG
Sbjct: 487 VLDAMPSSNLKPENEEEELDEKKIYRTTYMFSATMPPGVERLARKYLRNPVVVTIGTAGK 546
Query: 367 ANLDVIQEVEYVKQEAKIVYLLECLQKT-PPPVLIFCENKADVDDIHEYLLLKGVEAVAI 425
+ Q V +K+ K L + L + ++F K + D I + L G +
Sbjct: 547 TTDLISQHVIMMKESEKFFRLQKLLDELGEKTAIVFVNTKKNCDSIAKNLDKAGYRVTTL 606
Query: 426 HGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYVHRIGR 485
HGGK QE+RE ++ F+A + +VLVATDV +G+D PD+ HVINYDMP IE Y HRIGR
Sbjct: 607 HGGKSQEQREISLEGFRAKRYNVLVATDVVGRGIDIPDVAHVINYDMPKHIEMYTHRIGR 666
Query: 486 TGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLA 527
TGR GK+G+AT+F+ + +E DLK +L ++ +PP LA
Sbjct: 667 TGRAGKSGVATSFLTLHDTE-VFYDLKQMLVQSNSAVPPELA 707
>At3g58570 ATP-dependent RNA helicase-like protein
Length = 646
Score = 288 bits (738), Expect = 4e-78
Identities = 171/472 (36%), Positives = 260/472 (54%), Gaps = 34/472 (7%)
Query: 132 IIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTG 191
I +G+ +PPP+ F ++ + + ++ V+PTP+Q +P++ +GRD++ A TG
Sbjct: 134 IETSGDNVPPPVNTFAEIDLGEALNLNIQRCKYVKPTPVQRNAIPILAAGRDLMACAQTG 193
Query: 192 SGKTLVFVLPMIMMAMQEE-IMMPI-VPGEGPFGLIICPSRELARQTYEVIEEFLLPLKE 249
SGKT F P+I M+++ I P V G P +I+ P+RELA Q ++ +F +
Sbjct: 194 SGKTAAFCFPIISGIMKDQHIERPRGVRGVYPLAVILSPTRELACQIHDEARKFSY---Q 250
Query: 250 AGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDNCRYLTLDEA 309
G ++ ++ GG + Q+ +++GV I+VATPGRL D+L + +++L R+L LDEA
Sbjct: 251 TG---VKVVVAYGGTPVNQQIRELERGVDILVATPGRLNDLLERGRVSLQMVRFLALDEA 307
Query: 310 DRLVDLGFEDDIREVFDHFKAQ----RQTLLFSATMPTKIQNFARSALVKPIIVNVGRAG 365
DR++D+GFE IR++ RQT+LFSAT P +IQ A L I + VGR G
Sbjct: 308 DRMLDMGFEPQIRKIVQQMDMPPPGVRQTMLFSATFPREIQRLASDFLSNYIFLAVGRVG 367
Query: 366 AANLDVIQEVEYVKQEAKIVYLLECL--------QKTPPPVLIFCENKADVDDIHEYLLL 417
++ ++Q VE+V K +L++ L Q L+F E K D + +L +
Sbjct: 368 SSTDLIVQRVEFVHDSDKRSHLMDLLHAQRENGNQGKQALTLVFVETKKGADSLENWLCI 427
Query: 418 KGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIE 477
G A IHG + Q+ERE A+ SFK G+ +LVATDVA++GLD P + HV+N+D+P +I+
Sbjct: 428 NGFPATTIHGDRSQQEREVALRSFKTGRTPILVATDVAARGLDIPHVAHVVNFDLPNDID 487
Query: 478 NYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPMEDNE 537
+YVHRIGRTGR G +G+AT F N N + T L L+QEA Q +P L
Sbjct: 488 DYVHRIGRTGRAGNSGLATAFFNDNNT-TMAKPLAELMQEANQEVPDWLTRYASR----- 541
Query: 538 EITGISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDYFGSGGYRGEI 589
G K G G R ++S + DY+G GG G +
Sbjct: 542 --ASFGGGKNRRSGGRFGGRD------FRRESFSRGGGGADYYGGGGGYGGV 585
>At3g58510 ATP-dependent RNA helicase-like protein
Length = 612
Score = 288 bits (737), Expect = 6e-78
Identities = 166/465 (35%), Positives = 258/465 (54%), Gaps = 26/465 (5%)
Query: 135 NGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGK 194
+G ++PPP+ F D+ D + ++ V+PTP+Q +P++L+ RD++ A TGSGK
Sbjct: 142 SGGDVPPPVNTFADIDLGDALNLNIRRCKYVRPTPVQRHAIPILLAERDLMACAQTGSGK 201
