

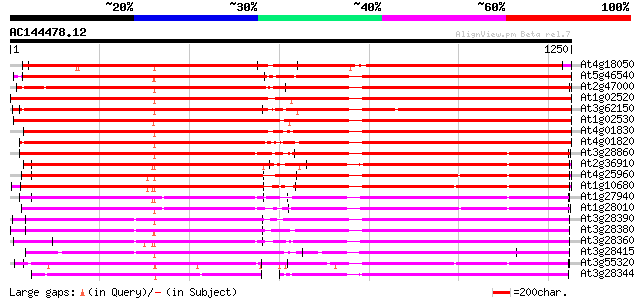
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144478.12 + phase: 0 /pseudo
(1250 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g18050 multidrug resistance protein/P-glycoprotein - like 1571 0.0
At5g46540 multidrug resistance p-glycoprotein 1487 0.0
At2g47000 putative ABC transporter 1476 0.0
At1g02520 P-glycoprotein, putative 1475 0.0
At3g62150 P-glycoprotein-like proetin 1456 0.0
At1g02530 hypothetical protein 1437 0.0
At4g01830 putative P-glycoprotein-like protein 1409 0.0
At4g01820 P-glycoprotein-like protein pgp3 1402 0.0
At3g28860 P-glycoprotein, putative 1004 0.0
At2g36910 putative ABC transporter 986 0.0
At4g25960 P-glycoprotein-2 (pgp2) 984 0.0
At1g10680 putative P-glycoprotein-2 emb|CAA71277 980 0.0
At1g27940 hypothetical protein 938 0.0
At1g28010 hypothetical protein 919 0.0
At3g28390 P-glycoprotein, putative 889 0.0
At3g28380 P-glycoprotein, putative 882 0.0
At3g28360 P-glycoprotein like protein 858 0.0
At3g28415 putative protein 732 0.0
At3g55320 P-glycoprotein - like 695 0.0
At3g28344 P-glycoprotein, 5' partial 431 e-120
>At4g18050 multidrug resistance protein/P-glycoprotein - like
Length = 1323
Score = 1571 bits (4067), Expect = 0.0
Identities = 815/1269 (64%), Positives = 990/1269 (77%), Gaps = 87/1269 (6%)
Query: 28 QKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSSNPADAIKQV 87
QKV F+ LF+FAD DV LM +GTI+A NGL P MTL G +INAFG+++P +++V
Sbjct: 14 QKVSFFKLFSFADKTDVVLMTVGTIAAAGNGLTQPFMTLIFGQLINAFGTTDPDHMVREV 73
Query: 88 SKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTETNTGE 147
KV++ F+YLA+ S + +FLQV+CWMVTGERQ+A IR LYLKTIL+QDI +FDTETNTGE
Sbjct: 74 WKVAVKFIYLAVYSCVVAFLQVSCWMVTGERQSATIRGLYLKTILRQDIGYFDTETNTGE 133
Query: 148 VIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAVAGA 207
VIGRMSGDTILIQ+AMGEKVGKF QL F GGF +AF KG LA VL +C+P + +AGA
Sbjct: 134 VIGRMSGDTILIQDAMGEKVGKFTQLLCTFLGGFAIAFYKGPLLAGVLCSCIPLIVIAGA 193
Query: 208 FMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTTMVK 267
MS++M+KM+ RGQ+AYAEAGNVV+QTVGAIRTV +FTGEK+A EKY SK++IAY T+V+
Sbjct: 194 AMSLIMSKMAGRGQVAYAEAGNVVEQTVGAIRTVVAFTGEKQATEKYESKLEIAYKTVVQ 253
Query: 268 QGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI------- 320
QG++SGFG+G + + FC+YGLA+WYG+KL++EKGYNGG V+ VI A++TGG+
Sbjct: 254 QGLISGFGLGTMLAVIFCSYGLAVWYGAKLIMEKGYNGGQVINVIFAVLTGGMSLGQTSP 313
Query: 321 -------------------------------------IKGDIELRDVSFRYPARPDVQIF 343
I+GDIEL+DV FRYPARPDVQIF
Sbjct: 314 SLNAFAAGRAAAFKMFETIKRSPKIDAYDMSGSVLEDIRGDIELKDVYFRYPARPDVQIF 373
Query: 344 DGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQI 403
GFSLFVP+G T ALVGQSGSGKSTVISL+ERFYDP++G+VLID ++LK LQL+WIR +I
Sbjct: 374 AGFSLFVPNGKTVALVGQSGSGKSTVISLIERFYDPESGQVLIDNIDLKKLQLKWIRSKI 433
Query: 404 GLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQ 463
GLVSQEP+LF T+I+ENIAYGKE ATD+EI TAI LANA KFIDKLPQGLDTM G++GTQ
Sbjct: 434 GLVSQEPVLFATTIKENIAYGKEDATDQEIRTAIELANAAKFIDKLPQGLDTMVGEHGTQ 493
Query: 464 LSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLT 523
+SGGQKQR+AIARAILKNPKILLLDEATSALDAESERIVQ+AL ++ RTTVVVAHRLT
Sbjct: 494 MSGGQKQRLAIARAILKNPKILLLDEATSALDAESERIVQDALVNLMSNRTTVVVAHRLT 553
Query: 524 TIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNE-AEGSRKSEADKLGD 582
TIR AD+IAVV QGKIVE+GTH + DP+GAYSQL+RLQEG E A S + E
Sbjct: 554 TIRTADVIAVVHQGKIVEKGTHDEMIQDPEGAYSQLVRLQEGSKEEATESERPET----- 608
Query: 583 NLNIDSHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGL----SGEIVESDIEQGQLDNK 638
++D +GS ++ RS+S+ SS S RHS SL + ++D + + +N
Sbjct: 609 --SLDVERSGSLRLSSAMRRSVSRNSS-SSRHSFSLASNMFFPGVNVNQTDEMEDEENNV 665
Query: 639 KKPKVSIWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKES 698
+ KVS+ RLA LNKPEIPV++LG+IAA+V+G VFPIFG L S+ I+MFY+P + +K+S
Sbjct: 666 RHKKVSLKRLAHLNKPEIPVLVLGSIAAMVHGTVFPIFGLLLSSSINMFYEPAKILKKDS 725
Query: 699 RFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRY 758
FW+L+++ LGL V++P+ N+FFGIAGGKLI+RIRS+ F+K+VHQEISWFDD ++SRY
Sbjct: 726 HFWALIYIALGLTNFVMIPVPNYFFGIAGGKLIKRIRSMCFDKVVHQEISWFDDTANSRY 785
Query: 759 ---------------VIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVG 803
++ +C + R E CS DASTV+SLVG
Sbjct: 786 YNFIYIINRRILYVLILIFICVLLPP-------VRLERECS--------TDASTVRSLVG 830
Query: 804 DTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMY 863
D +ALIVQNI+TV GL+IAFTANWILA IVL L+P I++QG Q KFL GFSADAK MY
Sbjct: 831 DALALIVQNIATVTTGLIIAFTANWILALIVLALSPFIVIQGYAQTKFLTGFSADAKAMY 890
Query: 864 EEASQVANDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLY 923
EEASQVANDAVSSIRTVASFCAE KVMD+Y +KC GP K GVRLGL+SG GFG SF LY
Sbjct: 891 EEASQVANDAVSSIRTVASFCAEEKVMDLYQQKCDGPKKNGVRLGLLSGAGFGFSFFFLY 950
Query: 924 CTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFE 983
C N F G+ L+Q GKATF EVF+VFFALT+ AI VSQT+ +APD+NKAKDSAASIF+
Sbjct: 951 CINCVCFVSGAGLIQIGKATFGEVFKVFFALTIMAIGVSQTSAMAPDSNKAKDSAASIFD 1010
Query: 984 IIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVG 1043
I+DS P IDSSS+ G T + V GDIE +HV+F YP RPD+QIF+DL L+IPS KT+ALVG
Sbjct: 1011 ILDSTPKIDSSSDEGTTLQNVNGDIEFRHVSFRYPMRPDVQIFRDLCLTIPSGKTVALVG 1070
Query: 1044 ESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRAN 1103
ESGSGKSTVIS++ERFY+P+SG+IL+D V+++TF+LSWLRQQMGLV QEPILFNE+IR+N
Sbjct: 1071 ESGSGKSTVISMIERFYNPDSGKILIDQVEIQTFKLSWLRQQMGLVSQEPILFNETIRSN 1130
Query: 1104 IGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTML 1163
I YGK GGATE+EIIAAA AANAH+FIS+LP GYDTSVGERG QLSGGQKQRIAIAR +L
Sbjct: 1131 IAYGKTGGATEEEIIAAAKAANAHNFISSLPQGYDTSVGERGVQLSGGQKQRIAIARAIL 1190
Query: 1164 KNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAV 1223
K+PKILLLDEATSALDAESER+VQ+ALDRV VNRTTVVVAHRLTTI+ AD IAV+KNG +
Sbjct: 1191 KDPKILLLDEATSALDAESERVVQDALDRVMVNRTTVVVAHRLTTIKNADVIAVVKNGVI 1250
Query: 1224 AEKGRHDEL 1232
AEKGRH+ L
Sbjct: 1251 AEKGRHETL 1259
Score = 429 bits (1102), Expect = e-120
Identities = 249/611 (40%), Positives = 350/611 (56%), Gaps = 31/611 (5%)
Query: 642 KVSIWRLAKL-NKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMF-YKPPEQQRKESR 699
KVS ++L +K ++ ++ +G IAA NG+ P +F +I+ F P+ +E
Sbjct: 15 KVSFFKLFSFADKTDVVLMTVGTIAAAGNGLTQPFMTLIFGQLINAFGTTDPDHMVREVW 74
Query: 700 FWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRYV 759
++ F+ L + + V+ LQ + + G + IR L + I+ Q+I +FD
Sbjct: 75 KVAVKFIYLAVYSCVVAFLQVSCWMVTGERQSATIRGLYLKTILRQDIGYFD-------- 126
Query: 760 IQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAG 819
TE N +G V R+S D ++ +G+ + Q + T + G
Sbjct: 127 ----------------TETN----TGEVIGRMSGDTILIQDAMGEKVGKFTQLLCTFLGG 166
Query: 820 LVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRT 879
IAF +LA ++ P+I++ G + + +V Y EA V V +IRT
Sbjct: 167 FAIAFYKGPLLAGVLCSCIPLIVIAGAAMSLIMSKMAGRGQVAYAEAGNVVEQTVGAIRT 226
Query: 880 VASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQH 939
V +F E + + Y K K V+ GL+SG G G V++C+ + G+ L+
Sbjct: 227 VVAFTGEKQATEKYESKLEIAYKTVVQQGLISGFGLGTMLAVIFCSYGLAVWYGAKLIME 286
Query: 940 GKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGV 999
+V V FA+ +++ QT+ + +A +FE I P ID+ +G
Sbjct: 287 KGYNGGQVINVIFAVLTGGMSLGQTSPSLNAFAAGRAAAFKMFETIKRSPKIDAYDMSGS 346
Query: 1000 TRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERF 1059
E + GDIEL+ V F YP RPD+QIF SL +P+ KT+ALVG+SGSGKSTVISL+ERF
Sbjct: 347 VLEDIRGDIELKDVYFRYPARPDVQIFAGFSLFVPNGKTVALVGQSGSGKSTVISLIERF 406
Query: 1060 YDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGATEDEIIA 1119
YDP SG++L+D +DLK +L W+R ++GLV QEP+LF +I+ NI YGKE AT+ EI
Sbjct: 407 YDPESGQVLIDNIDLKKLQLKWIRSKIGLVSQEPVLFATTIKENIAYGKE-DATDQEIRT 465
Query: 1120 AANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALD 1179
A ANA FI LP G DT VGE GTQ+SGGQKQR+AIAR +LKNPKILLLDEATSALD
Sbjct: 466 AIELANAAKFIDKLPQGLDTMVGEHGTQMSGGQKQRLAIARAILKNPKILLLDEATSALD 525
Query: 1180 AESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGV 1239
AESERIVQ+AL + NRTTVVVAHRLTTIR AD IAV+ G + EKG HDE+++ +G
Sbjct: 526 AESERIVQDALVNLMSNRTTVVVAHRLTTIRTADVIAVVHQGKIVEKGTHDEMIQDPEGA 585
Query: 1240 YASLVALHSSA 1250
Y+ LV L +
Sbjct: 586 YSQLVRLQEGS 596
Score = 389 bits (999), Expect = e-108
Identities = 236/586 (40%), Positives = 327/586 (55%), Gaps = 82/586 (13%)
Query: 43 DVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSSNPADAIKQVSKVSLLFVYLAIGSG 102
++ ++++G+I+A+ +G P+ L L + IN F PA +K+ S L +Y+A+G
Sbjct: 682 EIPVLVLGSIAAMVHGTVFPIFGLLLSSSINMF--YEPAKILKKDSHFWAL-IYIALG-- 736
Query: 103 IASFLQVTC----WMVTGERQAARIRSLYLKTILQQDIAFFDTETNTGE-----VIGRM- 152
+ +F+ + + + G + RIRS+ ++ Q+I++FD N+ +I R
Sbjct: 737 LTNFVMIPVPNYFFGIAGGKLIKRIRSMCFDKVVHQEISWFDDTANSRYYNFIYIINRRI 796
Query: 153 ----------------------SGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWR 190
S D ++ +G+ + Q + G ++AF W
Sbjct: 797 LYVLILIFICVLLPPVRLERECSTDASTVRSLVGDALALIVQNIATVTTGLIIAFTANWI 856
Query: 191 LAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKA 250
LA+++LA P + + G + + S+ + Y EA V + V +IRTVASF E+K
Sbjct: 857 LALIVLALSPFIVIQGYAQTKFLTGFSADAKAMYEEASQVANDAVSSIRTVASFCAEEKV 916
Query: 251 IEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMT 310
++ Y K V+ G++SG G G F +C + G+ L+ G V
Sbjct: 917 MDLYQQKCDGPKKNGVRLGLLSGAGFGFSFFFLYCINCVCFVSGAGLIQIGKATFGEVFK 976
Query: 311 VIIALMTGGI--------------------------------------------IKGDIE 326
V AL I + GDIE
Sbjct: 977 VFFALTIMAIGVSQTSAMAPDSNKAKDSAASIFDILDSTPKIDSSSDEGTTLQNVNGDIE 1036
Query: 327 LRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLI 386
R VSFRYP RPDVQIF L +PSG T ALVG+SGSGKSTVIS++ERFY+PD+G++LI
Sbjct: 1037 FRHVSFRYPMRPDVQIFRDLCLTIPSGKTVALVGESGSGKSTVISMIERFYNPDSGKILI 1096
Query: 387 DGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEG-ATDEEITTAITLANAKKF 445
D V ++ +L W+R+Q+GLVSQEPILF +IR NIAYGK G AT+EEI A ANA F
Sbjct: 1097 DQVEIQTFKLSWLRQQMGLVSQEPILFNETIRSNIAYGKTGGATEEEIIAAAKAANAHNF 1156
Query: 446 IDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEA 505
I LPQG DT G+ G QLSGGQKQRIAIARAILK+PKILLLDEATSALDAESER+VQ+A
Sbjct: 1157 ISSLPQGYDTSVGERGVQLSGGQKQRIAIARAILKDPKILLLDEATSALDAESERVVQDA 1216
Query: 506 LEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMD 551
L+++++ RTTVVVAHRLTTI+NAD+IAVV+ G I E+G H L D
Sbjct: 1217 LDRVMVNRTTVVVAHRLTTIKNADVIAVVKNGVIAEKGRHETLDED 1262
>At5g46540 multidrug resistance p-glycoprotein
Length = 1248
Score = 1487 bits (3849), Expect = 0.0
Identities = 770/1267 (60%), Positives = 957/1267 (74%), Gaps = 79/1267 (6%)
Query: 28 QKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSSNPADAIKQV 87
Q++ FY LF FAD D+ LM+IGT+SA+ANGL P M++ +G +IN FG S+ K+V
Sbjct: 16 QRIAFYKLFTFADRYDIVLMVIGTLSAMANGLTQPFMSILMGQLINVFGFSDHDHVFKEV 75
Query: 88 SKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTETNTGE 147
SKV++ F+YLA +G+ SFLQV+CWMVTGERQ+ RIR LYLKTIL+QDI FFDTETNTGE
Sbjct: 76 SKVAVKFLYLAAYAGVVSFLQVSCWMVTGERQSTRIRRLYLKTILRQDIGFFDTETNTGE 135
Query: 148 VIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAVAGA 207
VIGRMSGDTILIQ++MGEKVGKF QL S+F GGF +AFI G +L + LL CVP + G
Sbjct: 136 VIGRMSGDTILIQDSMGEKVGKFTQLVSSFVGGFTVAFIVGMKLTLALLPCVPLIVGTGG 195
Query: 208 FMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTTMVK 267
M+ +M+K + R Q+AY EAGNVV Q VG+IRTV +FTGEK+++ KY K++IAY +MVK
Sbjct: 196 AMTYIMSKKAQRVQLAYTEAGNVVQQAVGSIRTVVAFTGEKQSMGKYEKKLEIAYKSMVK 255
Query: 268 QGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI------- 320
QG+ SG GIG++ + +CTYG A+WYG++ +IEKGY GG VM VI +++TGG+
Sbjct: 256 QGLYSGLGIGIMMVVVYCTYGFAIWYGARQIIEKGYTGGQVMNVITSILTGGMALGQTLP 315
Query: 321 -------------------------------------IKGDIELRDVSFRYPARPDVQIF 343
IKGDIELRDV FRYPARPDVQIF
Sbjct: 316 SLNSFAAGTAAAYKMFETIKRKPKIDAYDMSGEVLEEIKGDIELRDVYFRYPARPDVQIF 375
Query: 344 DGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQI 403
GFSL VP+G T ALVGQSGSGKSTVISL+ERFYDP++GEVLIDG++LK Q++WIR +I
Sbjct: 376 VGFSLTVPNGMTVALVGQSGSGKSTVISLIERFYDPESGEVLIDGIDLKKFQVKWIRSKI 435
Query: 404 GLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQ 463
GLVSQEPILF T+IRENI YGK+ A+D+EI TA+ LANA FIDKLPQGL+TM G++GTQ
Sbjct: 436 GLVSQEPILFATTIRENIVYGKKDASDQEIRTALKLANASNFIDKLPQGLETMVGEHGTQ 495
Query: 464 LSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLT 523
LSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQ+AL K++L RTTVVVAHRLT
Sbjct: 496 LSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQDALVKLMLSRTTVVVAHRLT 555
Query: 524 TIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEGSRKSEADKLGDN 583
TIR AD+IAVVQQGK++E+GTH + DP+G YSQL+RLQEG + E K E +K +
Sbjct: 556 TIRTADMIAVVQQGKVIEKGTHDEMIKDPEGTYSQLVRLQEGSKKEEAIDK-EPEKCEMS 614
Query: 584 LNIDSHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQLDNKKKPKV 643
L I+S S +Q +++ S + S E + S Q KK +V
Sbjct: 615 LEIES----SDSQNGIHSGTLTSPSGLPGVISLDQTEEFHENISSTKTQTV---KKGKEV 667
Query: 644 SIWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKESRFWSL 703
S+ RLA LNKPEI V+LLG++AA+++G+VFP+ G L S I +F++P + + +S FW+L
Sbjct: 668 SLRRLAHLNKPEISVLLLGSLAAVIHGIVFPVQGLLLSRTIRIFFEPSNKLKNDSLFWAL 727
Query: 704 LFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQNV 763
+FV LGL L+++PLQN+ F IAG KLI+RIRSL+F++++HQ+ISWFDD +S
Sbjct: 728 IFVALGLTDLIVIPLQNYLFAIAGAKLIKRIRSLSFDRVLHQDISWFDDTKNS------- 780
Query: 764 CYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIA 823
SG +GARLS DASTVKS+VGD + LI+QN++T+I +IA
Sbjct: 781 --------------------SGVIGARLSTDASTVKSIVGDVLGLIMQNMATIIGAFIIA 820
Query: 824 FTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASF 883
FTANW+LA + L++ P++ QG Q+KF+ GF A A+ YEEASQVA+DAVSSIRTVASF
Sbjct: 821 FTANWLLALMALLVAPVMFFQGYYQIKFITGFGAKARGKYEEASQVASDAVSSIRTVASF 880
Query: 884 CAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKAT 943
CAE KVMD+Y +KC P +QG +LGLVSG+ +G S+L LY + F GS L+Q+ +AT
Sbjct: 881 CAEDKVMDLYQEKCDEPKQQGFKLGLVSGLCYGGSYLALYVIESVCFLGGSWLIQNRRAT 940
Query: 944 FTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRET 1003
F E F+VFFALT+TA+ V+QT+T+APD NKAKDSAASIF+I+DSKP IDSSS G
Sbjct: 941 FGEFFQVFFALTLTAVGVTQTSTMAPDINKAKDSAASIFDILDSKPKIDSSSEKGTILPI 1000
Query: 1004 VVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPN 1063
V GDIELQHV+F YP RPDIQIF DL L+I S +T+ALVGESGSGKSTVISLLERFYDP+
Sbjct: 1001 VHGDIELQHVSFRYPMRPDIQIFSDLCLTISSGQTVALVGESGSGKSTVISLLERFYDPD 1060
Query: 1064 SGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGATEDEIIAAANA 1123
SG+ILLD V++++ +LSWLR+QMGLV QEP+LFNE+I +NI YGK GGATE+EII AA A
Sbjct: 1061 SGKILLDQVEIQSLKLSWLREQMGLVSQEPVLFNETIGSNIAYGKIGGATEEEIITAAKA 1120
Query: 1124 ANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESE 1183
AN H+FIS+LP GY+TSVGERG QLSGGQKQRIAIAR +LK+PKILLLDEATSALDAESE
Sbjct: 1121 ANVHNFISSLPQGYETSVGERGVQLSGGQKQRIAIARAILKDPKILLLDEATSALDAESE 1180
Query: 1184 RIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGVYASL 1243
R+VQ+ALD+V VNRTTVVVAH LTTI+ AD IAV+KNG +AE GRH+ LM I+ G YASL
Sbjct: 1181 RVVQDALDQVMVNRTTVVVAHLLTTIKDADMIAVVKNGVIAESGRHETLMEISGGAYASL 1240
Query: 1244 VALHSSA 1250
VA + SA
Sbjct: 1241 VAFNMSA 1247
Score = 404 bits (1038), Expect = e-112
Identities = 249/607 (41%), Positives = 340/607 (55%), Gaps = 55/607 (9%)
Query: 8 HDNSSSSPTQQHGIRDNKTKQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLF 67
H+N SS+ TQ K ++V L + + +++++++G+++AV +G+ P+ L
Sbjct: 650 HENISSTKTQTV-----KKGKEVSLRRLAHL-NKPEISVLLLGSLAAVIHGIVFPVQGLL 703
Query: 68 LGNVINAFGSSNPADAIKQVSKV-SLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSL 126
L I F P++ +K S +L+FV L + I LQ + + G + RIRSL
Sbjct: 704 LSRTIRIF--FEPSNKLKNDSLFWALIFVALGLTDLIVIPLQNYLFAIAGAKLIKRIRSL 761
Query: 127 YLKTILQQDIAFFDTETNTGEVIG-RMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAF 185
+L QDI++FD N+ VIG R+S D ++ +G+ +G Q + G F++AF
Sbjct: 762 SFDRVLHQDISWFDDTKNSSGVIGARLSTDASTVKSIVGDVLGLIMQNMATIIGAFIIAF 821
Query: 186 IKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFT 245
W LA++ L P + G + + ++ + Y EA V V +IRTVASF
Sbjct: 822 TANWLLALMALLVAPVMFFQGYYQIKFITGFGAKARGKYEEASQVASDAVSSIRTVASFC 881
Query: 246 GEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNG 305
E K ++ Y K K G+VSG G + + GS L+ +
Sbjct: 882 AEDKVMDLYQEKCDEPKQQGFKLGLVSGLCYGGSYLALYVIESVCFLGGSWLIQNRRATF 941
Query: 306 GTVMTVIIALMTGG--------------------------------------------II 321
G V AL I+
Sbjct: 942 GEFFQVFFALTLTAVGVTQTSTMAPDINKAKDSAASIFDILDSKPKIDSSSEKGTILPIV 1001
Query: 322 KGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDA 381
GDIEL+ VSFRYP RPD+QIF L + SG T ALVG+SGSGKSTVISLLERFYDPD+
Sbjct: 1002 HGDIELQHVSFRYPMRPDIQIFSDLCLTISSGQTVALVGESGSGKSTVISLLERFYDPDS 1061
Query: 382 GEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGK-EGATDEEITTAITLA 440
G++L+D V +++L+L W+REQ+GLVSQEP+LF +I NIAYGK GAT+EEI TA A
Sbjct: 1062 GKILLDQVEIQSLKLSWLREQMGLVSQEPVLFNETIGSNIAYGKIGGATEEEIITAAKAA 1121
Query: 441 NAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESER 500
N FI LPQG +T G+ G QLSGGQKQRIAIARAILK+PKILLLDEATSALDAESER
Sbjct: 1122 NVHNFISSLPQGYETSVGERGVQLSGGQKQRIAIARAILKDPKILLLDEATSALDAESER 1181
Query: 501 IVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLI 560
+VQ+AL+++++ RTTVVVAH LTTI++AD+IAVV+ G I E G H L GAY+ L+
Sbjct: 1182 VVQDALDQVMVNRTTVVVAHLLTTIKDADMIAVVKNGVIAESGRHETLMEISGGAYASLV 1241
Query: 561 RLQEGDN 567
N
Sbjct: 1242 AFNMSAN 1248
>At2g47000 putative ABC transporter
Length = 1286
Score = 1476 bits (3822), Expect = 0.0
Identities = 764/1287 (59%), Positives = 968/1287 (74%), Gaps = 87/1287 (6%)
Query: 16 TQQHGIRDNKTKQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAF 75
T++ KTK VPFY LF FAD D LMI+GT+ ++ NGL PLMTL G++I+AF
Sbjct: 33 TEKKDEEHEKTKT-VPFYKLFAFADSFDFLLMILGTLGSIGNGLGFPLMTLLFGDLIDAF 91
Query: 76 GSSNPADAIKQVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQD 135
G N + +VSKV+L FV+L IG+ A+FLQ++ WM++GERQAARIRSLYLKTIL+QD
Sbjct: 92 GE-NQTNTTDKVSKVALKFVWLGIGTFAAAFLQLSGWMISGERQAARIRSLYLKTILRQD 150
Query: 136 IAFFDTETNTGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVL 195
IAFFD +TNTGEV+GRMSGDT+LIQ+AMGEKVGK QL + F GGFV+AF++GW L +V+
Sbjct: 151 IAFFDIDTNTGEVVGRMSGDTVLIQDAMGEKVGKAIQLLATFVGGFVIAFVRGWLLTLVM 210
Query: 196 LACVPCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYN 255
L+ +P + +AGA ++IV+AK +SRGQ AYA+A VV+QT+G+IRTVASFTGEK+AI YN
Sbjct: 211 LSSIPLLVMAGALLAIVIAKTASRGQTAYAKAATVVEQTIGSIRTVASFTGEKQAISNYN 270
Query: 256 SKIKIAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIAL 315
+ AY V +G +G G+G L + FC+Y LA+WYG KL+++KGY GG V+ +IIA+
Sbjct: 271 KHLVTAYKAGVIEGGSTGLGLGTLFLVVFCSYALAVWYGGKLILDKGYTGGQVLNIIIAV 330
Query: 316 MTGGI--------------------------------------------IKGDIELRDVS 331
+TG + IKGDIEL+DV
Sbjct: 331 LTGSMSLGQTSPCLSAFAAGQAAAYKMFETIERRPNIDSYSTNGKVLDDIKGDIELKDVY 390
Query: 332 FRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNL 391
F YPARPD QIF GFSLF+ SGTT ALVGQSGSGKSTV+SL+ERFYDP AG+VLIDG+NL
Sbjct: 391 FTYPARPDEQIFRGFSLFISSGTTVALVGQSGSGKSTVVSLIERFYDPQAGDVLIDGINL 450
Query: 392 KNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQ 451
K QL+WIR +IGLVSQEP+LFT SI++NIAYGKE AT EEI A LANA KF+DKLPQ
Sbjct: 451 KEFQLKWIRSKIGLVSQEPVLFTASIKDNIAYGKEDATTEEIKAAAELANASKFVDKLPQ 510
Query: 452 GLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIIL 511
GLDTM G++GTQLSGGQKQRIA+ARAILK+P+ILLLDEATSALDAESER+VQEAL++I++
Sbjct: 511 GLDTMVGEHGTQLSGGQKQRIAVARAILKDPRILLLDEATSALDAESERVVQEALDRIMV 570
Query: 512 KRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEG 571
RTTVVVAHRL+T+RNAD+IAV+ QGKIVE+G+H+ L DP+GAYSQLIRLQE E
Sbjct: 571 NRTTVVVAHRLSTVRNADMIAVIHQGKIVEKGSHTELLKDPEGAYSQLIRLQEEKKSDEN 630
Query: 572 SRKSEADKLGDNLNIDSHMAGSSTQRTSFVRSISQTSSV---SHRHSQSL----RGLSGE 624
+ +E K+ +I+S SS +++S RS+S+ S S RHS ++ G+ G
Sbjct: 631 A--AEEQKMS---SIESFKQ-SSLRKSSLGRSLSKGGSSRGNSSRHSFNMFGFPAGIDGN 684
Query: 625 IVESDIEQGQLDNKKKPK-VSIWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAV 683
+V+ E K +PK VSI+R+A LNKPEIPV++LG+I+A NGV+ PIFG L S+V
Sbjct: 685 VVQDQEEDDTTQPKTEPKKVSIFRIAALNKPEIPVLILGSISAAANGVILPIFGILISSV 744
Query: 684 ISMFYKPPEQQRKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIV 743
I F++PP++ ++++ FW+++F+ LG +++ P Q FFF IAG KL++RIRS+ FEK+V
Sbjct: 745 IKAFFQPPKKLKEDTSFWAIIFMVLGFASIIAYPAQTFFFAIAGCKLVQRIRSMCFEKVV 804
Query: 744 HQEISWFDDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVG 803
H E+ WFD+P +S SG +GARLS DA+T++ LVG
Sbjct: 805 HMEVGWFDEPENS---------------------------SGTIGARLSADAATIRGLVG 837
Query: 804 DTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMY 863
D++A VQN+S+++AGL+IAF A W LAF+VL + P+I + G + MKF+KGFSADAK MY
Sbjct: 838 DSLAQTVQNLSSILAGLIIAFLACWQLAFVVLAMLPLIALNGFLYMKFMKGFSADAKKMY 897
Query: 864 EEASQVANDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLY 923
EASQVANDAV SIRTVASFCAE KVM+MYSKKC GP K G+R G+VSGIGFG SF VL+
Sbjct: 898 GEASQVANDAVGSIRTVASFCAEDKVMNMYSKKCEGPMKNGIRQGIVSGIGFGFSFFVLF 957
Query: 924 CTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFE 983
+ A FY+G+ LV GK TF VFRVFFALTM A+A+SQ+++L+PD++KA +AASIF
Sbjct: 958 SSYAASFYVGARLVDDGKTTFDSVFRVFFALTMAAMAISQSSSLSPDSSKADVAAASIFA 1017
Query: 984 IIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVG 1043
I+D + ID S +G + V GDIEL+HV+F YP RPD+QIF+DL LSI + KT+ALVG
Sbjct: 1018 IMDRESKIDPSVESGRVLDNVKGDIELRHVSFKYPARPDVQIFQDLCLSIRAGKTVALVG 1077
Query: 1044 ESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRAN 1103
ESGSGKSTVI+LL+RFYDP+SG I LDGV++K+ RL WLRQQ GLV QEPILFNE+IRAN
Sbjct: 1078 ESGSGKSTVIALLQRFYDPDSGEITLDGVEIKSLRLKWLRQQTGLVSQEPILFNETIRAN 1137
Query: 1104 IGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTML 1163
I YGK G A+E EI+++A +NAH FIS L GYDT VGERG QLSGGQKQR+AIAR ++
Sbjct: 1138 IAYGKGGDASESEIVSSAELSNAHGFISGLQQGYDTMVGERGIQLSGGQKQRVAIARAIV 1197
Query: 1164 KNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAV 1223
K+PK+LLLDEATSALDAESER+VQ+ALDRV VNRTT+VVAHRL+TI+ AD IAV+KNG +
Sbjct: 1198 KDPKVLLLDEATSALDAESERVVQDALDRVMVNRTTIVVAHRLSTIKNADVIAVVKNGVI 1257
Query: 1224 AEKGRHDELMRITDGVYASLVALHSSA 1250
EKG+HD L+ I DGVYASLV LH +A
Sbjct: 1258 VEKGKHDTLINIKDGVYASLVQLHLTA 1284
Score = 452 bits (1164), Expect = e-127
Identities = 259/633 (40%), Positives = 376/633 (58%), Gaps = 30/633 (4%)
Query: 615 SQSLRGLSGEIVESDIEQGQLDNKKKPKVSIWRL-AKLNKPEIPVILLGAIAAIVNGVVF 673
S++ R E E+ +++K V ++L A + + +++LG + +I NG+ F
Sbjct: 18 SETKRDKEEEEEVKKTEKKDEEHEKTKTVPFYKLFAFADSFDFLLMILGTLGSIGNGLGF 77
Query: 674 PIFGFLFSAVISMFYKPPEQQRKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIER 733
P+ LF +I F + + +L FV LG+ T LQ + I+G + R
Sbjct: 78 PLMTLLFGDLIDAFGENQTNTTDKVSKVALKFVWLGIGTFAAAFLQLSGWMISGERQAAR 137
Query: 734 IRSLTFEKIVHQEISWFDDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSI 793
IRSL + I+ Q+I++FD +++ G V R+S
Sbjct: 138 IRSLYLKTILRQDIAFFDIDTNT----------------------------GEVVGRMSG 169
Query: 794 DASTVKSLVGDTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLK 853
D ++ +G+ + +Q ++T + G VIAF W+L ++L P+++M G + +
Sbjct: 170 DTVLIQDAMGEKVGKAIQLLATFVGGFVIAFVRGWLLTLVMLSSIPLLVMAGALLAIVIA 229
Query: 854 GFSADAKVMYEEASQVANDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGI 913
++ + Y +A+ V + SIRTVASF E + + Y+K + K GV G +G+
Sbjct: 230 KTASRGQTAYAKAATVVEQTIGSIRTVASFTGEKQAISNYNKHLVTAYKAGVIEGGSTGL 289
Query: 914 GFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNK 973
G G FLV++C+ A + G L+ T +V + A+ ++++ QT+
Sbjct: 290 GLGTLFLVVFCSYALAVWYGGKLILDKGYTGGQVLNIIIAVLTGSMSLGQTSPCLSAFAA 349
Query: 974 AKDSAASIFEIIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSI 1033
+ +A +FE I+ +P+IDS S G + + GDIEL+ V F YP RPD QIF+ SL I
Sbjct: 350 GQAAAYKMFETIERRPNIDSYSTNGKVLDDIKGDIELKDVYFTYPARPDEQIFRGFSLFI 409
Query: 1034 PSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEP 1093
S T+ALVG+SGSGKSTV+SL+ERFYDP +G +L+DG++LK F+L W+R ++GLV QEP
Sbjct: 410 SSGTTVALVGQSGSGKSTVVSLIERFYDPQAGDVLIDGINLKEFQLKWIRSKIGLVSQEP 469
Query: 1094 ILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQK 1153
+LF SI+ NI YGKE AT +EI AAA ANA F+ LP G DT VGE GTQLSGGQK
Sbjct: 470 VLFTASIKDNIAYGKE-DATTEEIKAAAELANASKFVDKLPQGLDTMVGEHGTQLSGGQK 528
Query: 1154 QRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGAD 1213
QRIA+AR +LK+P+ILLLDEATSALDAESER+VQEALDR+ VNRTTVVVAHRL+T+R AD
Sbjct: 529 QRIAVARAILKDPRILLLDEATSALDAESERVVQEALDRIMVNRTTVVVAHRLSTVRNAD 588
Query: 1214 TIAVIKNGAVAEKGRHDELMRITDGVYASLVAL 1246
IAVI G + EKG H EL++ +G Y+ L+ L
Sbjct: 589 MIAVIHQGKIVEKGSHTELLKDPEGAYSQLIRL 621