Query: 195 TLVFVLPMIMMAMQEEIMMPIVPGEG--PFGLIICPSRELARQTYEVIEEFLLPLKEAGY 252
T F P+I M+++ + PF +I+ P+RELA Q ++ ++F + G
Sbjct: 202 TAAFCFPIISGIMKDQHVERPRGSRAVYPFAVILSPTRELACQIHDEAKKFSY---QTG- 257
Query: 253 PELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDNCRYLTLDEADRL 312
++ ++ GG + QL +++G I+VATPGRL D+L + ++++ R+L LDEADR+
Sbjct: 258 --VKVVVAYGGTPIHQQLRELERGCDILVATPGRLNDLLERARVSMQMIRFLALDEADRM 315
Query: 313 VDLGFEDDIREVFDHF----KAQRQTLLFSATMPTKIQNFARSALVKPIIVNVGRAGAAN 368
+D+GFE IR++ + + RQT+LFSAT P++IQ A + I + VGR G++
Sbjct: 316 LDMGFEPQIRKIVEQMDMPPRGVRQTMLFSATFPSQIQRLAADFMSNYIFLAVGRVGSST 375
Query: 369 LDVIQEVEYVKQEAKIVYLLECL------QKTPPPVLIFCENKADVDDIHEYLLLKGVEA 422
+ Q VE+V++ K +L++ L Q L+F E K D + +L + A
Sbjct: 376 DLITQRVEFVQESDKRSHLMDLLHAQRETQDKQSLTLVFVETKRGADTLENWLCMNEFPA 435
Query: 423 VAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYVHR 482
+IHG + Q+ERE A+ SFK G+ +LVATDVA++GLD P + HV+N+D+P +I++YVHR
Sbjct: 436 TSIHGDRTQQEREVALRSFKTGRTPILVATDVAARGLDIPHVAHVVNFDLPNDIDDYVHR 495
Query: 483 IGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPMEDNEEITGI 542
IGRTGR GK+GIAT F N+N ++ L L+QEA Q +P L
Sbjct: 496 IGRTGRAGKSGIATAFFNENNAQLA-RSLAELMQEANQEVPEWLTRYASRASFGGGKKRS 554
Query: 543 SGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDYFGSGGYRG 587
G G G R DY+G GGY G
Sbjct: 555 GGRFG-------GRDFRREGSYSRGGGGGGGGGGSDYYGGGGYGG 592
>At2g42520 putative ATP-dependent RNA helicase
Length = 633
Score = 288 bits (736), Expect = 8e-78
Identities = 166/470 (35%), Positives = 262/470 (55%), Gaps = 35/470 (7%)
Query: 132 IIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTG 191
I +G+ +PPP+ F ++ + + ++ V+PTP+Q +P++L GRD++ A TG
Sbjct: 147 IETSGDNVPPPVNTFAEIDLGEALNLNIRRCKYVKPTPVQRHAIPILLEGRDLMACAQTG 206
Query: 192 SGKTLVFVLPMIMMAMQEEIMMPIVPGEG--PFGLIICPSRELARQTYEVIEEFLLPLKE 249
SGKT F P+I M+++ + P +I+ P+RELA Q ++ ++F +
Sbjct: 207 SGKTAAFCFPIISGIMKDQHVQRPRGSRTVYPLAVILSPTRELASQIHDEAKKFSY---Q 263
Query: 250 AGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDNCRYLTLDEA 309
G ++ ++ GG + QL +++GV I+VATPGRL D+L + ++++ R+L LDEA
Sbjct: 264 TG---VKVVVAYGGTPINQQLRELERGVDILVATPGRLNDLLERARVSMQMIRFLALDEA 320
Query: 310 DRLVDLGFEDDIREVFDHF----KAQRQTLLFSATMPTKIQNFARSALVKPIIVNVGRAG 365
DR++D+GFE IR++ + + RQTLLFSAT P +IQ A L I + VGR G
Sbjct: 321 DRMLDMGFEPQIRKIVEQMDMPPRGVRQTLLFSATFPREIQRLAADFLANYIFLAVGRVG 380
Query: 366 AANLDVIQEVEYVKQEAKIVYLLECL--------QKTPPPVLIFCENKADVDDIHEYLLL 417
++ ++Q VE+V K +L++ L Q L+F E K D + +L +
Sbjct: 381 SSTDLIVQRVEFVLDSDKRSHLMDLLHAQRENGIQGKQALTLVFVETKRGADSLENWLCI 440
Query: 418 KGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIE 477
G A +IHG + Q+ERE A+ +FK+G+ +LVATDVA++GLD P + HV+N+D+P +I+
Sbjct: 441 NGFPATSIHGDRTQQEREVALKAFKSGRTPILVATDVAARGLDIPHVAHVVNFDLPNDID 500
Query: 478 NYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPMEDNE 537
+YVHRIGRTGR GK+G+AT F N + + L L+QEA Q +P L
Sbjct: 501 DYVHRIGRTGRAGKSGLATAFFNDGNT-SLARPLAELMQEANQEVPEWLTRYASR----- 554
Query: 538 EITGISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDYFGSGGYRG 587
+ G K G G R + ++ + + R Y G GG G
Sbjct: 555 --SSFGGGKNRRSGGRFGGR-------DFRREGSFGSGRGGYGGGGGGYG 595
>At3g01540 AT3g01540/F4P13_9
Length = 619
Score = 276 bits (706), Expect = 2e-74
Identities = 162/409 (39%), Positives = 236/409 (57%), Gaps = 22/409 (5%)
Query: 128 KQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGI 187
++ I V+G ++PPP+ +F+ FP +L+ + + G PTPIQ Q P+ + GRD++ I
Sbjct: 142 RRHEITVSGGQVPPPLMSFEATGFPPELLREVLSAGFSAPTPIQAQSWPIAMQGRDIVAI 201
Query: 188 AFTGSGKTLVFVLP--MIMMAMQEEIMMPIVPGEGPFGLIICPSRELARQTYEVIEEFLL 245
A TGSGKTL +++P + + ++ + M GP L++ P+RELA Q E +F
Sbjct: 202 AKTGSGKTLGYLIPGFLHLQRIRNDSRM------GPTILVLSPTRELATQIQEEAVKF-- 253
Query: 246 PLKEAGYPELRPLLCI-GGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDNCRYL 304
G C+ GG QL +++G IVVATPGRL D+L ++++L YL
Sbjct: 254 -----GRSSRISCTCLYGGAPKGPQLRDLERGADIVVATPGRLNDILEMRRISLRQISYL 308
Query: 305 TLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALVKPIIVNVGRA 364
LDEADR++D+GFE IR++ +RQTL+++AT P ++ A LV P VN+G
Sbjct: 309 VLDEADRMLDMGFEPQIRKIVKEIPTKRQTLMYTATWPKGVRKIAADLLVNPAQVNIGNV 368
Query: 365 G--AANLDVIQEVEYVKQEAKIVYLLECLQKTPP--PVLIFCENKADVDDIHEYLLLKGV 420
AN + Q +E V K L + L+ P V+IFC K D + L +
Sbjct: 369 DELVANKSITQHIEVVAPMEKQRRLEQILRSQEPGSKVIIFCSTKRMCDQLTRNLT-RQF 427
Query: 421 EAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYV 480
A AIHG K Q ER+ ++ F++G+ VLVATDVA++GLD DI+ V+NYD P +E+YV
Sbjct: 428 GAAAIHGDKSQPERDNVLNQFRSGRTPVLVATDVAARGLDVKDIRAVVNYDFPNGVEDYV 487
Query: 481 HRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAEL 529
HRIGRTGR G TG A TF +Q DL +L+ A QR+PP + E+
Sbjct: 488 HRIGRTGRAGATGQAFTFFG-DQDSKHASDLIKILEGANQRVPPQIREM 535
>At3g06480 putative RNA helicase
Length = 1088
Score = 271 bits (692), Expect = 1e-72
Identities = 164/411 (39%), Positives = 233/411 (55%), Gaps = 18/411 (4%)
Query: 124 DLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRD 183
++ +KQ + GE IP P F+ P IL+ L + G PTPIQ Q P+ L RD
Sbjct: 415 EIYRKQHEVTTTGENIPAPYITFESSGLPPEILRELLSAGFPSPTPIQAQTWPIALQSRD 474
Query: 184 MIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELARQTYEVIEEF 243
++ IA TGSGKTL +++P ++ GP LI+ P+RELA Q + F
Sbjct: 475 IVAIAKTGSGKTLGYLIPAFILLRH----CRNDSRNGPTVLILAPTRELATQIQDEALRF 530
Query: 244 LLPLKEAGYPELRPLLCI-GGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDNCR 302
G C+ GG QL+ +++G IVVATPGRL D+L K ++
Sbjct: 531 -------GRSSRISCTCLYGGAPKGPQLKELERGADIVVATPGRLNDILEMKMIDFQQVS 583
Query: 303 YLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALVKPIIVNVG 362
L LDEADR++D+GFE IR++ + +RQTL+++AT P +++ A LV P+ VN+G
Sbjct: 584 LLVLDEADRMLDMGFEPQIRKIVNEIPPRRQTLMYTATWPKEVRKIASDLLVNPVQVNIG 643