>At1g02520 P-glycoprotein, putative
Length = 1278
Score = 1475 bits (3819), Expect = 0.0
Identities = 767/1311 (58%), Positives = 965/1311 (73%), Gaps = 105/1311 (8%)
Query: 3 ENPNVHDNSSSSPTQQHGI---RDNKTKQK---VPFYMLFNFADHLDVTLMIIGTISAVA 56
E +V S+S + + G ++ K+++K VPFY LF FAD DV LMI G+I A+
Sbjct: 8 EGDSVSHEPSTSKSPKEGEETKKEEKSEEKANTVPFYKLFAFADSSDVLLMICGSIGAIG 67
Query: 57 NGLASPLMTLFLGNVINAFGSS-NPADAIKQVSKVSLLFVYLAIGSGIASFLQVTCWMVT 115
NG++ P MTL G++I++FG + N D + VSKV L FVYL +G+ A+FLQV CWM+T
Sbjct: 68 NGMSLPFMTLLFGDLIDSFGKNQNNKDIVDVVSKVCLKFVYLGLGTLGAAFLQVACWMIT 127
Query: 116 GERQAARIRSLYLKTILQQDIAFFDTETNTGEVIGRMSGDTILIQEAMGEKVGKFFQLAS 175
GERQAARIRS YLKTIL+QDI FFD ETNTGEV+GRMSGDT+LIQ+AMGEKVGKF QL S
Sbjct: 128 GERQAARIRSTYLKTILRQDIGFFDVETNTGEVVGRMSGDTVLIQDAMGEKVGKFIQLVS 187
Query: 176 NFCGGFVMAFIKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTV 235
F GGFV+AFIKGW L +V+L +P +A+AGA M++++ + SSRGQ AYA+A VV+QT+
Sbjct: 188 TFVGGFVLAFIKGWLLTLVMLTSIPLLAMAGAAMALIVTRASSRGQAAYAKAATVVEQTI 247
Query: 236 GAIRTVASFTGEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGS 295
G+IRTVASFTGEK+AI Y I AY + ++QG +G G+G++ F+ F +Y LA+W+G
Sbjct: 248 GSIRTVASFTGEKQAINSYKKFITSAYKSSIQQGFSTGLGLGVMFFVFFSSYALAIWFGG 307
Query: 296 KLVIEKGYNGGTVMTVIIALMTGGI----------------------------------- 320
K+++EKGY GG V+ VII ++ G +
Sbjct: 308 KMILEKGYTGGAVINVIIIVVAGSMSLGQTSPCVTAFAAGQAAAYKMFETIKRKPLIDAY 367
Query: 321 ---------IKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVIS 371
I+GDIEL+DV F YPARPD +IFDGFSLF+PSG T ALVG+SGSGKSTVIS
Sbjct: 368 DVNGKVLEDIRGDIELKDVHFSYPARPDEEIFDGFSLFIPSGATAALVGESGSGKSTVIS 427
Query: 372 LLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEGATDE 431
L+ERFYDP +G VLIDGVNLK QL+WIR +IGLVSQEP+LF++SI ENIAYGKE AT E
Sbjct: 428 LIERFYDPKSGAVLIDGVNLKEFQLKWIRSKIGLVSQEPVLFSSSIMENIAYGKENATVE 487
Query: 432 EITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEAT 491
EI A LANA KFIDKLPQGLDTM G++GTQLSGGQKQRIAIARAILK+P+ILLLDEAT
Sbjct: 488 EIKAATELANAAKFIDKLPQGLDTMVGEHGTQLSGGQKQRIAIARAILKDPRILLLDEAT 547
Query: 492 SALDAESERIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMD 551
SALDAESER+VQEAL+++++ RTTV+VAHRL+T+RNAD+IAV+ +GK+VE+G+HS L D
Sbjct: 548 SALDAESERVVQEALDRVMVNRTTVIVAHRLSTVRNADMIAVIHRGKMVEKGSHSELLKD 607
Query: 552 PDGAYSQLIRLQEGDNEAEGSRKSEADKLGDNLNIDSHMAGSSTQRTSFVRSISQTSSV- 610
+GAYSQLIRLQE + + + S S +GSS + ++ +S+ TSSV
Sbjct: 608 SEGAYSQLIRLQEINKDVKTSELS---------------SGSSFRNSNLKKSMEGTSSVG 652
Query: 611 --SHRHSQSLRGLSGEIV-------ESDIEQGQLDNKKKPKVSIWRLAKLNKPEIPVILL 661
S HS ++ GL+ + E G + PKVS+ R+A LNKPEIPV+LL
Sbjct: 653 NSSRHHSLNVLGLTTGLDLGSHSQRAGQDETGTASQEPLPKVSLTRIAALNKPEIPVLLL 712
Query: 662 GAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKESRFWSLLFVGLGLVTLVILPLQNF 721
G +AA +NG +FP+FG L S VI F+KP + +++SRFW+++FV LG+ +L++ P Q +
Sbjct: 713 GTVAAAINGAIFPLFGILISRVIEAFFKPAHELKRDSRFWAIIFVALGVTSLIVSPTQMY 772
Query: 722 FFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQNVCYFMTK**KNYETERNES 781
F +AGGKLI RIRS+ FEK VH E++WFD+P +S
Sbjct: 773 LFAVAGGKLIRRIRSMCFEKAVHMEVAWFDEPQNS------------------------- 807
Query: 782 SCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTPMI 841
SG +GARLS DA+ +++LVGD ++L VQN+++ +GL+IAFTA+W LA I+LV+ P+I
Sbjct: 808 --SGTMGARLSADATLIRALVGDALSLAVQNVASAASGLIIAFTASWELALIILVMLPLI 865
Query: 842 LMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFCAESKVMDMYSKKCLGPA 901
+ G VQ+KF+KGFSADAK YEEASQVANDAV SIRTVASFCAE KVM MY K+C GP
Sbjct: 866 GINGFVQVKFMKGFSADAKSKYEEASQVANDAVGSIRTVASFCAEEKVMQMYKKQCEGPI 925
Query: 902 KQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAIAV 961
K G++ G +SG+GFG SF +L+C A FY G+ LV+ GK TF VF+VFFALTM AI +
Sbjct: 926 KDGIKQGFISGLGFGFSFFILFCVYATSFYAGARLVEDGKTTFNNVFQVFFALTMAAIGI 985
Query: 962 SQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPTRP 1021
SQ++T APD++KAK +AASIF IID K IDSS G E V GDIEL+H++F YP RP
Sbjct: 986 SQSSTFAPDSSKAKVAAASIFAIIDRKSKIDSSDETGTVLENVKGDIELRHLSFTYPARP 1045
Query: 1022 DIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSW 1081
DIQIF+DL L+I + KT+ALVGESGSGKSTVISLL+RFYDP+SG I LDGV+LK +L W
Sbjct: 1046 DIQIFRDLCLTIRAGKTVALVGESGSGKSTVISLLQRFYDPDSGHITLDGVELKKLQLKW 1105
Query: 1082 LRQQMGLVGQEPILFNESIRANIGYGK--EGGATEDEIIAAANAANAHSFISNLPDGYDT 1139
LRQQMGLVGQEP+LFN++IRANI YGK E ATE EIIAAA ANAH FIS++ GYDT
Sbjct: 1106 LRQQMGLVGQEPVLFNDTIRANIAYGKGSEEAATESEIIAAAELANAHKFISSIQQGYDT 1165
Query: 1140 SVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTT 1199
VGERG QLSGGQKQR+AIAR ++K PKILLLDEATSALDAESER+VQ+ALDRV VNRTT
Sbjct: 1166 VVGERGIQLSGGQKQRVAIARAIVKEPKILLLDEATSALDAESERVVQDALDRVMVNRTT 1225
Query: 1200 VVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGVYASLVALHSSA 1250
+VVAHRL+TI+ AD IAV+KNG +AEKG H+ L++I GVYASLV LH +A
Sbjct: 1226 IVVAHRLSTIKNADVIAVVKNGVIAEKGTHETLIKIEGGVYASLVQLHMTA 1276
>At3g62150 P-glycoprotein-like proetin
Length = 1292
Score = 1456 bits (3768), Expect = 0.0
Identities = 759/1278 (59%), Positives = 959/1278 (74%), Gaps = 94/1278 (7%)
Query: 23 DNKTKQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSS-NPA 81
D KTK VPF+ LF FAD D+ LMI+GTI AV NGL P+MT+ G+VI+ FG + N +
Sbjct: 57 DEKTKT-VPFHKLFAFADSFDIILMILGTIGAVGNGLGFPIMTILFGDVIDVFGQNQNSS 115
Query: 82 DAIKQVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDT 141
D +++KV+L FVYL +G+ +A+ LQV+ WM++GERQA RIRSLYL+TIL+QDIAFFD
Sbjct: 116 DVSDKIAKVALKFVYLGLGTLVAALLQVSGWMISGERQAGRIRSLYLQTILRQDIAFFDV 175
Query: 142 ETNTGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPC 201
ETNTGEV+GRMSGDT+LIQ+AMGEKVGK QL S F GGFV+AF +GW L +V+++ +P
Sbjct: 176 ETNTGEVVGRMSGDTVLIQDAMGEKVGKAIQLVSTFIGGFVIAFTEGWLLTLVMVSSIPL 235
Query: 202 VAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIA 261
+ ++GA ++IV++KM+SRGQ +YA+A VV+QTVG+IRTVASFTGEK+AI YN + A
Sbjct: 236 LVMSGAALAIVISKMASRGQTSYAKAAVVVEQTVGSIRTVASFTGEKQAISNYNKHLVSA 295
Query: 262 YTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI- 320
Y V +G +G G+G L + FCTY LA+WYG K+++EKGY GG V+ +I A++TG +
Sbjct: 296 YRAGVFEGASTGLGLGTLNIVIFCTYALAVWYGGKMILEKGYTGGQVLIIIFAVLTGSMS 355
Query: 321 -------------------------------------------IKGDIELRDVSFRYPAR 337
I+GDIEL +V+F YPAR
Sbjct: 356 LGQASPCLSAFAAGQAAAYKMFEAIKRKPEIDASDTTGKVLDDIRGDIELNNVNFSYPAR 415
Query: 338 PDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLR 397
P+ QIF GFSL + SG+T ALVGQSGSGKSTV+SL+ERFYDP +GEV IDG+NLK QL+
Sbjct: 416 PEEQIFRGFSLSISSGSTVALVGQSGSGKSTVVSLIERFYDPQSGEVRIDGINLKEFQLK 475
Query: 398 WIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMA 457
WIR +IGLVSQEP+LFT+SI+ENIAYGKE AT EEI A LANA KFIDKLPQGLDTM
Sbjct: 476 WIRSKIGLVSQEPVLFTSSIKENIAYGKENATVEEIRKATELANASKFIDKLPQGLDTMV 535
Query: 458 GQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVV 517
G++GTQLSGGQKQRIA+ARAILK+P+ILLLDEATSALDAESERIVQEAL++I++ RTTVV
Sbjct: 536 GEHGTQLSGGQKQRIAVARAILKDPRILLLDEATSALDAESERIVQEALDRIMVNRTTVV 595
Query: 518 VAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEGSRKSEA 577
VAHRL+T+RNAD+IAV+ QGKIVE+G+HS L DP+GAYSQLIRLQE + E S +
Sbjct: 596 VAHRLSTVRNADMIAVIHQGKIVEKGSHSELLRDPEGAYSQLIRLQEDTKQTEDSTDEQ- 654
Query: 578 DKLGDNLNIDSHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQLDN 637
L+++S M SS +++S RS+S+ SS S S+ G I ++ + D
Sbjct: 655 -----KLSMES-MKRSSLRKSSLSRSLSKRSS-----SFSMFGFPAGIDTNNEAIPEKDI 703
Query: 638 K-----KKPKVSIWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPPE 692
K K+ KVS +R+A LNKPEIP+++LG+IAA++NGV+ PIFG L S+VI F+KPPE
Sbjct: 704 KVSTPIKEKKVSFFRVAALNKPEIPMLILGSIAAVLNGVILPIFGILISSVIKAFFKPPE 763
Query: 693 QQRKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDD 752
Q + ++RFW+++F+ LG+ ++V+ P Q FF IAG KL++RIRS+ FEK+V E+ WFD+
Sbjct: 764 QLKSDTRFWAIIFMLLGVASMVVFPAQTIFFSIAGCKLVQRIRSMCFEKVVRMEVGWFDE 823
Query: 753 PSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQN 812
+S SGA+GARLS DA+TV+ LVGD +A VQN
Sbjct: 824 TENS---------------------------SGAIGARLSADAATVRGLVGDALAQTVQN 856
Query: 813 ISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVAND 872
+++V AGLVIAF A+W LAFIVL + P+I + G + MKF+ GFSADAK EASQVAND
Sbjct: 857 LASVTAGLVIAFVASWQLAFIVLAMLPLIGLNGYIYMKFMVGFSADAK----EASQVAND 912
Query: 873 AVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYI 932
AV SIRTVASFCAE KVM MY KKC GP + G+R G+VSGIGFG SF VL+ + A FY
Sbjct: 913 AVGSIRTVASFCAEEKVMKMYKKKCEGPMRTGIRQGIVSGIGFGVSFFVLFSSYAASFYA 972
Query: 933 GSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDID 992
G+ LV GK TF VFRVFFALTM A+A+SQ+++L+PD++KA ++AASIF +ID + ID
Sbjct: 973 GARLVDDGKTTFDSVFRVFFALTMAAVAISQSSSLSPDSSKASNAAASIFAVIDRESKID 1032
Query: 993 SSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTV 1052
S +G + V GDIEL+H++F YP+RPD+QIF+DL LSI + KTIALVGESGSGKSTV
Sbjct: 1033 PSDESGRVLDNVKGDIELRHISFKYPSRPDVQIFQDLCLSIRAGKTIALVGESGSGKSTV 1092
Query: 1053 ISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGA 1112
I+LL+RFYDP+SG+I LDGV++KT +L WLRQQ GLV QEP+LFNE+IRANI YGK G A
Sbjct: 1093 IALLQRFYDPDSGQITLDGVEIKTLQLKWLRQQTGLVSQEPVLFNETIRANIAYGKGGDA 1152
Query: 1113 TEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLD 1172
TE EI++AA +NAH FIS L GYDT VGERG QLSGGQKQR+AIAR ++K+PK+LLLD
Sbjct: 1153 TETEIVSAAELSNAHGFISGLQQGYDTMVGERGVQLSGGQKQRVAIARAIVKDPKVLLLD 1212
Query: 1173 EATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDEL 1232
EATSALDAESER+VQ+ALDRV VNRTTVVVAHRL+TI+ AD IAV+KNG + EKG+H+ L
Sbjct: 1213 EATSALDAESERVVQDALDRVMVNRTTVVVAHRLSTIKNADVIAVVKNGVIVEKGKHETL 1272
Query: 1233 MRITDGVYASLVALHSSA 1250
+ I DGVYASLV LH SA
Sbjct: 1273 INIKDGVYASLVQLHLSA 1290
Score = 440 bits (1132), Expect = e-123
Identities = 244/603 (40%), Positives = 369/603 (60%), Gaps = 54/603 (8%)
Query: 7 VHDNSSSSPTQQHGIRDNKTKQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTL 66
+ N+ + P + + ++KV F+ + + ++ ++I+G+I+AV NG+ P+ +
Sbjct: 691 IDTNNEAIPEKDIKVSTPIKEKKVSFFRVAAL-NKPEIPMLILGSIAAVLNGVILPIFGI 749
Query: 67 FLGNVINAFGSSNPADAIKQVSKV-SLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRS 125
+ +VI AF P + +K ++ +++F+ L + S + Q + + G + RIRS
Sbjct: 750 LISSVIKAF--FKPPEQLKSDTRFWAIIFMLLGVASMVVFPAQTIFFSIAGCKLVQRIRS 807
Query: 126 LYLKTILQQDIAFFD-TETNTGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMA 184
+ + +++ ++ +FD TE ++G + R+S D ++ +G+ + + Q ++ G V+A
Sbjct: 808 MCFEKVVRMEVGWFDETENSSGAIGARLSADAATVRGLVGDALAQTVQNLASVTAGLVIA 867
Query: 185 FIKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASF 244
F+ W+LA ++LA +P + + G M S+ + EA V + VG+IRTVASF
Sbjct: 868 FVASWQLAFIVLAMLPLIGLNGYIYMKFMVGFSADAK----EASQVANDAVGSIRTVASF 923
Query: 245 TGEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYN 304
E+K ++ Y K + T ++QGIVSG G G+ F+ F +Y + + G++LV +
Sbjct: 924 CAEEKVMKMYKKKCEGPMRTGIRQGIVSGIGFGVSFFVLFSSYAASFYAGARLVDDGKTT 983
Query: 305 GGTVMTVIIALMTGGI-------------------------------------------- 320
+V V AL +
Sbjct: 984 FDSVFRVFFALTMAAVAISQSSSLSPDSSKASNAAASIFAVIDRESKIDPSDESGRVLDN 1043
Query: 321 IKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPD 380
+KGDIELR +SF+YP+RPDVQIF L + +G T ALVG+SGSGKSTVI+LL+RFYDPD
Sbjct: 1044 VKGDIELRHISFKYPSRPDVQIFQDLCLSIRAGKTIALVGESGSGKSTVIALLQRFYDPD 1103
Query: 381 AGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEG-ATDEEITTAITL 439
+G++ +DGV +K LQL+W+R+Q GLVSQEP+LF +IR NIAYGK G AT+ EI +A L
Sbjct: 1104 SGQITLDGVEIKTLQLKWLRQQTGLVSQEPVLFNETIRANIAYGKGGDATETEIVSAAEL 1163
Query: 440 ANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESE 499
+NA FI L QG DTM G+ G QLSGGQKQR+AIARAI+K+PK+LLLDEATSALDAESE
Sbjct: 1164 SNAHGFISGLQQGYDTMVGERGVQLSGGQKQRVAIARAIVKDPKVLLLDEATSALDAESE 1223
Query: 500 RIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQL 559
R+VQ+AL+++++ RTTVVVAHRL+TI+NAD+IAVV+ G IVE+G H L DG Y+ L
Sbjct: 1224 RVVQDALDRVMVNRTTVVVAHRLSTIKNADVIAVVKNGVIVEKGKHETLINIKDGVYASL 1283
Query: 560 IRL 562
++L
Sbjct: 1284 VQL 1286
>At1g02530 hypothetical protein
Length = 1273
Score = 1437 bits (3720), Expect = 0.0
Identities = 751/1302 (57%), Positives = 941/1302 (71%), Gaps = 94/1302 (7%)
Query: 9 DNSSSSPTQQHGIRDNKTKQK---VPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMT 65
D + + H +KT +K VP Y LF FAD DV LMI G++ A+ NG+ PLMT