Query: 363 RAG--AANLDVIQEVEYVKQEAKIVYLLECL--QKTPPPVLIFCENKADVDDIHEYLLLK 418
R AAN + Q VE V Q K L + L Q+ V+IFC K D + + +
Sbjct: 644 RVDELAANKAITQYVEVVPQMEKERRLEQILRSQERGSKVIIFCSTKRLCDHLARSVG-R 702
Query: 419 GVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIEN 478
AV IHG K Q ER++ ++ F++GK VL+ATDVA++GLD DI+ VINYD P +E+
Sbjct: 703 HFGAVVIHGDKTQGERDWVLNQFRSGKSCVLIATDVAARGLDIKDIRVVINYDFPTGVED 762
Query: 479 YVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAEL 529
YVHRIGRTGR G TG+A TF + Q DL +L+ A Q++PP + ++
Sbjct: 763 YVHRIGRTGRAGATGVAFTFFTE-QDWKYAPDLIKVLEGANQQVPPQVRDI 812
>At5g14610 DRH1 DEAD box protein - like
Length = 713
Score = 254 bits (649), Expect = 9e-68
Identities = 152/395 (38%), Positives = 223/395 (55%), Gaps = 20/395 (5%)
Query: 141 PPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKTLVFVL 200
PP F +M+ + G P+PIQ Q P+ + RD++ IA TGSGKTL +++
Sbjct: 226 PPAAGFNSYLVLPANGRMVYSAGFSAPSPIQAQSWPIAMQNRDIVAIAKTGSGKTLGYLI 285
Query: 201 P--MIMMAMQEEIMMPIVPGEGPFGLIICPSRELARQTYEVIEEFLLPLKEAGYPELRPL 258
P M + + + M GP L++ P+RELA Q + LK ++
Sbjct: 286 PGFMHLQRIHNDSRM------GPTILVLSPTRELATQIQ------VEALKFGKSSKISCA 333
Query: 259 LCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDNCRYLTLDEADRLVDLGFE 318
GG QL+ +++GV IVVATPGRL D+L K+++L YL LDEADR++D+GFE
Sbjct: 334 CLYGGAPKGPQLKEIERGVDIVVATPGRLNDILEMKRISLHQVSYLVLDEADRMLDMGFE 393
Query: 319 DDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALVKPIIVNVGRAG--AANLDVIQEVE 376
IR++ + +RQTL+++AT P +++ A LV P VN+G AN + Q +E
Sbjct: 394 PQIRKIVNEVPTKRQTLMYTATWPKEVRKIAADLLVNPAQVNIGNVDELVANKSITQTIE 453
Query: 377 YVKQEAKIVYLLECLQKTPP--PVLIFCENKADVDDIHEYLLLKGVEAVAIHGGKDQEER 434
+ K L + L+ P ++IFC K D + L + A AIHG K Q ER
Sbjct: 454 VLAPMEKHSRLEQILRSQEPGSKIIIFCSTKRMCDQLARNLT-RTFGAAAIHGDKSQAER 512
Query: 435 EYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYVHRIGRTGRCGKTGI 494
+ ++ F++G+ VLVATDVA++GLD DI+ V+NYD P +E+YVHRIGRTGR G TG+
Sbjct: 513 DDVLNQFRSGRTPVLVATDVAARGLDVKDIRVVVNYDFPNGVEDYVHRIGRTGRAGATGL 572
Query: 495 ATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAEL 529
A TF +Q DL +L+ A Q++PP + E+
Sbjct: 573 AYTFFG-DQDAKHASDLIKILEGANQKVPPQVREM 606
>At1g31970 unknown protein
Length = 537
Score = 253 bits (647), Expect = 2e-67
Identities = 160/415 (38%), Positives = 241/415 (57%), Gaps = 36/415 (8%)
Query: 128 KQWHIIVNGEEIPPP----IKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRD 183
+Q ++V G+ + +K F + P+ +L KT +P+PIQ P +L GRD
Sbjct: 96 EQQKVVVTGKGVEEAKYAALKTFAESNLPENVLDCCKT--FEKPSPIQSHTWPFLLDGRD 153
Query: 184 MIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEG-----PFGLIICPSRELARQTYE 238
+IGIA TGSGKTL F +P IM +++ + G G P L++ P+RELA Q +
Sbjct: 154 LIGIAKTGSGKTLAFGIPAIMHVLKKNKKI----GGGSKKVNPTCLVLSPTRELAVQISD 209
Query: 239 VIEEFLLPLKEAGYP-ELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMN 297
V L+EAG P L+ + GG Q+ ++ GV IV+ TPGRL+D++ +
Sbjct: 210 V-------LREAGEPCGLKSICVYGGSSKGPQISAIRSGVDIVIGTPGRLRDLIESNVLR 262
Query: 298 LDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSAL-VKP 356
L + ++ LDEADR++D+GFE+ +R + + RQ ++FSAT P + A+ + P
Sbjct: 263 LSDVSFVVLDEADRMLDMGFEEPVRFILSNTNKVRQMVMFSATWPLDVHKLAQEFMDPNP 322
Query: 357 IIVNVGRAG-AANLDVIQEVEYVKQEAK---IVYLLECLQKTPPP-VLIFCENKADVDDI 411
I V +G AAN DV+Q +E + + A+ ++ LLE K+ VL+F K + + +
Sbjct: 323 IKVIIGSVDLAANHDVMQIIEVLDERARDQRLIALLEKYHKSQKNRVLVFALYKVEAERL 382
Query: 412 HEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYD 471
+L +G +AV+IHG K Q ER ++S FK G +LVATDVA++GLD PD++ VINY
Sbjct: 383 ERFLQQRGWKAVSIHGNKAQSERTRSLSLFKEGSCPLLVATDVAARGLDIPDVEVVINYT 442
Query: 472 MPAEIENYVHRIGRTGRCGKTGIATTF---INKNQSETTLLDLKHLLQEAKQRIP 523
P E+YVHRIGRTGR GK G+A TF +NK + +L ++L+EA Q +P
Sbjct: 443 FPLTTEDYVHRIGRTGRAGKKGVAHTFFTPLNKGLAG----ELVNVLREAGQVVP 493
>At1g28180 hypothetical protein
Length = 622
Score = 228 bits (580), Expect = 9e-60
Identities = 145/433 (33%), Positives = 217/433 (49%), Gaps = 55/433 (12%)
Query: 115 IRRMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQG 174
+ M+++D + ++ ++I G +IP P++N+++
Sbjct: 206 LEEMNERDWRIFKEDFNISYRGSKIPHPMRNWEET------------------------- 240
Query: 175 LPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELAR 234
+P+ L RD+IGI+ TGSGKT FVLPM+ + M EGP+ L++ P+RELA
Sbjct: 241 IPLGLEQRDVIGISATGSGKTAAFVLPMLAYISRLPPMREENQTEGPYALVMVPTRELAH 300
Query: 235 QTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKK 294
Q E +F + Y + + G + Q + +G IV+ATPGRL D L ++
Sbjct: 301 QIEEETVKF------SRYLGFKAVSITGWESIEKQALKLSQGCEIVIATPGRLLDCLERR 354
Query: 295 KMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHF-----------------KAQRQTLLF 337
+ L+ C YL LDEADR++D+ FE + EV D K R T +F
Sbjct: 355 YVVLNQCNYLVLDEADRMIDMDFEPQVSEVLDVMPCSNLKPEKEDEELEEKKIYRTTYMF 414
Query: 338 SATMPTKIQNFARSALVKPIIVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKT--P 395
SATM ++ AR L P++V +G + Q+V K+ K L + +
Sbjct: 415 SATMLLSVERLARKFLRNPVVVTIGETTKF---ITQQVIMTKESDKFSRLKKLIDDLGDD 471
Query: 396 PPVLIFCENKADVDDIHEYLLLKG-VEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDV 454
++F + VD I + L G +H GK QE+R+Y++ FK + +VLV TDV
Sbjct: 472 KTAIVFVNTRNKVDYIVKNLEKVGRCRVTTLHAGKSQEQRDYSLEEFKKKRFNVLVTTDV 531
Query: 455 ASKGLDFPDIQHVINYDMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHL 514
+GLD D+ VINYDMP ++ Y HRIGRTGR GKTG+ATTF+ + + LK
Sbjct: 532 LGRGLDILDLAQVINYDMPNTMDLYTHRIGRTGRAGKTGVATTFLTL-EDKDVFYGLKQK 590
Query: 515 LQEAKQRIPPVLA 527
L E +PP LA
Sbjct: 591 LNECNSLVPPELA 603
>At1g77050
Length = 513
Score = 224 bits (570), Expect = 1e-58
Identities = 134/394 (34%), Positives = 213/394 (54%), Gaps = 18/394 (4%)
Query: 146 FKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKTLVFVLPMIMM 205