Sbjct: 4 DGAGEGDSVSHEHSTSKTDEKAKTVPLYKLFAFADSFDVFLMICGSLGAIGNGVCLPLMT 63
Query: 66 LFLGNVINAFGSS-NPADAIKQVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIR 124
L G++I++FG + N D + VSKV L FVYL +G A+FLQV CWM+TGERQAA+IR
Sbjct: 64 LLFGDLIDSFGKNQNNKDIVDVVSKVCLKFVYLGLGRLGAAFLQVACWMITGERQAAKIR 123
Query: 125 SLYLKTILQQDIAFFDTETNTGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMA 184
S YLKTIL+QDI FFD ETNTGEV+GRMSGDT+ IQ+AMGEKVGKF QL S F GGF +A
Sbjct: 124 SNYLKTILRQDIGFFDVETNTGEVVGRMSGDTVHIQDAMGEKVGKFIQLVSTFVGGFALA 183
Query: 185 FIKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASF 244
F KGW L +V+L +P +A+AGA M++++ + SSRGQ AYA+A VV+QT+G+IRTVASF
Sbjct: 184 FAKGWLLTLVMLTSIPFLAMAGAAMALLVTRASSRGQAAYAKAATVVEQTIGSIRTVASF 243
Query: 245 TGEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYN 304
TGEK+AI Y I AY + ++QG +G G+G++ ++ F +Y LA+W+G K+++EKGY
Sbjct: 244 TGEKQAINSYKKYITSAYKSSIQQGFSTGLGLGVMIYVFFSSYALAIWFGGKMILEKGYT 303
Query: 305 GGTVMTVIIALMTG--------------------------------------------GI 320
GG+V+ VII ++ G G
Sbjct: 304 GGSVINVIIIVVAGSMSLGQTSPCVTAFAAGQAAAYKMFETIKRKPLIDAYDVNGKVLGD 363
Query: 321 IKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPD 380
I+GDIEL+DV F YPARPD +IFDGFSLF+PSG T ALVG+SGSGKSTVI+L+ERFYDP
Sbjct: 364 IRGDIELKDVHFSYPARPDEEIFDGFSLFIPSGATAALVGESGSGKSTVINLIERFYDPK 423
Query: 381 AGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLA 440
AGEVLIDG+NLK QL+WIR +IGLV QEP+LF++SI ENIAYGKE AT +EI A LA
Sbjct: 424 AGEVLIDGINLKEFQLKWIRSKIGLVCQEPVLFSSSIMENIAYGKENATLQEIKVATELA 483
Query: 441 NAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESER 500
NA KFI+ LPQGLDT G++GTQLSGGQKQRIAIARAILK+P++LLLDEATSALD ESER
Sbjct: 484 NAAKFINNLPQGLDTKVGEHGTQLSGGQKQRIAIARAILKDPRVLLLDEATSALDTESER 543
Query: 501 IVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLI 560
+VQEAL+++++ RTTVVVAHRL+T+RNAD+IAV+ GK+VE+G+HS L D GAYSQLI
Sbjct: 544 VVQEALDRVMVNRTTVVVAHRLSTVRNADMIAVIHSGKMVEKGSHSELLKDSVGAYSQLI 603
Query: 561 RLQE---GDNEAEGSRKSEADKLGDNLNIDSHMAGSSTQRTSFVRSISQTSSVSHRHSQS 617
R QE G + S + NLNI + S +SF S S HS +
Sbjct: 604 RCQEINKGHDAKPSDMASGSSFRNSNLNISREGSVISGGTSSFGNS-------SRHHSLN 656
Query: 618 LRGL-------SGEIVESDIEQGQLDNKKKPKVSIWRLAKLNKPEIPVILLGAIAAIVNG 670
+ GL SG E G + KVS+ R+A LNKPEIPV+LLG + A +NG
Sbjct: 657 VLGLFAGLDLGSGSQRVGQEETGTTSQEPLRKVSLTRIAALNKPEIPVLLLGTVVAAING 716
Query: 671 VVFPIFGFLFSAVISMFYKPPEQQRKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKL 730
+FP+FG L S VI F+KP +Q +K+SRFW+++FV LG+ +L++ P Q + F +AGGKL
Sbjct: 717 AIFPLFGILISRVIEAFFKPADQLKKDSRFWAIIFVALGVTSLIVSPSQMYLFAVAGGKL 776
Query: 731 IERIRSLTFEKIVHQEISWFDDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGAR 790
I RI+S+ FEK VH E+SWFD+P +S SG +GAR
Sbjct: 777 IRRIQSMCFEKAVHMEVSWFDEPENS---------------------------SGTMGAR 809
Query: 791 LSIDASTVKSLVGDTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMK 850
LS DA+ +++LVGD ++L VQN ++ +GL+IAFTA+W LA I+LV+ P+I + G +Q+K
Sbjct: 810 LSTDAALIRALVGDALSLAVQNAASAASGLIIAFTASWELALIILVMLPLIGINGFLQVK 869
Query: 851 FLKGFSADAKVMYEEASQVANDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLV 910
F+KGFSADAK YEEASQVANDAV SIRTVASFCAE KVM MY+K+C GP K GV+ G +
Sbjct: 870 FMKGFSADAKSKYEEASQVANDAVGSIRTVASFCAEEKVMQMYNKQCEGPIKDGVKQGFI 929
Query: 911 SGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPD 970
SG+GFG SF +L+C A FY + LV+ GK TF +VF+VFFALTM AI +SQ++T APD
Sbjct: 930 SGLGFGFSFFILFCVYATSFYAAARLVEDGKTTFIDVFQVFFALTMAAIGISQSSTFAPD 989
Query: 971 TNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLS 1030
++KAK +AASIF IID K IDSS G E V GDIEL+H++F YP RP IQIF+DL
Sbjct: 990 SSKAKVAAASIFAIIDRKSKIDSSDETGTVLENVKGDIELRHLSFTYPARPGIQIFRDLC 1049
Query: 1031 LSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVG 1090
L+I + KT+ALVGESGSGKSTVISLL+RFYDP+SG+I LDGV+LK +L WLRQQMGLVG
Sbjct: 1050 LTIRAGKTVALVGESGSGKSTVISLLQRFYDPDSGQITLDGVELKKLQLKWLRQQMGLVG 1109
Query: 1091 QEPILFNESIRANIGYGK--EGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQL 1148
QEP+LFN++IRANI YGK E ATE EIIAAA ANAH FIS++ GYDT VGE+G QL
Sbjct: 1110 QEPVLFNDTIRANIAYGKGSEEAATESEIIAAAELANAHKFISSIQQGYDTVVGEKGIQL 1169
Query: 1149 SGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTT 1208
SGGQKQR+AIAR ++K PKILLLDEATSALDAESER+VQ+ALDRV VNRTTVVVAHRL+T
Sbjct: 1170 SGGQKQRVAIARAIVKEPKILLLDEATSALDAESERLVQDALDRVIVNRTTVVVAHRLST 1229
Query: 1209 IRGADTIAVIKNGAVAEKGRHDELMRITDGVYASLVALHSSA 1250
I+ AD IA++KNG +AE G H+ L++I GVYASLV LH +A
Sbjct: 1230 IKNADVIAIVKNGVIAENGTHETLIKIDGGVYASLVQLHMTA 1271
>At4g01830 putative P-glycoprotein-like protein
Length = 1230
Score = 1409 bits (3646), Expect = 0.0
Identities = 724/1267 (57%), Positives = 932/1267 (73%), Gaps = 97/1267 (7%)
Query: 30 VPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFG-SSNPADAIKQVS 88
VPFY LF F+D DV LMI+G+I A+ANG+ SPLMTL G +I+A G + N + +++VS
Sbjct: 13 VPFYKLFFFSDSTDVLLMIVGSIGAIANGVCSPLMTLLFGELIDAMGPNQNNEEIVERVS 72
Query: 89 KVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTETNTGEV 148
KV L VYL +G+ A+FLQV CWM+TGERQAARIRSLYLKTIL+QDI FFD E TGEV
Sbjct: 73 KVCLSLVYLGLGALGAAFLQVACWMITGERQAARIRSLYLKTILRQDIGFFDVEMTTGEV 132
Query: 149 IGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAVAGAF 208
+GRMSGDT+LI +AMGEKVGKF QL S F GGFV+AF++GW L +V+L +P +A++GA
Sbjct: 133 VGRMSGDTVLILDAMGEKVGKFIQLISTFVGGFVIAFLRGWLLTLVMLTSIPLLAMSGAA 192
Query: 209 MSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTTMVKQ 268
++I++ + SS+ Q AYA+A NVV+QT+G+IRTVASFTGEK+A+ Y I +AY + VKQ
Sbjct: 193 IAIIVTRASSQEQAAYAKASNVVEQTLGSIRTVASFTGEKQAMSSYKELINLAYKSNVKQ 252
Query: 269 GIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI-------- 320
G V+G G+G++ + F TY L W+G ++++ KGY GG V+ V++ +++ I
Sbjct: 253 GFVTGLGLGVMFLVFFSTYALGTWFGGEMILRKGYTGGAVINVMVTVVSSSIALGQASPC 312
Query: 321 ------------------------------------IKGDIELRDVSFRYPARPDVQIFD 344
I+G+IELRDV F YPARP ++F
Sbjct: 313 LTAFTAGKAAAYKMFETIEREPLIDTFDLNGKVLEDIRGEIELRDVCFSYPARPKEEVFG 372
Query: 345 GFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIG 404
GFSL +PSGTTTALVG+SGSGKSTVISL+ERFYDP++G+VLIDGV+LK QL+WIR +IG
Sbjct: 373 GFSLLIPSGTTTALVGESGSGKSTVISLIERFYDPNSGQVLIDGVDLKEFQLKWIRGKIG 432
Query: 405 LVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQL 464
LVSQEP+LF++SI ENI YGKEGAT EEI A LANA KFIDKLP GL+T+ G++GTQL
Sbjct: 433 LVSQEPVLFSSSIMENIGYGKEGATVEEIQAASKLANAAKFIDKLPLGLETLVGEHGTQL 492
Query: 465 SGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLTT 524
SGGQKQRIAIARAILK+P+ILLLDEATSALDAESER+VQEAL++I++ RTTV+VAHRL+T
Sbjct: 493 SGGQKQRIAIARAILKDPRILLLDEATSALDAESERVVQEALDRIMVNRTTVIVAHRLST 552
Query: 525 IRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEGSRKSEADKLGDNL 584
+RNADIIAV+ +GKIVE G+HS L D +GAYSQL+RLQE + E++ S
Sbjct: 553 VRNADIIAVIHRGKIVEEGSHSELLKDHEGAYSQLLRLQEINKESKRLEIS--------- 603
Query: 585 NIDSHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQLDNKKKPKVS 644
D ++ S++ + R + SV L L+G+ + ++ + KVS
Sbjct: 604 --DGSISSGSSRGNNSTRQDDDSFSV-------LGLLAGQ------DSTKMSQELSQKVS 648
Query: 645 IWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKESRFWSLL 704
R+A LNKPEIP+++LG + VNG +FPIFG LF+ VI F+K P + +++SRFWS++
Sbjct: 649 FTRIAALNKPEIPILILGTLVGAVNGTIFPIFGILFAKVIEAFFKAPHELKRDSRFWSMI 708
Query: 705 FVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQNVC 764
FV LG+ +++ P N+ F IAGG+LI RIRS+ FEK+VH E+ WFD+P +S
Sbjct: 709 FVLLGVAAVIVYPTTNYLFAIAGGRLIRRIRSMCFEKVVHMEVGWFDEPGNS-------- 760
Query: 765 YFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIAF 824
SGA+GARLS DA+ +++LVGD++ L V+N+++++ GL+IAF
Sbjct: 761 -------------------SGAMGARLSADAALIRTLVGDSLCLSVKNVASLVTGLIIAF 801
Query: 825 TANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFC 884
TA+W +A I+LV+ P I + G +Q+KF+KGFSADAK YEEASQVANDAV SIRTVASFC
Sbjct: 802 TASWEVAIIILVIIPFIGINGYIQIKFMKGFSADAKAKYEEASQVANDAVGSIRTVASFC 861
Query: 885 AESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATF 944
AE KVM+MY K+C K G++ GL+SG+GFG SF VLY A FY+G+ LV+ G+ F
Sbjct: 862 AEEKVMEMYKKRCEDTIKSGIKQGLISGVGFGISFFVLYSVYASCFYVGARLVKAGRTNF 921
Query: 945 TEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETV 1004
+VF+VF ALT+TA+ +SQ ++ APD++K K +A SIF IID IDS +G+ E V
Sbjct: 922 NDVFQVFLALTLTAVGISQASSFAPDSSKGKGAAVSIFRIIDRISKIDSRDESGMVLENV 981
Query: 1005 VGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNS 1064
GDIEL H++F Y TRPD+Q+F+DL LSI + +T+ALVGESGSGKSTVISLL+RFYDP+S
Sbjct: 982 KGDIELCHISFTYQTRPDVQVFRDLCLSIRAGQTVALVGESGSGKSTVISLLQRFYDPDS 1041
Query: 1065 GRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEG-GATEDEIIAAANA 1123
G I LDGV+LK RL WLRQQMGLVGQEP+LFN++IRANI YGK G ATE EIIAA+
Sbjct: 1042 GHITLDGVELKKLRLKWLRQQMGLVGQEPVLFNDTIRANIAYGKGGEEATEAEIIAASEL 1101
Query: 1124 ANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESE 1183
ANAH FIS++ GYDT VGERG QLSGGQKQR+AIAR ++K PKILLLDEATSALDAESE
Sbjct: 1102 ANAHRFISSIQKGYDTVVGERGIQLSGGQKQRVAIARAIVKEPKILLLDEATSALDAESE 1161
Query: 1184 RIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGVYASL 1243
R+VQ+ALDRV VNRTT+VVAHRL+TI+ AD IAV+KNG +AEKG H+ L+ I GVYASL
Sbjct: 1162 RVVQDALDRVMVNRTTIVVAHRLSTIKNADVIAVVKNGVIAEKGTHETLINIEGGVYASL 1221
Query: 1244 VALHSSA 1250
V LH +A
Sbjct: 1222 VQLHINA 1228
>At4g01820 P-glycoprotein-like protein pgp3
Length = 1229
Score = 1402 bits (3629), Expect = 0.0
Identities = 729/1274 (57%), Positives = 934/1274 (73%), Gaps = 94/1274 (7%)
Query: 23 DNKTKQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSSNP-A 81
+ KTK VPFY LF+F+D DV LMI+G+I A+ NG+ PLMTL G++I++ G +
Sbjct: 2 EEKTKT-VPFYKLFSFSDSTDVLLMIVGSIGAIGNGVGFPLMTLLFGDLIDSIGQNQSNK 60
Query: 82 DAIKQVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDT 141
D ++ VSKV L FVYL +G+ A+FLQV CWM+TGERQAARIRSLYLKTIL+QDI FFD
Sbjct: 61 DIVEIVSKVCLKFVYLGLGTLGAAFLQVACWMITGERQAARIRSLYLKTILRQDIGFFDV 120
Query: 142 ETNTGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPC 201
ET+TGEV+GRMSGDT+LI EAMGEKVGKF QL + F GGFV+AF+KGW L +V+L +P
Sbjct: 121 ETSTGEVVGRMSGDTVLILEAMGEKVGKFIQLIATFVGGFVLAFVKGWLLTLVMLVSIPL 180
Query: 202 VAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIA 261
+A+AGA M I++ + SSR Q AYA+A VV+QT+G+IRTVASFTGEK+A++ Y I +A
Sbjct: 181 LAIAGAAMPIIVTRASSREQAAYAKASTVVEQTLGSIRTVASFTGEKQAMKSYREFINLA 240
Query: 262 YTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI- 320
Y VKQG G G+G++ F+ FC+Y LA+W+G +++++KGY GG V+ V++ ++ +
Sbjct: 241 YRASVKQGFSMGLGLGVVFFVFFCSYALAIWFGGEMILKKGYTGGEVVNVMVTVVASSMS 300
Query: 321 -------------------------------------------IKGDIELRDVSFRYPAR 337
I+G+IELRDV F YPAR
Sbjct: 301 LGQTTPCLTAFAAGKAAAYKMFETIERKPSIDAFDLNGKVLEDIRGEIELRDVCFSYPAR 360
Query: 338 PDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLR 397
P ++F GFSL +PSG T ALVG+SGSGKS+VISL+ERFYDP +G VLIDGVNLK QL+
Sbjct: 361 PMEEVFGGFSLLIPSGATAALVGESGSGKSSVISLIERFYDPSSGSVLIDGVNLKEFQLK 420
Query: 398 WIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMA 457
WIR +IGLVSQEP+LF++SI ENI YGKE AT EEI A LANA FIDKLP+GL+T+
Sbjct: 421 WIRGKIGLVSQEPVLFSSSIMENIGYGKENATVEEIQAAAKLANAANFIDKLPRGLETLV 480
Query: 458 GQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVV 517
G++GTQLSGGQKQRIAIARAILK+P+ILLLDEATSALDAESER+VQEAL+++++ RTTV+
Sbjct: 481 GEHGTQLSGGQKQRIAIARAILKDPRILLLDEATSALDAESERVVQEALDRVMMSRTTVI 540
Query: 518 VAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEGSRKSEA 577
VAHRL+T+RNAD+IAV+ +GKIVE G+HS L D +GAY+QLIRLQ+ E + R +
Sbjct: 541 VAHRLSTVRNADMIAVIHRGKIVEEGSHSELLKDHEGAYAQLIRLQKIKKEPK--RLESS 598
Query: 578 DKLGDNLNIDSHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQLDN 637
++L D S GSS R+I + V S S+ GL G ++I + Q N
Sbjct: 599 NELRDR----SINRGSS-------RNIR--TRVHDDDSVSVLGLLGRQENTEISREQSRN 645
Query: 638 KKKPKVSIWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKE 697
VSI R+A LNKPE +++LG + VNG +FPIFG LF+ VI F+KPP +++
Sbjct: 646 -----VSITRIAALNKPETTILILGTLLGAVNGTIFPIFGILFAKVIEAFFKPPHDMKRD 700
Query: 698 SRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSR 757
SRFWS++FV LG+ +L++ P+ + F +AGG+LI+RIR + FEK+VH E+ WFDDP +S
Sbjct: 701 SRFWSMIFVLLGVASLIVYPMHTYLFAVAGGRLIQRIRVMCFEKVVHMEVGWFDDPENS- 759
Query: 758 YVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVI 817