F+ + + +K KG PTPIQ + +P+ILSG D++ +A TGSGKT F++PM+
Sbjct: 30 FESLNLGPNVFNAIKKKGYKVPTPIQRKTMPLILSGVDVVAMARTGSGKTAAFLIPMLEK 89
Query: 206 AMQEEIMMPIVPGEGPFGLIICPSRELARQTYEVIEEFLLPLKEAGYPELRPLLCIGGID 265
Q VP G LI+ P+R+LA QT + +E + +LR L +GG
Sbjct: 90 LKQH------VPQGGVRALILSPTRDLAEQTLKFTKEL------GKFTDLRVSLLVGGDS 137
Query: 266 MRSQLEIVKKGVHIVVATPGRLKDMLAK-KKMNLDNCRYLTLDEADRLVDLGFEDDIREV 324
M Q E + KG +++ATPGRL +L++ M L Y+ DEAD L +GF + + ++
Sbjct: 138 MEDQFEELTKGPDVIIATPGRLMHLLSEVDDMTLRTVEYVVFDEADSLFGMGFAEQLHQI 197
Query: 325 FDHFKAQRQTLLFSATMPTKIQNFARSALVKPIIVNVGRAGAANLDVIQEVEYVKQEAK- 383
RQTLLFSAT+P+ + FA++ L +P +V + + D+ V+ E K
Sbjct: 198 LTQLSENRQTLLFSATLPSALAEFAKAGLREPQLVRLDVENKISPDLKLSFLTVRPEEKY 257
Query: 384 --IVYLLECLQKTPPPVLIFCENKADVDDIHEYLLLKGVEAVAIHGGKDQEEREYAISSF 441
++YL+ + LIF K V+ ++ L+ +E +G DQ+ R+ +S F
Sbjct: 258 SALLYLVREHISSDQQTLIFVSTKHHVEFVNSLFKLENIEPSVCYGDMDQDARKIHVSRF 317
Query: 442 KAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYVHRIGRTGRCGKTGIATTFINK 501
+A K +L+ TD+A++G+D P + +VIN+D P + +VHR+GR R G+TG A +F+
Sbjct: 318 RARKTMLLIVTDIAARGIDIPLLDNVINWDFPPRPKIFVHRVGRAARAGRTGCAYSFVTP 377
Query: 502 NQSETTLLDLKHLLQEAKQRIPPVLAELVDPMED 535
+ +LDL HL R P E++ ME+
Sbjct: 378 -EDMPYMLDL-HLFLSKPVRPAPTEDEVLKNMEE 409
>At5g60990 replication protein A1 - like
Length = 456
Score = 214 bits (546), Expect = 8e-56
Identities = 133/415 (32%), Positives = 217/415 (52%), Gaps = 13/415 (3%)
Query: 137 EEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKTL 196
EE +K F ++ + ++K + G P+ IQ + LP L G+D+IG+A TGSGKT
Sbjct: 2 EEENEVVKTFAELGVREELVKACERLGWKNPSKIQAEALPFALEGKDVIGLAQTGSGKTG 61
Query: 197 VFVLPMIMMAMQEEIMMPIVPGEGP----FGLIICPSRELARQTYEVIEEFLLPLKEAGY 252
F +P++ ++ G P F ++ P+RELA Q E E +
Sbjct: 62 AFAIPILQALLEYVYDSEPKKGRRPDPAFFACVLSPTRELAIQIAEQFEALGADIS---- 117
Query: 253 PELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKK-MNLDNCRYLTLDEADR 311
LR + +GGID Q + K H++VATPGRL D ++ K +L + +YL LDEADR
Sbjct: 118 --LRCAVLVGGIDRMQQTIALGKRPHVIVATPGRLWDHMSDTKGFSLKSLKYLVLDEADR 175
Query: 312 LVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALVKPIIVNVGRAGAANLDV 371
L++ FE + ++ + +R+T LFSATM K++ R+ L P+ + + +
Sbjct: 176 LLNEDFEKSLNQILEEIPLERKTFLFSATMTKKVRKLQRACLRNPVKIEAASKYSTVDTL 235
Query: 372 IQEVEYVKQEAKIVYLLECLQKTPPPV-LIFCENKADVDDIHEYLLLKGVEAVAIHGGKD 430
Q+ +V + K YL+ L + P +IF + L G A+ I G
Sbjct: 236 KQQYRFVAAKYKDCYLVYILSEMPESTSMIFTRTCDGTRFLALVLRSLGFRAIPISGQMT 295
Query: 431 QEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYVHRIGRTGRCG 490
Q +R A++ FKAG+ ++LV TDVAS+GLD P + VINYD+P ++Y+HR+GRT R G
Sbjct: 296 QSKRLGALNKFKAGECNILVCTDVASRGLDIPSVDVVINYDIPTNSKDYIHRVGRTARAG 355
Query: 491 KTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPMEDNEEITGISGV 545
++G+ + +N+ + E + ++ L+ + P E++ +E E +S +
Sbjct: 356 RSGVGISLVNQYELE-WYIQIEKLIGKKLPEYPAEEDEVLSLLERVAEAKKLSAM 409
>At1g16280 putative ATP-dependent RNA helicase
Length = 491
Score = 214 bits (545), Expect = 1e-55
Identities = 144/436 (33%), Positives = 233/436 (53%), Gaps = 36/436 (8%)
Query: 145 NFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKTLVFVLPMIM 204
NF+ + + ++ K G+ +PTP+Q +P IL+GRD++G+A TGSGKT F LP++
Sbjct: 59 NFEGLGLAEWAVETCKELGMRKPTPVQTHCVPKILAGRDVLGLAQTGSGKTAAFALPILH 118
Query: 205 MAMQEEIMMPIVPGEGPFGLIICPSRELARQTYEVIEEFLLPLKEAGYP-ELRPLLCIGG 263
++ G F L++ P+RELA +++ E+F K G LR + +GG
Sbjct: 119 RLAEDPY--------GVFALVVTPTRELA---FQLAEQF----KALGSCLNLRCSVIVGG 163
Query: 264 IDMRSQLEIVKKGVHIVVATPGRLKDMLAKKK---MNLDNCRYLTLDEADRLVDLGFEDD 320
+DM +Q + HIV+ TPGR+K +L ++L LDEADR++D+GF+D+
Sbjct: 164 MDMLTQTMSLVSRPHIVITTPGRIKVLLENNPDVPPVFSRTKFLVLDEADRVLDVGFQDE 223
Query: 321 IREVFDHFKAQRQTLLFSATMPTKIQNFARSALVKPIIVNVGRAGAANLDVI-QEVEYVK 379
+R +F RQTLLFSATM + +Q + K G +D + Q+ +
Sbjct: 224 LRTIFQCLPKSRQTLLFSATMTSNLQALLEHSSNKAYFYEAYE-GLKTVDTLTQQFIFED 282
Query: 380 QEAKIVYLLECLQKTPPP----VLIFCENKADVDDIHEYLLLKGVEAVAIHGGKDQEERE 435
++AK +YL+ L + +IF + L VE +A+H Q R
Sbjct: 283 KDAKELYLVHILSQMEDKGIRSAMIFVSTCRTCQRLSLMLDELEVENIAMHSLNSQSMRL 342
Query: 436 YAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYVHRIGRTGRCGKTGIA 495
A+S FK+GK +L+ATDVAS+GLD P + VINYD+P + +YVHR+GRT R G+ G+A
Sbjct: 343 SALSKFKSGKVPILLATDVASRGLDIPTVDLVINYDIPRDPRDYVHRVGRTARAGRGGLA 402
Query: 496 TTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPMEDNEEITGISGVKGCAYC---- 551
+ I +ET + + + +E +++ P +++ D+ E+T +S K A
Sbjct: 403 VSII----TETDVKLIHKIEEEVGKKMEPYNKKVI---TDSLEVTKVSKAKRVAMMKMLD 455
Query: 552 GGLGHRIRDCPKLEHQ 567
G +++D KL+ +
Sbjct: 456 NGFEDKVKDRRKLKRK 471
>At3g09720 RNA helicase like protein
Length = 541
Score = 209 bits (532), Expect = 3e-54
Identities = 140/430 (32%), Positives = 224/430 (51%), Gaps = 32/430 (7%)
Query: 111 PPLHIRRMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMRF----PDPILKMLKTKGIVQ 166
P + R ++D L +KQ+ I V+G IPPP+K+F ++ IL+ L G +
Sbjct: 105 PKKELNRQMERDA-LSRKQYSIHVSGNNIPPPLKSFAELSSRYGCEGYILRNLAELGFKE 163
Query: 167 PTPIQVQGLPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLII 226
PTPIQ Q +P++LSGR+ A TGSGKT F+ PM++ + +G +I+
Sbjct: 164 PTPIQRQAIPILLSGRECFACAPTGSGKTFAFICPMLIKLKRPST-------DGIRAVIL 216
Query: 227 CPSRELARQTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGR 286
P+RELA QT ++ + +PL+ + K ++++TP R
Sbjct: 217 SPARELAAQTAREGKKLIKGSNFHIRLMTKPLV--------KTADFSKLWCDVLISTPMR 268
Query: 287 LKDMLAKKKMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQT-----LLFSATM 341
LK + KK++L YL LDE+D+L FE + + D LFSAT+
Sbjct: 269 LKRAIKAKKIDLSKVEYLVLDESDKL----FEQSLLKQIDCVVKACSNPSIIRSLFSATL 324
Query: 342 PTKIQNFARSALVKPIIVNVGRAGAANLDVIQEVEYV-KQEAKIVYLLECLQKT-PPPVL 399
P ++ ARS + + V +GR A+ V Q++ + +E K++ L + ++ PPVL