SG +G+RLS DA+ +K+LVGD+++L V+N + +
Sbjct: 760 --------------------------SGTIGSRLSADAALIKTLVGDSLSLSVKNAAAAV 793
Query: 818 AGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSI 877
+GL+IAFTA+W LA I+LV+ P+I + G +Q+KF+KGF+ADAK YEEASQVANDAV SI
Sbjct: 794 SGLIIAFTASWKLAVIILVMIPLIGINGYLQIKFIKGFTADAKAKYEEASQVANDAVGSI 853
Query: 878 RTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLV 937
RTVASFCAE KVM+MY K+C K G++ GL+SG+GFG SF VLY A FY+G+ LV
Sbjct: 854 RTVASFCAEEKVMEMYKKRCEDTIKSGIKQGLISGVGFGISFFVLYSVYASCFYVGARLV 913
Query: 938 QHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNA 997
+ G+ F +VF+VF ALTMTAI +SQ ++ APD++KAK +AASIF IID K IDS +
Sbjct: 914 KAGRTNFNDVFQVFLALTMTAIGISQASSFAPDSSKAKGAAASIFGIIDGKSMIDSRDES 973
Query: 998 GVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLE 1057
G+ E V GDIEL H++F Y TRPD+QIF+DL +I + +T+ALVGESGSGKSTVISLL+
Sbjct: 974 GLVLENVKGDIELCHISFTYQTRPDVQIFRDLCFAIRAGQTVALVGESGSGKSTVISLLQ 1033
Query: 1058 RFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGG-ATEDE 1116
RFYDP+SG I LD V+LK +L W+RQQMGLVGQEP+LFN++IR+NI YGK G A+E E
Sbjct: 1034 RFYDPDSGHITLDRVELKKLQLKWVRQQMGLVGQEPVLFNDTIRSNIAYGKGGDEASEAE 1093
Query: 1117 IIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATS 1176
IIAAA ANAH FIS++ GYDT VGERG QLSGGQKQR+AIAR ++K PKILLLDEATS
Sbjct: 1094 IIAAAELANAHGFISSIQQGYDTVVGERGIQLSGGQKQRVAIARAIVKEPKILLLDEATS 1153
Query: 1177 ALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRIT 1236
ALDAESER+VQ+ALDRV VNRTTVVVAHRL+TI+ AD IAV+KNG + EKG H+ L+ I
Sbjct: 1154 ALDAESERVVQDALDRVMVNRTTVVVAHRLSTIKNADVIAVVKNGVIVEKGTHETLINIE 1213
Query: 1237 DGVYASLVALHSSA 1250
GVYASLV LH SA
Sbjct: 1214 GGVYASLVQLHISA 1227
>At3g28860 P-glycoprotein, putative
Length = 1252
Score = 1004 bits (2597), Expect = 0.0
Identities = 535/1278 (41%), Positives = 790/1278 (60%), Gaps = 97/1278 (7%)
Query: 23 DNKTKQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSSNPA- 81
+ K +Q +PF+ LF+FAD D LM +G++ A+ +G + P+ L G ++N FG +
Sbjct: 17 EKKKEQSLPFFKLFSFADKFDYLLMFVGSLGAIVHGSSMPVFFLLFGQMVNGFGKNQMDL 76
Query: 82 -DAIKQVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFD 140
+ +VS+ SL FVYL + +S+ ++ CWM +GERQ A +R YL+ +L+QD+ FFD
Sbjct: 77 HQMVHEVSRYSLYFVYLGLVVCFSSYAEIACWMYSGERQVAALRKKYLEAVLKQDVGFFD 136
Query: 141 TETNTGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVP 200
T+ TG+++ +S DT+L+Q+A+ EKVG F S F G V+ F+ W+LA++ +A +P
Sbjct: 137 TDARTGDIVFSVSTDTLLVQDAISEKVGNFIHYLSTFLAGLVVGFVSAWKLALLSVAVIP 196
Query: 201 CVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKI 260
+A AG + + ++S+ + +YA AG + +Q + +RTV S+ GE KA+ Y+ I+
Sbjct: 197 GIAFAGGLYAYTLTGITSKSRESYANAGVIAEQAIAQVRTVYSYVGESKALNAYSDAIQY 256
Query: 261 AYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI 320
K G+ G G+G IA ++ L WY + +GG T I + + GG+
Sbjct: 257 TLKLGYKAGMAKGLGLGCTYGIACMSWALVFWYAGVFIRNGQTDGGKAFTAIFSAIVGGM 316
Query: 321 --------------------------------------------IKGDIELRDVSFRYPA 336
+ G+IE +DV+F YP+
Sbjct: 317 SLGQSFSNLGAFSKGKAAGYKLMEIINQRPTIIQDPLDGKCLDQVHGNIEFKDVTFSYPS 376
Query: 337 RPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQL 396
RPDV IF F++F PSG T A+VG SGSGKSTV+SL+ERFYDP++G++L+DGV +K LQL
Sbjct: 377 RPDVMIFRNFNIFFPSGKTVAVVGGSGSGKSTVVSLIERFYDPNSGQILLDGVEIKTLQL 436
Query: 397 RWIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTM 456
+++REQIGLV+QEP LF T+I ENI YGK AT E+ A + ANA FI LP+G DT
Sbjct: 437 KFLREQIGLVNQEPALFATTILENILYGKPDATMVEVEAAASAANAHSFITLLPKGYDTQ 496
Query: 457 AGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTV 516
G+ G QLSGGQKQRIAIARA+LK+PKILLLDEATSALDA SE IVQEAL+++++ RTTV
Sbjct: 497 VGERGVQLSGGQKQRIAIARAMLKDPKILLLDEATSALDASSESIVQEALDRVMVGRTTV 556
Query: 517 VVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQE----GDNEAEGS 572
VVAHRL TIRN D IAV+QQG++VE GTH L + GAY+ LIR QE D +
Sbjct: 557 VVAHRLCTIRNVDSIAVIQQGQVVETGTHEEL-IAKSGAYASLIRFQEMVGTRDFSNPST 615
Query: 573 RKSEADKLGDNLNIDSHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQ 632
R++ + +L +L+ S + R+ +R++S + S G G I I
Sbjct: 616 RRTRSTRLSHSLS-----TKSLSLRSGSLRNLSYSYST---------GADGRI--EMISN 659
Query: 633 GQLDNK-KKPKVSIWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPP 691
+ D K + P+ +RL KLN PE P ++GA+ +I++G + P F + S +I +FY
Sbjct: 660 AETDRKTRAPENYFYRLLKLNSPEWPYSIMGAVGSILSGFIGPTFAIVMSNMIEVFYYTD 719
Query: 692 -EQQRKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWF 750
+ ++++ + +++G GL + +Q++FF I G L R+R + I+ E+ WF
Sbjct: 720 YDSMERKTKEYVFIYIGAGLYAVGAYLIQHYFFSIMGENLTTRVRRMMLSAILRNEVGWF 779
Query: 751 DDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIV 810
D+ H+ S + ARL+ DA+ VKS + + +++I+
Sbjct: 780 DEDEHN---------------------------SSLIAARLATDAADVKSAIAERISVIL 812
Query: 811 QNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVA 870
QN+++++ ++AF W ++ ++L P++++ Q LKGF+ D + + S +A
Sbjct: 813 QNMTSLLTSFIVAFIVEWRVSLLILGTFPLLVLANFAQQLSLKGFAGDTAKAHAKTSMIA 872
Query: 871 NDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIF 930
+ VS+IRTVA+F A+SK++ ++ + P K+ + SG FG S L LY + A I
Sbjct: 873 GEGVSNIRTVAAFNAQSKILSLFCHELRVPQKRSLYRSQTSGFLFGLSQLALYGSEALIL 932
Query: 931 YIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPD 990
+ G+ LV G +TF++V +VF L +TA +V++T +LAP+ + ++ S+F ++D +
Sbjct: 933 WYGAHLVSKGVSTFSKVIKVFVVLVITANSVAETVSLAPEIIRGGEAVGSVFSVLDRQTR 992
Query: 991 IDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKS 1050
ID ET+ GDIE +HV+F YP+RPD+ +F+D +L I + + ALVG SGSGKS
Sbjct: 993 IDPDDADADPVETIRGDIEFRHVDFAYPSRPDVMVFRDFNLRIRAGHSQALVGASGSGKS 1052
Query: 1051 TVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEG 1110
+VI+++ERFYDP +G++++DG D++ L LR ++GLV QEP LF +I NI YGK+
Sbjct: 1053 SVIAMIERFYDPLAGKVMIDGKDIRRLNLKSLRLKIGLVQQEPALFAATIFDNIAYGKD- 1111
Query: 1111 GATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILL 1170
GATE E+I AA AANAH FIS LP+GY T VGERG QLSGGQKQRIAIAR +LKNP +LL
Sbjct: 1112 GATESEVIDAARAANAHGFISGLPEGYKTPVGERGVQLSGGQKQRIAIARAVLKNPTVLL 1171
Query: 1171 LDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHD 1230
LDEATSALDAESE ++QEAL+R+ RTTVVVAHRL+TIRG D I VI++G + E+G H
Sbjct: 1172 LDEATSALDAESECVLQEALERLMRGRTTVVVAHRLSTIRGVDCIGVIQDGRIVEQGSHS 1231
Query: 1231 ELMRITDGVYASLVALHS 1248
EL+ +G Y+ L+ L +
Sbjct: 1232 ELVSRPEGAYSRLLQLQT 1249
Score = 430 bits (1105), Expect = e-120
Identities = 249/615 (40%), Positives = 365/615 (58%), Gaps = 34/615 (5%)
Query: 634 QLDNKKKPKVSIWRLAKL-NKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPP- 691
+ + KK+ + ++L +K + ++ +G++ AIV+G P+F LF +++ F K
Sbjct: 15 EAEKKKEQSLPFFKLFSFADKFDYLLMFVGSLGAIVHGSSMPVFFLLFGQMVNGFGKNQM 74
Query: 692 --EQQRKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISW 749
Q E +SL FV LGLV + + +G + + +R E ++ Q++ +
Sbjct: 75 DLHQMVHEVSRYSLYFVYLGLVVCFSSYAEIACWMYSGERQVAALRKKYLEAVLKQDVGF 134
Query: 750 FDDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALI 809
FD + + ++ +V S D V+ + + +
Sbjct: 135 FDTDARTGDIVFSV----------------------------STDTLLVQDAISEKVGNF 166
Query: 810 VQNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQV 869
+ +ST +AGLV+ F + W LA + + + P I G + L G ++ ++ Y A +
Sbjct: 167 IHYLSTFLAGLVVGFVSAWKLALLSVAVIPGIAFAGGLYAYTLTGITSKSRESYANAGVI 226
Query: 870 ANDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFI 929
A A++ +RTV S+ ESK ++ YS K G + G+ G+G GC++ + + A +
Sbjct: 227 AEQAIAQVRTVYSYVGESKALNAYSDAIQYTLKLGYKAGMAKGLGLGCTYGIACMSWALV 286
Query: 930 FYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKP 989
F+ V +++G+ + F F+ + +++ Q+ + +K K + + EII+ +P
Sbjct: 287 FWYAGVFIRNGQTDGGKAFTAIFSAIVGGMSLGQSFSNLGAFSKGKAAGYKLMEIINQRP 346
Query: 990 DIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGK 1049
I G + V G+IE + V F+YP+RPD+ IF++ ++ PS KT+A+VG SGSGK
Sbjct: 347 TIIQDPLDGKCLDQVHGNIEFKDVTFSYPSRPDVMIFRNFNIFFPSGKTVAVVGGSGSGK 406
Query: 1050 STVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKE 1109
STV+SL+ERFYDPNSG+ILLDGV++KT +L +LR+Q+GLV QEP LF +I NI YGK
Sbjct: 407 STVVSLIERFYDPNSGQILLDGVEIKTLQLKFLREQIGLVNQEPALFATTILENILYGKP 466
Query: 1110 GGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKIL 1169
AT E+ AAA+AANAHSFI+ LP GYDT VGERG QLSGGQKQRIAIAR MLK+PKIL
Sbjct: 467 -DATMVEVEAAASAANAHSFITLLPKGYDTQVGERGVQLSGGQKQRIAIARAMLKDPKIL 525
Query: 1170 LLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRH 1229
LLDEATSALDA SE IVQEALDRV V RTTVVVAHRL TIR D+IAVI+ G V E G H
Sbjct: 526 LLDEATSALDASSESIVQEALDRVMVGRTTVVVAHRLCTIRNVDSIAVIQQGQVVETGTH 585
Query: 1230 DELMRITDGVYASLV 1244
+EL+ G YASL+
Sbjct: 586 EELI-AKSGAYASLI 599
>At2g36910 putative ABC transporter
Length = 1286
Score = 986 bits (2549), Expect = 0.0
Identities = 539/1276 (42%), Positives = 791/1276 (61%), Gaps = 99/1276 (7%)
Query: 30 VPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSS--NPADAIKQV 87
V F LF FAD LD LM IG++ A +G + PL F +++N+FGS+ N +++V
Sbjct: 27 VAFKELFRFADGLDYVLMGIGSVGAFVHGCSLPLFLRFFADLVNSFGSNSNNVEKMMEEV 86
Query: 88 SKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTETNTGE 147
K +L F+ + +S+ +++CWM +GERQ ++R YL+ L QDI FFDTE T +
Sbjct: 87 LKYALYFLVVGAAIWASSWAEISCWMWSGERQTTKMRIKYLEAALNQDIQFFDTEVRTSD 146
Query: 148 VIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAVAGA 207
V+ ++ D +++Q+A+ EK+G F + F GF++ F W+LA+V LA VP +AV G
Sbjct: 147 VVFAINTDAVMVQDAISEKLGNFIHYMATFVSGFIVGFTAVWQLALVTLAVVPLIAVIGG 206
Query: 208 FMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTTMVK 267
+ ++K+S++ Q + ++AGN+V+QTV IR V +F GE +A + Y+S +KIA K
Sbjct: 207 IHTTTLSKLSNKSQESLSQAGNIVEQTVVQIRVVMAFVGESRASQAYSSALKIAQKLGYK 266
Query: 268 QGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI------- 320
G+ G G+G F+ FC Y L +WYG LV NGG + + A+M GG+
Sbjct: 267 TGLAKGMGLGATYFVVFCCYALLLWYGGYLVRHHLTNGGLAIATMFAVMIGGLALGQSAP 326
Query: 321 -------------------------------------IKGDIELRDVSFRYPARPDVQIF 343
+ G +EL++V F YP+RPDV+I
Sbjct: 327 SMAAFAKAKVAAAKIFRIIDHKPTIERNSESGVELDSVTGLVELKNVDFSYPSRPDVKIL 386
Query: 344 DGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQI 403
+ F L VP+G T ALVG SGSGKSTV+SL+ERFYDP++G+VL+DG +LK L+LRW+R+QI
Sbjct: 387 NNFCLSVPAGKTIALVGSSGSGKSTVVSLIERFYDPNSGQVLLDGQDLKTLKLRWLRQQI 446
Query: 404 GLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQ 463
GLVSQEP LF TSI+ENI G+ A EI A +ANA FI KLP G DT G+ G Q
Sbjct: 447 GLVSQEPALFATSIKENILLGRPDADQVEIEEAARVANAHSFIIKLPDGFDTQVGERGLQ 506
Query: 464 LSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLT 523
LSGGQKQRIAIARA+LKNP ILLLDEATSALD+ESE++VQEAL++ ++ RTT+++AHRL+
Sbjct: 507 LSGGQKQRIAIARAMLKNPAILLLDEATSALDSESEKLVQEALDRFMIGRTTLIIAHRLS 566
Query: 524 TIRNADIIAVVQQGKIVERGTHSGL-TMDPDGAYSQLIRLQEGDNEA--EGSRKSEADKL 580
TIR AD++AV+QQG + E GTH L + +G Y++LI++QE +E +RKS A
Sbjct: 567 TIRKADLVAVLQQGSVSEIGTHDELFSKGENGVYAKLIKMQEAAHETAMSNARKSSARPS 626
Query: 581 GDNLNIDSHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQLDNKKK 640
++ S + T+ +S+ RS +S+ L S I+ N +
Sbjct: 627 SARNSVSSPI---MTRNSSYGRS---------PYSRRLSDFSTSDFSLSIDASSYPNYRN 674
Query: 641 PKV-------SIWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPP-E 692
K+ S WRLAK+N PE LLG++ +++ G + F ++ SAV+S++Y P E
Sbjct: 675 EKLAFKDQANSFWRLAKMNSPEWKYALLGSVGSVICGSLSAFFAYVLSAVLSVYYNPDHE 734
Query: 693 QQRKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDD 752
K+ + L +GL LV LQ+ F+ I G L +R+R ++ E++WFD
Sbjct: 735 YMIKQIDKYCYLLIGLSSAALVFNTLQHSFWDIVGENLTKRVREKMLSAVLKNEMAWFDQ 794
Query: 753 PSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQN 812
E NES+ + ARL++DA+ V+S +GD +++IVQN
Sbjct: 795 ------------------------EENESA---RIAARLALDANNVRSAIGDRISVIVQN 827
Query: 813 ISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVAND 872
+ ++ F W LA +++ + P+++ ++Q F+ GFS D + + + +Q+A +
Sbjct: 828 TALMLVACTAGFVLQWRLALVLVAVFPVVVAATVLQKMFMTGFSGDLEAAHAKGTQLAGE 887
Query: 873 AVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYI 932
A++++RTVA+F +E+K++ +Y+ P K+ G ++G G+G + LY + A +
Sbjct: 888 AIANVRTVAAFNSEAKIVRLYTANLEPPLKRCFWKGQIAGSGYGVAQFCLYASYALGLWY 947
Query: 933 GSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDID 992
S LV+HG + F++ RVF L ++A ++T TLAPD K + S+FE++D K +I+
Sbjct: 948 ASWLVKHGISDFSKTIRVFMVLMVSANGAAETLTLAPDFIKGGQAMRSVFELLDRKTEIE 1007