Sbjct: 325 PDSVEELARSIMHDAVRVIIGRKNTASETVKQKLVFAGSEEGKLLALRQSFAESLNPPVL 384
Query: 400 IFCENKADVDDIHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGL 459
IF ++K ++++ L + + A IH ERE A+ F+AG+K VL+ATDV ++G+
Sbjct: 385 IFVQSKERAKELYDELKCENIRAGVIHSDLPPGERENAVDQFRAGEKWVLIATDVIARGM 444
Query: 460 DFPDIQHVINYDMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAK 519
DF I VINYD P Y+HRIGR+GR G++G A TF + Q L ++ + + +
Sbjct: 445 DFKGINCVINYDFPDSASAYIHRIGRSGRAGRSGEAITFYTE-QDVPFLRNIANTMMSSG 503
Query: 520 QRIPPVLAEL 529
+P + L
Sbjct: 504 CEVPSWIMSL 513
>At3g22330 DEAD-Box RNA helicase like protein
Length = 616
Score = 207 bits (526), Expect = 2e-53
Identities = 132/407 (32%), Positives = 211/407 (51%), Gaps = 20/407 (4%)
Query: 155 ILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMP 214
I+K L +KGI + PIQ L + GRDMIG A TG+GKTL F +P+I ++
Sbjct: 115 IVKALSSKGIEKLFPIQKAVLEPAMEGRDMIGRARTGTGKTLAFGIPIIDKIIKYNAKHG 174
Query: 215 IVPGEGPFGLIICPSRELARQTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVK 274
G P L++ P+RELARQ + E P L + GG + Q+ +
Sbjct: 175 --RGRNPLCLVLAPTRELARQVEKEFRE--------SAPSLDTICLYGGTPIGQQMRQLD 224
Query: 275 KGVHIVVATPGRLKDMLAKKKMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQT 334
GV + V TPGR+ D++ + +NL +++ LDEAD+++ +GF +D+ + + +RQ+
Sbjct: 225 YGVDVAVGTPGRVIDLMKRGALNLSEVQFVVLDEADQMLQVGFAEDVEIILEKLPEKRQS 284
Query: 335 LLFSATMPTKIQNFARSALVKPIIVN-VGRAGAANLDVIQEVEYVKQE---AKIVYLLEC 390
++FSATMP+ I++ + L P+ V+ VG + D I + A I+ L
Sbjct: 285 MMFSATMPSWIRSLTKKYLNNPLTVDLVGDSDQKLADGITTYSIIADSYGRASIIGPLVT 344
Query: 391 LQKTPPPVLIFCENKADVDDIHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLV 450
++F + K D D + Y L + + A+HG Q +RE ++ F+ G ++LV
Sbjct: 345 EHAKGGKCIVFTQTKRDADRL-SYALARSFKCEALHGDISQSQRERTLAGFRDGHFNILV 403
Query: 451 ATDVASKGLDFPDIQHVINYDMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLD 510
ATDVA++GLD P++ +I+Y++P E +VHR GRTGR GK G A +++QS
Sbjct: 404 ATDVAARGLDVPNVDLIIHYELPNNTETFVHRTGRTGRAGKKGSAILIYSQDQSRA---- 459
Query: 511 LKHLLQEAKQRIPPVLAELVDPMEDNEEITGISGVKGCAYCGGLGHR 557
+K + +E R L + GI G ++ GG+ R
Sbjct: 460 VKIIEREVGSRFTE-LPSIAVERGSASMFEGIGSRSGGSFGGGMRDR 505
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.138 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,350,688
Number of Sequences: 26719
Number of extensions: 599380
Number of successful extensions: 2225
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 88
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 1885
Number of HSP's gapped (non-prelim): 182
length of query: 589
length of database: 11,318,596
effective HSP length: 105
effective length of query: 484
effective length of database: 8,513,101
effective search space: 4120340884
effective search space used: 4120340884
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144484.10