Query: 993 -SSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKST 1051
+ + + G++EL+H++F+YP+RPDIQIF+DLSL + KT+ALVG SG GKS+
Sbjct: 1008 PDDPDTTPVPDRLRGEVELKHIDFSYPSRPDIQIFRDLSLRARAGKTLALVGPSGCGKSS 1067
Query: 1052 VISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGG 1111
VISL++RFY+P+SGR+++DG D++ + L +R+ + +V QEP LF +I NI YG E
Sbjct: 1068 VISLIQRFYEPSSGRVMIDGKDIRKYNLKAIRKHIAIVPQEPCLFGTTIYENIAYGHE-C 1126
Query: 1112 ATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLL 1171
ATE EII AA A+AH FIS LP+GY T VGERG QLSGGQKQRIAIAR +++ +I+LL
Sbjct: 1127 ATEAEIIQAATLASAHKFISALPEGYKTYVGERGVQLSGGQKQRIAIARALVRKAEIMLL 1186
Query: 1172 DEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDE 1231
DEATSALDAESER VQEALD+ RT++VVAHRL+TIR A IAVI +G VAE+G H
Sbjct: 1187 DEATSALDAESERSVQEALDQACSGRTSIVVAHRLSTIRNAHVIAVIDDGKVAEQGSHSH 1246
Query: 1232 LMR-ITDGVYASLVAL 1246
L++ DG+YA ++ L
Sbjct: 1247 LLKNHPDGIYARMIQL 1262
Score = 429 bits (1103), Expect = e-120
Identities = 245/594 (41%), Positives = 353/594 (59%), Gaps = 33/594 (5%)
Query: 661 LGAIAAIVNGVVFPIFGFLFSAVISMFYKPP---EQQRKESRFWSLLFVGLGLVTLVILP 717
+G++ A V+G P+F F+ +++ F E+ +E ++L F+ +G
Sbjct: 46 IGSVGAFVHGCSLPLFLRFFADLVNSFGSNSNNVEKMMEEVLKYALYFLVVGAAIWASSW 105
Query: 718 LQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQNVCYFMTK**KNYETE 777
+ + +G + ++R E ++Q+I +FD
Sbjct: 106 AEISCWMWSGERQTTKMRIKYLEAALNQDIQFFD-------------------------- 139
Query: 778 RNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIAFTANWILAFIVLVL 837
E S V A ++ DA V+ + + + + ++T ++G ++ FTA W LA + L +
Sbjct: 140 -TEVRTSDVVFA-INTDAVMVQDAISEKLGNFIHYMATFVSGFIVGFTAVWQLALVTLAV 197
Query: 838 TPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFCAESKVMDMYSKKC 897
P+I + G + L S ++ +A + V IR V +F ES+ YS
Sbjct: 198 VPLIAVIGGIHTTTLSKLSNKSQESLSQAGNIVEQTVVQIRVVMAFVGESRASQAYSSAL 257
Query: 898 LGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMT 957
K G + GL G+G G ++ V++C A + + G LV+H FA+ +
Sbjct: 258 KIAQKLGYKTGLAKGMGLGATYFVVFCCYALLLWYGGYLVRHHLTNGGLAIATMFAVMIG 317
Query: 958 AIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNY 1017
+A+ Q+ KAK +AA IF IID KP I+ +S +GV ++V G +EL++V+F+Y
Sbjct: 318 GLALGQSAPSMAAFAKAKVAAAKIFRIIDHKPTIERNSESGVELDSVTGLVELKNVDFSY 377
Query: 1018 PTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTF 1077
P+RPD++I + LS+P+ KTIALVG SGSGKSTV+SL+ERFYDPNSG++LLDG DLKT
Sbjct: 378 PSRPDVKILNNFCLSVPAGKTIALVGSSGSGKSTVVSLIERFYDPNSGQVLLDGQDLKTL 437
Query: 1078 RLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGY 1137
+L WLRQQ+GLV QEP LF SI+ NI G+ A + EI AA ANAHSFI LPDG+
Sbjct: 438 KLRWLRQQIGLVSQEPALFATSIKENILLGRP-DADQVEIEEAARVANAHSFIIKLPDGF 496
Query: 1138 DTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNR 1197
DT VGERG QLSGGQKQRIAIAR MLKNP ILLLDEATSALD+ESE++VQEALDR + R
Sbjct: 497 DTQVGERGLQLSGGQKQRIAIARAMLKNPAILLLDEATSALDSESEKLVQEALDRFMIGR 556
Query: 1198 TTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELM-RITDGVYASLVALHSSA 1250
TT+++AHRL+TIR AD +AV++ G+V+E G HDEL + +GVYA L+ + +A
Sbjct: 557 TTLIIAHRLSTIRKADLVAVLQQGSVSEIGTHDELFSKGENGVYAKLIKMQEAA 610
Score = 372 bits (954), Expect = e-103
Identities = 217/585 (37%), Positives = 324/585 (55%), Gaps = 54/585 (9%)
Query: 48 IIGTISAVANGLASPLMTLFLGNVINAFGSSNPADAIKQVSKVSLLFVYLAIGSGIASFL 107
++G++ +V G S L V++ + + + IKQ+ K L + L+ + + + L
Sbjct: 701 LLGSVGSVICGSLSAFFAYVLSAVLSVYYNPDHEYMIKQIDKYCYLLIGLSSAALVFNTL 760
Query: 108 QVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTETN-TGEVIGRMSGDTILIQEAMGEK 166
Q + W + GE R+R L +L+ ++A+FD E N + + R++ D ++ A+G++
Sbjct: 761 QHSFWDIVGENLTKRVREKMLSAVLKNEMAWFDQEENESARIAARLALDANNVRSAIGDR 820
Query: 167 VGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAE 226
+ Q + F+ WRLA+VL+A P V A + M S + A+A+
Sbjct: 821 ISVIVQNTALMLVACTAGFVLQWRLALVLVAVFPVVVAATVLQKMFMTGFSGDLEAAHAK 880
Query: 227 AGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCT 286
+ + + +RTVA+F E K + Y + ++ +G ++G G G+ F + +
Sbjct: 881 GTQLAGEAIANVRTVAAFNSEAKIVRLYTANLEPPLKRCFWKGQIAGSGYGVAQFCLYAS 940
Query: 287 YGLAMWYGSKLVIEKGYNGGTVMTVIIALMT-----------------GGI--------- 320
Y L +WY S LV + + V + LM GG
Sbjct: 941 YALGLWYASWLVKHGISDFSKTIRVFMVLMVSANGAAETLTLAPDFIKGGQAMRSVFELL 1000
Query: 321 -------------------IKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQ 361
++G++EL+ + F YP+RPD+QIF SL +G T ALVG
Sbjct: 1001 DRKTEIEPDDPDTTPVPDRLRGEVELKHIDFSYPSRPDIQIFRDLSLRARAGKTLALVGP 1060
Query: 362 SGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENI 421
SG GKS+VISL++RFY+P +G V+IDG +++ L+ IR+ I +V QEP LF T+I ENI
Sbjct: 1061 SGCGKSSVISLIQRFYEPSSGRVMIDGKDIRKYNLKAIRKHIAIVPQEPCLFGTTIYENI 1120
Query: 422 AYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKN 481
AYG E AT+ EI A TLA+A KFI LP+G T G+ G QLSGGQKQRIAIARA+++
Sbjct: 1121 AYGHECATEAEIIQAATLASAHKFISALPEGYKTYVGERGVQLSGGQKQRIAIARALVRK 1180
Query: 482 PKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVE 541
+I+LLDEATSALDAESER VQEAL++ RT++VVAHRL+TIRNA +IAV+ GK+ E
Sbjct: 1181 AEIMLLDEATSALDAESERSVQEALDQACSGRTSIVVAHRLSTIRNAHVIAVIDDGKVAE 1240
Query: 542 RGTHSGLTMD-PDGAYSQLIRLQE-------GDNEAEGSRKSEAD 578
+G+HS L + PDG Y+++I+LQ G SR E D
Sbjct: 1241 QGSHSHLLKNHPDGIYARMIQLQRFTHTQVIGMTSGSSSRVKEDD 1285
>At4g25960 P-glycoprotein-2 (pgp2)
Length = 1233
Score = 984 bits (2544), Expect = 0.0
Identities = 531/1268 (41%), Positives = 776/1268 (60%), Gaps = 103/1268 (8%)
Query: 26 TKQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSSN--PADA 83
T+ KV LF+FAD D LM +G++ A +G + P+ +F G +IN G + P A
Sbjct: 16 TQPKVSLLKLFSFADFYDCVLMTLGSVGACIHGASVPIFFIFFGKLINIIGLAYLFPKQA 75
Query: 84 IKQVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTET 143
+V+K SL FVYL++ +S+L+V CWM TGERQAA++R YL+++L QDI+ FDTE
Sbjct: 76 SHRVAKYSLDFVYLSVAILFSSWLEVACWMHTGERQAAKMRRAYLRSMLSQDISLFDTEA 135
Query: 144 NTGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVA 203
+TGEVI ++ D +++Q+A+ EKVG F S F GF + F W++++V L+ VP +A
Sbjct: 136 STGEVISAITSDILVVQDALSEKVGNFLHYISRFIAGFAIGFTSVWQISLVTLSIVPLIA 195
Query: 204 VAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYT 263
+AG + V + +R + +Y +AG + ++ +G +RTV +FTGE++A+ Y ++ Y
Sbjct: 196 LAGGIYAFVAIGLIARVRKSYIKAGEIAEEVIGNVRTVQAFTGEERAVRLYREALENTYK 255
Query: 264 TMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI--- 320
K G+ G G+G + + F ++ L +W+ S +V + +GG T ++ ++ G+
Sbjct: 256 YGRKAGLTKGLGLGSMHCVLFLSWALLVWFTSVVVHKDIADGGKSFTTMLNVVIAGLSLG 315
Query: 321 -----------------------------------------IKGDIELRDVSFRYPARPD 339
+ G I+ +D +F YP+RPD
Sbjct: 316 QAAPDISAFVRAKAAAYPIFKMIERNTVTKTSAKSGRKLGKVDGHIQFKDATFSYPSRPD 375
Query: 340 VQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWI 399
V IFD +L +P+G ALVG SGSGKSTVISL+ERFY+P +G VL+DG N+ L ++W+
Sbjct: 376 VVIFDRLNLAIPAGKIVALVGGSGSGKSTVISLIERFYEPISGAVLLDGNNISELDIKWL 435
Query: 400 REQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQ 459
R QIGLV+QEP LF T+IRENI YGK+ AT EEIT A L+ A FI+ LP+G +T G+
Sbjct: 436 RGQIGLVNQEPALFATTIRENILYGKDDATAEEITRAAKLSEAISFINNLPEGFETQVGE 495
Query: 460 NGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVA 519
G QLSGGQKQRIAI+RAI+KNP ILLLDEATSALDAESE+ VQEAL+++++ RTTVVVA
Sbjct: 496 RGIQLSGGQKQRIAISRAIVKNPSILLLDEATSALDAESEKSVQEALDRVMVGRTTVVVA 555
Query: 520 HRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEGSRKSEADK 579
HRL+T+RNADIIAVV +GKIVE G H L +PDGAYS L+RLQE
Sbjct: 556 HRLSTVRNADIIAVVHEGKIVEFGNHENLISNPDGAYSSLLRLQE--------------- 600
Query: 580 LGDNLNIDSHMAGSSTQRTSFVRSISQTSSVSH-RHSQSLRGLSGEIVESDIEQGQLDNK 638
S + S R++S+ S+ + R R ES D
Sbjct: 601 -----------TASLQRNPSLNRTLSRPHSIKYSRELSRTRSSFCSERESVTRPDGADPS 649
Query: 639 KKPKVSIWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKES 698
KK KV++ RL + +P+ + G I A + G P+F S + +Y ++ +KE
Sbjct: 650 KKVKVTVGRLYSMIRPDWMYGVCGTICAFIAGSQMPLFALGVSQALVSYYSGWDETQKEI 709
Query: 699 RFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRY 758
+ ++LF ++TL++ +++ FG G +L R+R F I+ EI WFD+ ++
Sbjct: 710 KKIAILFCCASVITLIVYTIEHICFGTMGERLTLRVRENMFRAILKNEIGWFDEVDNT-- 767
Query: 759 VIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIA 818
S + +RL DA+ +K++V D +++QN+ V+
Sbjct: 768 -------------------------SSMLASRLESDATLLKTIVVDRSTILLQNLGLVVT 802
Query: 819 GLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIR 878
+IAF NW L +VL P+++ I + F++G+ D Y +A+ +A ++VS+IR
Sbjct: 803 SFIIAFILNWRLTLVVLATYPLVISGHISEKLFMQGYGGDLNKAYLKANMLAGESVSNIR 862
Query: 879 TVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQ 938
TVA+FCAE K++++YS++ L P+K R G ++G+ +G S ++ + + GS L+
Sbjct: 863 TVAAFCAEEKILELYSRELLEPSKSSFRRGQIAGLFYGVSQFFIFSSYGLALWYGSTLMD 922
Query: 939 HGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAG 998
G A F V + F L +TA+A+ +T LAPD K AS+FEI+D K I ++
Sbjct: 923 KGLAGFKSVMKTFMVLIVTALAMGETLALAPDLLKGNQMVASVFEILDRKTQIVGETSEE 982
Query: 999 VTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLER 1058
+ V G IEL+ V+F+YP+RPD+ IF+D L + + K++ALVG+SGSGKS+VISL+ R
Sbjct: 983 L--NNVEGTIELKGVHFSYPSRPDVVIFRDFDLIVRAGKSMALVGQSGSGKSSVISLILR 1040
Query: 1059 FYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGATEDEII 1118
FYDP +G+++++G D+K L LR+ +GLV QEP LF +I NI YG E GA++ E++
Sbjct: 1041 FYDPTAGKVMIEGKDIKKLDLKALRKHIGLVQQEPALFATTIYENILYGNE-GASQSEVV 1099
Query: 1119 AAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSAL 1178
+A ANAHSFI++LP+GY T VGERG Q+SGGQ+QRIAIAR +LKNP ILLLDEATSAL
Sbjct: 1100 ESAMLANAHSFITSLPEGYSTKVGERGVQMSGGQRQRIAIARAILKNPAILLLDEATSAL 1159
Query: 1179 DAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDG 1238
D ESER+VQ+ALDR+ NRTTVVVAHRL+TI+ ADTI+V+ G + E+G H +L+ G
Sbjct: 1160 DVESERVVQQALDRLMANRTTVVVAHRLSTIKNADTISVLHGGKIVEQGSHRKLVLNKSG 1219
Query: 1239 VYASLVAL 1246
Y L++L
Sbjct: 1220 PYFKLISL 1227
Score = 390 bits (1001), Expect = e-108
Identities = 237/618 (38%), Positives = 352/618 (56%), Gaps = 39/618 (6%)
Query: 640 KPKVSIWRLAKL-NKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMF---YKPPEQQR 695
+PKVS+ +L + + ++ LG++ A ++G PIF F +I++ Y P+Q
Sbjct: 17 QPKVSLLKLFSFADFYDCVLMTLGSVGACIHGASVPIFFIFFGKLINIIGLAYLFPKQAS 76
Query: 696 KESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSH 755
+SL FV L + L L+ + G + ++R ++ Q+IS FD
Sbjct: 77 HRVAKYSLDFVYLSVAILFSSWLEVACWMHTGERQAAKMRRAYLRSMLSQDISLFDT--- 133
Query: 756 SRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNIST 815
+ +G V + ++ D V+ + + + + IS
Sbjct: 134 -------------------------EASTGEVISAITSDILVVQDALSEKVGNFLHYISR 168
Query: 816 VIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVS 875
IAG I FT+ W ++ + L + P+I + G + G A + Y +A ++A + +
Sbjct: 169 FIAGFAIGFTSVWQISLVTLSIVPLIALAGGIYAFVAIGLIARVRKSYIKAGEIAEEVIG 228
Query: 876 SIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSV 935
++RTV +F E + + +Y + K G + GL G+G G VL+ + A + + SV
Sbjct: 229 NVRTVQAFTGEERAVRLYREALENTYKYGRKAGLTKGLGLGSMHCVLFLSWALLVWFTSV 288
Query: 936 LVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTN---KAKDSAASIFEIIDSKPDID 992
+V A + F + + +++ Q APD + +AK +A IF++I+
Sbjct: 289 VVHKDIADGGKSFTTMLNVVIAGLSLGQA---APDISAFVRAKAAAYPIFKMIERNTVTK 345
Query: 993 SSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTV 1052
+S+ +G V G I+ + F+YP+RPD+ IF L+L+IP+ K +ALVG SGSGKSTV
Sbjct: 346 TSAKSGRKLGKVDGHIQFKDATFSYPSRPDVVIFDRLNLAIPAGKIVALVGGSGSGKSTV 405
Query: 1053 ISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGA 1112
ISL+ERFY+P SG +LLDG ++ + WLR Q+GLV QEP LF +IR NI YGK+ A
Sbjct: 406 ISLIERFYEPISGAVLLDGNNISELDIKWLRGQIGLVNQEPALFATTIRENILYGKD-DA 464
Query: 1113 TEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLD 1172
T +EI AA + A SFI+NLP+G++T VGERG QLSGGQKQRIAI+R ++KNP ILLLD
Sbjct: 465 TAEEITRAAKLSEAISFINNLPEGFETQVGERGIQLSGGQKQRIAISRAIVKNPSILLLD 524
Query: 1173 EATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDEL 1232
EATSALDAESE+ VQEALDRV V RTTVVVAHRL+T+R AD IAV+ G + E G H+ L
Sbjct: 525 EATSALDAESEKSVQEALDRVMVGRTTVVVAHRLSTVRNADIIAVVHEGKIVEFGNHENL 584
Query: 1233 MRITDGVYASLVALHSSA 1250
+ DG Y+SL+ L +A
Sbjct: 585 ISNPDGAYSSLLRLQETA 602
Score = 388 bits (997), Expect = e-107
Identities = 227/561 (40%), Positives = 325/561 (57%), Gaps = 46/561 (8%)
Query: 48 IIGTISAVANGLASPLMTLFLGNVINAFGSSNPADAIKQVSKVSLLFVYLAIGSGIASFL 107
+ GTI A G PL L + + ++ S + K++ K+++LF ++ + I +
Sbjct: 671 VCGTICAFIAGSQMPLFALGVSQALVSYYSGWD-ETQKEIKKIAILFCCASVITLIVYTI 729
Query: 108 QVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTETNTGEVIG-RMSGDTILIQEAMGEK 166
+ C+ GER R+R + IL+ +I +FD NT ++ R+ D L++ + ++
Sbjct: 730 EHICFGTMGERLTLRVRENMFRAILKNEIGWFDEVDNTSSMLASRLESDATLLKTIVVDR 789
Query: 167 VGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAE 226
Q F++AFI WRL +V+LA P V + M AY +
Sbjct: 790 STILLQNLGLVVTSFIIAFILNWRLTLVVLATYPLVISGHISEKLFMQGYGGDLNKAYLK 849
Query: 227 AGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCT 286
A + ++V IRTVA+F E+K +E Y+ ++ + ++G ++G G+ F F +
Sbjct: 850 ANMLAGESVSNIRTVAAFCAEEKILELYSRELLEPSKSSFRRGQIAGLFYGVSQFFIFSS 909
Query: 287 YGLAMWYGSKLVIEKGYNG-----GTVMTVIIALMTGGI--------------------- 320
YGLA+WYGS L+ +KG G T M +I+ + G
Sbjct: 910 YGLALWYGSTLM-DKGLAGFKSVMKTFMVLIVTALAMGETLALAPDLLKGNQMVASVFEI 968
Query: 321 -----------------IKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSG 363
++G IEL+ V F YP+RPDV IF F L V +G + ALVGQSG
Sbjct: 969 LDRKTQIVGETSEELNNVEGTIELKGVHFSYPSRPDVVIFRDFDLIVRAGKSMALVGQSG 1028
Query: 364 SGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAY 423
SGKS+VISL+ RFYDP AG+V+I+G ++K L L+ +R+ IGLV QEP LF T+I ENI Y
Sbjct: 1029 SGKSSVISLILRFYDPTAGKVMIEGKDIKKLDLKALRKHIGLVQQEPALFATTIYENILY 1088
Query: 424 GKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPK 483
G EGA+ E+ + LANA FI LP+G T G+ G Q+SGGQ+QRIAIARAILKNP
Sbjct: 1089 GNEGASQSEVVESAMLANAHSFITSLPEGYSTKVGERGVQMSGGQRQRIAIARAILKNPA 1148
Query: 484 ILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERG 543
ILLLDEATSALD ESER+VQ+AL++++ RTTVVVAHRL+TI+NAD I+V+ GKIVE+G
Sbjct: 1149 ILLLDEATSALDVESERVVQQALDRLMANRTTVVVAHRLSTIKNADTISVLHGGKIVEQG 1208
Query: 544 THSGLTMDPDGAYSQLIRLQE 564
+H L ++ G Y +LI LQ+
Sbjct: 1209 SHRKLVLNKSGPYFKLISLQQ 1229
>At1g10680 putative P-glycoprotein-2 emb|CAA71277
Length = 1227
Score = 980 bits (2533), Expect = 0.0
Identities = 533/1265 (42%), Positives = 788/1265 (62%), Gaps = 107/1265 (8%)
Query: 25 KTKQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSSN--PAD 82
K + V F LF+FAD D LM +G+I A +G + P+ +F G +IN G + P +
Sbjct: 19 KKRPSVSFLKLFSFADFYDCVLMALGSIGACIHGASVPVFFIFFGKLINIIGLAYLFPQE 78
Query: 83 AIKQVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTE 142
A +V+K SL FVYL++ +S+L+V CWM TGERQAA+IR YL+++L QDI+ FDTE
Sbjct: 79 ASHKVAKYSLDFVYLSVVILFSSWLEVACWMHTGERQAAKIRKAYLRSMLSQDISLFDTE 138
Query: 143 TNTGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCV 202
+TGEVI ++ + +++Q+A+ EKVG F S F GF + F W++++V L+ VP +
Sbjct: 139 ISTGEVISAITSEILVVQDAISEKVGNFMHFISRFIAGFAIGFASVWQISLVTLSIVPFI 198
Query: 203 AVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAY 262
A+AG + V + + R + +Y +A + ++ +G +RTV +FTGE+KA+ Y ++ Y
Sbjct: 199 ALAGGIYAFVSSGLIVRVRKSYVKANEIAEEVIGNVRTVQAFTGEEKAVSSYQGALRNTY 258
Query: 263 TTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGY-NGG----TVMTVIIALM- 316
K G+ G G+G L F+ F ++ L +W+ S +V+ KG NGG T++ V+IA +
Sbjct: 259 NYGRKAGLAKGLGLGSLHFVLFLSWALLIWFTS-IVVHKGIANGGESFTTMLNVVIAGLS 317
Query: 317 --------------------------------TG---GIIKGDIELRDVSFRYPARPDVQ 341
TG G + GDI +DV+F YP+RPDV
Sbjct: 318 LGQAAPDISTFMRASAAAYPIFQMIERNTEDKTGRKLGNVNGDILFKDVTFTYPSRPDVV 377
Query: 342 IFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIRE 401
IFD + +P+G ALVG SGSGKST+ISL+ERFY+P G V++DG +++ L L+W+R
Sbjct: 378 IFDKLNFVIPAGKVVALVGGSGSGKSTMISLIERFYEPTDGAVMLDGNDIRYLDLKWLRG 437
Query: 402 QIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNG 461
IGLV+QEP+LF T+IRENI YGK+ AT EEIT A L+ A FI+ LP+G +T G+ G
Sbjct: 438 HIGLVNQEPVLFATTIRENIMYGKDDATSEEITNAAKLSEAISFINNLPEGFETQVGERG 497
Query: 462 TQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHR 521
QLSGGQKQRI+I+RAI+KNP ILLLDEATSALDAESE+IVQEAL+++++ RTTVVVAHR
Sbjct: 498 IQLSGGQKQRISISRAIVKNPSILLLDEATSALDAESEKIVQEALDRVMVGRTTVVVAHR 557
Query: 522 LTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEGSRKSEADKLG 581
L+T+RNADIIAVV GKI+E G+H L +PDGAYS L+R+QE +
Sbjct: 558 LSTVRNADIIAVVGGGKIIESGSHDELISNPDGAYSSLLRIQEAASP------------- 604
Query: 582 DNLNIDSHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQLDNKKKP 641
NLN + S+ I++T+S H+ Q D K+
Sbjct: 605 -NLNHTPSLPVSTKPLPEL--PITETTSSIHQ-----------------SVNQPDTTKQA 644
Query: 642 KVSIWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKESRFW 701
KV++ RL + +P+ L G + + + G P+F + + +Y E + E +
Sbjct: 645 KVTVGRLYSMIRPDWKYGLCGTLGSFIAGSQMPLFALGIAQALVSYYMDWETTQNEVKRI 704
Query: 702 SLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQ 761
S+LF ++T+++ +++ FGI G +L R+R F I+ EI WFD ++
Sbjct: 705 SILFCCGSVITVIVHTIEHTTFGIMGERLTLRVRQKMFSAILRNEIGWFDKVDNT----- 759
Query: 762 NVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAGLV 821
S + +RL DA+ ++++V D ++++N+ V+ +
Sbjct: 760 ----------------------SSMLASRLESDATLLRTIVVDRSTILLENLGLVVTAFI 797
Query: 822 IAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVA 881
I+F NW L +VL P+I+ I + F++G+ + Y +A+ +A +++S+IRTV
Sbjct: 798 ISFILNWRLTLVVLATYPLIISGHISEKIFMQGYGGNLSKAYLKANMLAGESISNIRTVV 857
Query: 882 SFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGK 941
+FCAE KV+D+YSK+ L P+++ R G ++GI +G S ++ + + GS+L++ G
Sbjct: 858 AFCAEEKVLDLYSKELLEPSERSFRRGQMAGILYGVSQFFIFSSYGLALWYGSILMEKGL 917
Query: 942 ATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTR 1001
++F V + F L +TA+ + + LAPD K S+FE++D + + + G
Sbjct: 918 SSFESVMKTFMVLIVTALVMGEVLALAPDLLKGNQMVVSVFELLDRRTQV--VGDTGEEL 975
Query: 1002 ETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYD 1061
V G IEL+ V+F+YP+RPD+ IF D +L +PS K++ALVG+SGSGKS+V+SL+ RFYD
Sbjct: 976 SNVEGTIELKGVHFSYPSRPDVTIFSDFNLLVPSGKSMALVGQSGSGKSSVLSLVLRFYD 1035
Query: 1062 PNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGATEDEIIAAA 1121
P +G I++DG D+K +L LR+ +GLV QEP LF +I NI YGKE GA+E E++ AA
Sbjct: 1036 PTAGIIMIDGQDIKKLKLKSLRRHIGLVQQEPALFATTIYENILYGKE-GASESEVMEAA 1094
Query: 1122 NAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAE 1181
ANAHSFIS+LP+GY T VGERG Q+SGGQ+QRIAIAR +LKNP+ILLLDEATSALD E
Sbjct: 1095 KLANAHSFISSLPEGYSTKVGERGIQMSGGQRQRIAIARAVLKNPEILLLDEATSALDVE 1154
Query: 1182 SERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGVYA 1241
SER+VQ+ALDR+ +RTTVVVAHRL+TI+ +D I+VI++G + E+G H+ L+ +G Y+
Sbjct: 1155 SERVVQQALDRLMRDRTTVVVAHRLSTIKNSDMISVIQDGKIIEQGSHNILVENKNGPYS 1214
Query: 1242 SLVAL 1246
L++L
Sbjct: 1215 KLISL 1219
Score = 393 bits (1009), Expect = e-109
Identities = 238/620 (38%), Positives = 346/620 (55%), Gaps = 43/620 (6%)
Query: 638 KKKPKVSIWRLAKL-NKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMF---YKPPEQ 693
KK+P VS +L + + ++ LG+I A ++G P+F F +I++ Y P++
Sbjct: 19 KKRPSVSFLKLFSFADFYDCVLMALGSIGACIHGASVPVFFIFFGKLINIIGLAYLFPQE 78
Query: 694 QRKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDP 753
+ +SL FV L +V L L+ + G + +IR ++ Q+IS FD
Sbjct: 79 ASHKVAKYSLDFVYLSVVILFSSWLEVACWMHTGERQAAKIRKAYLRSMLSQDISLFD-- 136
Query: 754 SHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNI 813
TE + A+ + + + + VG+ M I
Sbjct: 137 ----------------------TEISTGEVISAITSEILVVQDAISEKVGNFMHFI---- 170
Query: 814 STVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDA 873
S IAG I F + W ++ + L + P I + G + G + Y +A+++A +
Sbjct: 171 SRFIAGFAIGFASVWQISLVTLSIVPFIALAGGIYAFVSSGLIVRVRKSYVKANEIAEEV 230
Query: 874 VSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIG 933
+ ++RTV +F E K + Y G + GL G+G G VL+ + A + +
Sbjct: 231 IGNVRTVQAFTGEEKAVSSYQGALRNTYNYGRKAGLAKGLGLGSLHFVLFLSWALLIWFT 290
Query: 934 SVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTN---KAKDSAASIFEIIDSKPD 990
S++V G A E F + + +++ Q APD + +A +A IF++I+ +
Sbjct: 291 SIVVHKGIANGGESFTTMLNVVIAGLSLGQA---APDISTFMRASAAAYPIFQMIERNTE 347
Query: 991 IDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKS 1050
+ G V GDI + V F YP+RPD+ IF L+ IP+ K +ALVG SGSGKS
Sbjct: 348 DKTGRKLG----NVNGDILFKDVTFTYPSRPDVVIFDKLNFVIPAGKVVALVGGSGSGKS 403
Query: 1051 TVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEG 1110
T+ISL+ERFY+P G ++LDG D++ L WLR +GLV QEP+LF +IR NI YGK+
Sbjct: 404 TMISLIERFYEPTDGAVMLDGNDIRYLDLKWLRGHIGLVNQEPVLFATTIRENIMYGKDD 463
Query: 1111 GATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILL 1170
AT +EI AA + A SFI+NLP+G++T VGERG QLSGGQKQRI+I+R ++KNP ILL
Sbjct: 464 -ATSEEITNAAKLSEAISFINNLPEGFETQVGERGIQLSGGQKQRISISRAIVKNPSILL 522
Query: 1171 LDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHD 1230
LDEATSALDAESE+IVQEALDRV V RTTVVVAHRL+T+R AD IAV+ G + E G HD
Sbjct: 523 LDEATSALDAESEKIVQEALDRVMVGRTTVVVAHRLSTVRNADIIAVVGGGKIIESGSHD 582
Query: 1231 ELMRITDGVYASLVALHSSA 1250
EL+ DG Y+SL+ + +A
Sbjct: 583 ELISNPDGAYSSLLRIQEAA 602
Score = 389 bits (1000), Expect = e-108
Identities = 232/605 (38%), Positives = 349/605 (57%), Gaps = 48/605 (7%)
Query: 5 PNVHDNSSSSPTQQHGIRDNKTKQ-KVPFYMLFNFADHLDVTLMIIGTISAVANGLASPL 63
P + ++S Q + + TKQ KV L++ D + GT+ + G PL
Sbjct: 620 PELPITETTSSIHQSVNQPDTTKQAKVTVGRLYSMI-RPDWKYGLCGTLGSFIAGSQMPL 678
Query: 64 MTLFLGNVINAFGSSNPADAIKQVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARI 123
L + + ++ +V ++S+LF ++ + I ++ T + + GER R+
Sbjct: 679 FALGIAQALVSYYMDWETTQ-NEVKRISILFCCGSVITVIVHTIEHTTFGIMGERLTLRV 737
Query: 124 RSLYLKTILQQDIAFFDTETNTGEVIG-RMSGDTILIQEAMGEKVGKFFQLASNFCGGFV 182
R IL+ +I +FD NT ++ R+ D L++ + ++ + F+
Sbjct: 738 RQKMFSAILRNEIGWFDKVDNTSSMLASRLESDATLLRTIVVDRSTILLENLGLVVTAFI 797
Query: 183 MAFIKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVA 242
++FI WRL +V+LA P + I M AY +A + +++ IRTV
Sbjct: 798 ISFILNWRLTLVVLATYPLIISGHISEKIFMQGYGGNLSKAYLKANMLAGESISNIRTVV 857
Query: 243 SFTGEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKG 302
+F E+K ++ Y+ ++ ++G ++G G+ F F +YGLA+WYGS +++EKG
Sbjct: 858 AFCAEEKVLDLYSKELLEPSERSFRRGQMAGILYGVSQFFIFSSYGLALWYGS-ILMEKG 916
Query: 303 YNG-----GTVMTVII-ALMTGGI------------------------------------ 320
+ T M +I+ AL+ G +
Sbjct: 917 LSSFESVMKTFMVLIVTALVMGEVLALAPDLLKGNQMVVSVFELLDRRTQVVGDTGEELS 976
Query: 321 -IKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDP 379
++G IEL+ V F YP+RPDV IF F+L VPSG + ALVGQSGSGKS+V+SL+ RFYDP
Sbjct: 977 NVEGTIELKGVHFSYPSRPDVTIFSDFNLLVPSGKSMALVGQSGSGKSSVLSLVLRFYDP 1036
Query: 380 DAGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITL 439
AG ++IDG ++K L+L+ +R IGLV QEP LF T+I ENI YGKEGA++ E+ A L
Sbjct: 1037 TAGIIMIDGQDIKKLKLKSLRRHIGLVQQEPALFATTIYENILYGKEGASESEVMEAAKL 1096
Query: 440 ANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESE 499
ANA FI LP+G T G+ G Q+SGGQ+QRIAIARA+LKNP+ILLLDEATSALD ESE
Sbjct: 1097 ANAHSFISSLPEGYSTKVGERGIQMSGGQRQRIAIARAVLKNPEILLLDEATSALDVESE 1156
Query: 500 RIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQL 559
R+VQ+AL++++ RTTVVVAHRL+TI+N+D+I+V+Q GKI+E+G+H+ L + +G YS+L
Sbjct: 1157 RVVQQALDRLMRDRTTVVVAHRLSTIKNSDMISVIQDGKIIEQGSHNILVENKNGPYSKL 1216
Query: 560 IRLQE 564
I LQ+
Sbjct: 1217 ISLQQ 1221
>At1g27940 hypothetical protein
Length = 1245
Score = 938 bits (2424), Expect = 0.0
Identities = 521/1276 (40%), Positives = 761/1276 (58%), Gaps = 107/1276 (8%)
Query: 22 RDNKTKQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFG--SSN 79
+ N K+ V LF+ AD LD LM++G + A +G PL +F G ++++ G S++
Sbjct: 22 KKNIKKESVSLMGLFSAADKLDYFLMLLGGLGACIHGATLPLFFVFFGKMLDSLGNLSTD 81
Query: 80 PADAIKQVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFF 139
P +VS+ +L VYL + + +++++ V+CWM TGERQ AR+R YLK+IL +DI FF
Sbjct: 82 PKAISSRVSQNALYLVYLGLVNFVSAWIGVSCWMQTGERQTARLRINYLKSILAKDITFF 141
Query: 140 DTETNTGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACV 199
DTE +I +S D IL+Q+A+G+K + S F GFV+ F+ W+L ++ L V
Sbjct: 142 DTEARDSNLIFHISSDAILVQDAIGDKTDHVLRYLSQFIAGFVIGFLSVWQLTLLTLGVV 201
Query: 200 PCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIK 259
P +A+AG +IVM+ +S + + AYA+AG V ++ + +RTV +F GE+KA++ Y++ +K
Sbjct: 202 PLIAIAGGGYAIVMSTISEKSETAYADAGKVAEEVMSQVRTVYAFVGEEKAVKSYSNSLK 261
Query: 260 IAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGG 319
A + G+ G G+G+ + FC + L +WY S LV NG T I+ ++ G
Sbjct: 262 KALKLGKRSGLAKGLGVGLTYSLLFCAWALLLWYASLLVRHGKTNGAKAFTTILNVIFSG 321
Query: 320 I---------------------------------------------IKGDIELRDVSFRY 334
+ G IE + VSF Y
Sbjct: 322 FALGQAAPSLSAIAKGRVAAANIFRMIGNNNSESSQRLDEGTTLQNVAGRIEFQKVSFAY 381
Query: 335 PARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNL 394
P+RP++ +F+ S + SG T A VG SGSGKST+IS+++RFY+P++GE+L+DG ++K+L
Sbjct: 382 PSRPNM-VFENLSFTIRSGKTFAFVGPSGSGKSTIISMVQRFYEPNSGEILLDGNDIKSL 440
Query: 395 QLRWIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLD 454
+L+W REQ+GLVSQEP LF T+I NI GKE A ++I A ANA FI LP G +
Sbjct: 441 KLKWFREQLGLVSQEPALFATTIASNILLGKENANMDQIIEAAKAANADSFIKSLPNGYN 500
Query: 455 TMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRT 514
T G+ GTQLSGGQKQRIAIARA+L+NPKILLLDEATSALDAESE+IVQ+AL+ ++ KRT
Sbjct: 501 TQVGEGGTQLSGGQKQRIAIARAVLRNPKILLLDEATSALDAESEKIVQQALDNVMEKRT 560
Query: 515 TVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEGSRK 574
T+VVAHRL+TIRN D I V++ G++ E G+HS L M G Y+ L+ QE + + E SR
Sbjct: 561 TIVVAHRLSTIRNVDKIVVLRDGQVRETGSHSEL-MLRGGDYATLVNCQETEPQ-ENSRS 618
Query: 575 SEADKLGDNLNIDSHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQ 634
++ S SS++RTS R D E+ +
Sbjct: 619 IMSETCKSQAGSSSSRRVSSSRRTSSFR-------------------------VDQEKTK 653
Query: 635 LDNKKKPKVS---IWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKP- 690
D+ KK S IW L KLN PE P LLG+I A++ G P+F + V++ FY P
Sbjct: 654 NDDSKKDFSSSSMIWELIKLNSPEWPYALLGSIGAVLAGAQTPLFSMGIAYVLTAFYSPF 713
Query: 691 PEQQRKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWF 750
P +++ +++F G G+VT I LQ++F+ + G +L R+R F I+ EI WF
Sbjct: 714 PNVIKRDVEKVAIIFAGAGIVTAPIYLLQHYFYTLMGERLTSRVRLSLFSAILSNEIGWF 773
Query: 751 DDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIV 810
D + + +G++ + L+ DA+ V+S + D ++ IV
Sbjct: 774 D---------------------------LDENNTGSLTSILAADATLVRSALADRLSTIV 806
Query: 811 QNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVA 870
QN+S + L +AF +W +A +V P+++ + + FLKGF D Y A+ VA
Sbjct: 807 QNLSLTVTALALAFFYSWRVAAVVTACFPLLIAASLTEQLFLKGFGGDYTRAYSRATSVA 866
Query: 871 NDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIF 930
+A+++IRTVA++ AE ++ + ++ + P K G +SG G+G S + +C+ A
Sbjct: 867 REAIANIRTVAAYGAEKQISEQFTCELSKPTKNAFVRGHISGFGYGLSQFLAFCSYALGL 926
Query: 931 YIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPD 990
+ SVL+ H + F + + F L +TA +VS+T L PD K + S+F ++ +
Sbjct: 927 WYVSVLINHKETNFGDSIKSFMVLIVTAFSVSETLALTPDIVKGTQALGSVFRVLHRETK 986
Query: 991 IDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKS 1050
I V GDIE ++V+F YPTRP+I IFK+L+L + + K++A+VG SGSGKS
Sbjct: 987 ISPDQPNSRMVSQVKGDIEFRNVSFVYPTRPEIDIFKNLNLRVSAGKSLAVVGPSGSGKS 1046
Query: 1051 TVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEG 1110
TVI L+ RFYDP++G + +DG D+KT L LR+++ LV QEP LF+ +I NI YG E
Sbjct: 1047 TVIGLIMRFYDPSNGNLCIDGQDIKTLNLRSLRKKLALVQQEPALFSTTIYENIKYGNE- 1105
Query: 1111 GATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILL 1170
A+E EI+ AA AANAH FI + +GY T G++G QLSGGQKQR+AIAR +LK+P +LL
Sbjct: 1106 NASEAEIMEAAKAANAHEFIIKMEEGYKTHAGDKGVQLSGGQKQRVAIARAVLKDPSVLL 1165
Query: 1171 LDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHD 1230
LDEATSALD SE++VQEALD++ RTTV+VAHRL+TIR ADT+AV+ G V EKG H
Sbjct: 1166 LDEATSALDTSSEKLVQEALDKLMKGRTTVLVAHRLSTIRKADTVAVLHKGRVVEKGSHR 1225
Query: 1231 ELMRITDGVYASLVAL 1246
EL+ I +G Y L +L
Sbjct: 1226 ELVSIPNGFYKQLTSL 1241
Score = 407 bits (1045), Expect = e-113
Identities = 251/631 (39%), Positives = 362/631 (56%), Gaps = 36/631 (5%)
Query: 619 RGLSGEIVESDIEQGQLDNKKKPKVSIWRL-AKLNKPEIPVILLGAIAAIVNGVVFPIFG 677
R +G I + + N KK VS+ L + +K + ++LLG + A ++G P+F
Sbjct: 6 RSSNGNIQAETEAKEEKKNIKKESVSLMGLFSAADKLDYFLMLLGGLGACIHGATLPLFF 65
Query: 678 FLFSAVI-SMFYKPPEQQRKESRFW--SLLFVGLGLVTLVILPLQNFFFGIAGGKLIERI 734
F ++ S+ + + SR +L V LGLV V + + G + R+
Sbjct: 66 VFFGKMLDSLGNLSTDPKAISSRVSQNALYLVYLGLVNFVSAWIGVSCWMQTGERQTARL 125
Query: 735 RSLTFEKIVHQEISWFDDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSID 794
R + I+ ++I++FD + +I ++ S D
Sbjct: 126 RINYLKSILAKDITFFDTEARDSNLIFHI----------------------------SSD 157
Query: 795 ASTVKSLVGDTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKG 854
A V+ +GD +++ +S IAG VI F + W L + L + P+I + G +
Sbjct: 158 AILVQDAIGDKTDHVLRYLSQFIAGFVIGFLSVWQLTLLTLGVVPLIAIAGGGYAIVMST 217
Query: 855 FSADAKVMYEEASQVANDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIG 914
S ++ Y +A +VA + +S +RTV +F E K + YS K G R GL G+G
Sbjct: 218 ISEKSETAYADAGKVAEEVMSQVRTVYAFVGEEKAVKSYSNSLKKALKLGKRSGLAKGLG 277
Query: 915 FGCSFLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKA 974
G ++ +L+C A + + S+LV+HGK + F + + A+ Q K
Sbjct: 278 VGLTYSLLFCAWALLLWYASLLVRHGKTNGAKAFTTILNVIFSGFALGQAAPSLSAIAKG 337
Query: 975 KDSAASIFEII-DSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSI 1033
+ +AA+IF +I ++ + + G T + V G IE Q V+F YP+RP++ +F++LS +I
Sbjct: 338 RVAAANIFRMIGNNNSESSQRLDEGTTLQNVAGRIEFQKVSFAYPSRPNM-VFENLSFTI 396
Query: 1034 PSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEP 1093
S KT A VG SGSGKST+IS+++RFY+PNSG ILLDG D+K+ +L W R+Q+GLV QEP
Sbjct: 397 RSGKTFAFVGPSGSGKSTIISMVQRFYEPNSGEILLDGNDIKSLKLKWFREQLGLVSQEP 456
Query: 1094 ILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQK 1153
LF +I +NI GKE A D+II AA AANA SFI +LP+GY+T VGE GTQLSGGQK
Sbjct: 457 ALFATTIASNILLGKE-NANMDQIIEAAKAANADSFIKSLPNGYNTQVGEGGTQLSGGQK 515
Query: 1154 QRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGAD 1213
QRIAIAR +L+NPKILLLDEATSALDAESE+IVQ+ALD V RTT+VVAHRL+TIR D
Sbjct: 516 QRIAIARAVLRNPKILLLDEATSALDAESEKIVQQALDNVMEKRTTIVVAHRLSTIRNVD 575
Query: 1214 TIAVIKNGAVAEKGRHDELMRITDGVYASLV 1244
I V+++G V E G H ELM + G YA+LV
Sbjct: 576 KIVVLRDGQVRETGSHSELM-LRGGDYATLV 605
Score = 392 bits (1008), Expect = e-109
Identities = 222/562 (39%), Positives = 324/562 (57%), Gaps = 45/562 (8%)
Query: 48 IIGTISAVANGLASPLMTLFLGNVINAFGSSNPADAIKQVSKVSLLFVYLAIGSGIASFL 107
++G+I AV G +PL ++ + V+ AF S P + V KV+++F I + L
Sbjct: 682 LLGSIGAVLAGAQTPLFSMGIAYVLTAFYSPFPNVIKRDVEKVAIIFAGAGIVTAPIYLL 741
Query: 108 QVTCWMVTGERQAARIRSLYLKTILQQDIAFFDT-ETNTGEVIGRMSGDTILIQEAMGEK 166
Q + + GER +R+R IL +I +FD E NTG + ++ D L++ A+ ++
Sbjct: 742 QHYFYTLMGERLTSRVRLSLFSAILSNEIGWFDLDENNTGSLTSILAADATLVRSALADR 801
Query: 167 VGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAE 226
+ Q S +AF WR+A V+ AC P + A + + AY+
Sbjct: 802 LSTIVQNLSLTVTALALAFFYSWRVAAVVTACFPLLIAASLTEQLFLKGFGGDYTRAYSR 861
Query: 227 AGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCT 286
A +V + + IRTVA++ EK+ E++ ++ +G +SGFG G+ F+AFC+
Sbjct: 862 ATSVAREAIANIRTVAAYGAEKQISEQFTCELSKPTKNAFVRGHISGFGYGLSQFLAFCS 921
Query: 287 YGLAMWYGSKLVIEKGYN-GGTVMTVIIALMT---------------------GGI---- 320
Y L +WY S L+ K N G ++ + ++ ++T G +
Sbjct: 922 YALGLWYVSVLINHKETNFGDSIKSFMVLIVTAFSVSETLALTPDIVKGTQALGSVFRVL 981
Query: 321 ------------------IKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQS 362
+KGDIE R+VSF YP RP++ IF +L V +G + A+VG S
Sbjct: 982 HRETKISPDQPNSRMVSQVKGDIEFRNVSFVYPTRPEIDIFKNLNLRVSAGKSLAVVGPS 1041
Query: 363 GSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIA 422
GSGKSTVI L+ RFYDP G + IDG ++K L LR +R+++ LV QEP LF+T+I ENI
Sbjct: 1042 GSGKSTVIGLIMRFYDPSNGNLCIDGQDIKTLNLRSLRKKLALVQQEPALFSTTIYENIK 1101
Query: 423 YGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNP 482
YG E A++ EI A ANA +FI K+ +G T AG G QLSGGQKQR+AIARA+LK+P
Sbjct: 1102 YGNENASEAEIMEAAKAANAHEFIIKMEEGYKTHAGDKGVQLSGGQKQRVAIARAVLKDP 1161
Query: 483 KILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVER 542
+LLLDEATSALD SE++VQEAL+K++ RTTV+VAHRL+TIR AD +AV+ +G++VE+
Sbjct: 1162 SVLLLDEATSALDTSSEKLVQEALDKLMKGRTTVLVAHRLSTIRKADTVAVLHKGRVVEK 1221
Query: 543 GTHSGLTMDPDGAYSQLIRLQE 564
G+H L P+G Y QL LQE
Sbjct: 1222 GSHRELVSIPNGFYKQLTSLQE 1243
>At1g28010 hypothetical protein
Length = 1247
Score = 919 bits (2376), Expect = 0.0
Identities = 520/1277 (40%), Positives = 761/1277 (58%), Gaps = 112/1277 (8%)
Query: 27 KQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFG--SSNPADAI 84
K+ V LF+ AD++D LM +G + +G PL +F G ++++ G S++P
Sbjct: 28 KESVSLMGLFSAADNVDYFLMFLGGLGTCIHGGTLPLFFVFFGGMLDSLGKLSTDPNAIS 87
Query: 85 KQVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTETN 144
+VS+ +L VYL + + +++++ V CWM TGERQ AR+R YLK+IL +DI FFDTE
Sbjct: 88 SRVSQNALYLVYLGLVNLVSAWIGVACWMQTGERQTARLRINYLKSILAKDITFFDTEAR 147
Query: 145 TGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAV 204
I +S D IL+Q+A+G+K G + F GFV+ F+ W+L ++ L VP +A+
Sbjct: 148 DSNFIFHISSDAILVQDAIGDKTGHVLRYLCQFIAGFVIGFLSVWQLTLLTLGVVPLIAI 207
Query: 205 AGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTT 264
AG +IVM+ +S + + AYA+AG V ++ + +RTV +F GE+KA++ Y++ +K A
Sbjct: 208 AGGGYAIVMSTISEKSEAAYADAGKVAEEVMSQVRTVYAFVGEEKAVKSYSNSLKKALKL 267
Query: 265 MVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI---- 320
+ G+ G G+G+ + FC + L WY S LV NG T I+ ++ G
Sbjct: 268 SKRSGLAKGLGVGLTYSLLFCAWALLFWYASLLVRHGKTNGAKAFTTILNVIYSGFALGQ 327
Query: 321 -----------------------------------------IKGDIELRDVSFRYPARPD 339
+ G IE VSF YP+RP+
Sbjct: 328 AVPSLSAISKGRVAAANIFKMIGNNNLESSERLENGTTLQNVVGKIEFCGVSFAYPSRPN 387
Query: 340 VQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWI 399
+ +F+ S + SG T A VG SGSGKST+IS+++RFY+P +GE+L+DG ++KNL+L+W+
Sbjct: 388 M-VFENLSFTIHSGKTFAFVGPSGSGKSTIISMVQRFYEPRSGEILLDGNDIKNLKLKWL 446
Query: 400 REQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQ 459
REQ+GLVSQEP LF T+I NI GKE A ++I A ANA FI LP G +T G+
Sbjct: 447 REQMGLVSQEPALFATTIASNILLGKEKANMDQIIEAAKAANADSFIKSLPNGYNTQVGE 506
Query: 460 NGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVA 519
GTQLSGGQKQRIAIARA+L+NPKILLLDEATSALDAESE+IVQ+AL+ ++ KRTT+V+A
Sbjct: 507 GGTQLSGGQKQRIAIARAVLRNPKILLLDEATSALDAESEKIVQQALDNVMEKRTTIVIA 566
Query: 520 HRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEGSRKSEADK 579
HRL+TIRN D I V++ G++ E G+HS L + G Y+ L+ Q+ + +
Sbjct: 567 HRLSTIRNVDKIVVLRDGQVRETGSHSEL-ISRGGDYATLVNCQDTEPQE---------- 615
Query: 580 LGDNLNIDSHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQLDNKK 639
N+ S M S SQ S S R S R S D E+ + D+K
Sbjct: 616 -----NLRSVMYESCR---------SQAGSYSSRRVFSSRRTSS--FREDQEKTEKDSKG 659
Query: 640 KPKVS----IWRLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKP-PEQQ 694
+ +S IW L KLN PE LLG+I A++ G +F + V++ FY P P
Sbjct: 660 EDLISSSSMIWELIKLNAPEWLYALLGSIGAVLAGSQPALFSMGLAYVLTTFYSPFPSLI 719
Query: 695 RKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPS 754
++E +++FVG G+VT I LQ++F+ + G +L R+R F I+ EI WFD
Sbjct: 720 KREVDKVAIIFVGAGIVTAPIYILQHYFYTLMGERLTSRVRLSLFSAILSNEIGWFD--- 776
Query: 755 HSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNIS 814
+ + +G++ + L+ DA+ V+S + D ++ IVQN+S
Sbjct: 777 ------------------------LDENNTGSLTSILAADATLVRSAIADRLSTIVQNLS 812
Query: 815 TVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAV 874
I L +AF +W +A +V P+++ + + FLKGF D Y A+ +A +A+
Sbjct: 813 LTITALALAFFYSWRVAAVVTACFPLLIAASLTEQLFLKGFGGDYTRAYSRATSLAREAI 872
Query: 875 SSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGS 934
S+IRTVA+F AE ++ + ++ + P K + G +SG G+G S + +C+ A + S
Sbjct: 873 SNIRTVAAFSAEKQISEQFTCELSKPTKSALLRGHISGFGYGLSQCLAFCSYALGLWYIS 932
Query: 935 VLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDI--D 992
VL++ + F + + F L +TA +V++T L PD K + S+F ++ + +I D
Sbjct: 933 VLIKRNETNFEDSIKSFMVLLVTAYSVAETLALTPDIVKGTQALGSVFRVLHRETEIPPD 992
Query: 993 SSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTV 1052
++ VT + GDIE ++V+F YPTRP+I IFK+L+L + + K++A+VG SGSGKSTV
Sbjct: 993 QPNSRLVTH--IKGDIEFRNVSFAYPTRPEIAIFKNLNLRVSAGKSLAVVGPSGSGKSTV 1050
Query: 1053 ISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGA 1112
I L+ RFYDP++G + +DG D+K+ L LR+++ LV QEP LF+ SI NI YG E A
Sbjct: 1051 IGLIMRFYDPSNGNLCIDGHDIKSVNLRSLRKKLALVQQEPALFSTSIHENIKYGNE-NA 1109
Query: 1113 TEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLD 1172
+E EII AA AANAH FIS + +GY T VG++G QLSGGQKQR+AIAR +LK+P +LLLD
Sbjct: 1110 SEAEIIEAAKAANAHEFISRMEEGYMTHVGDKGVQLSGGQKQRVAIARAVLKDPSVLLLD 1169
Query: 1173 EATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDEL 1232
EATSALD +E+ VQEALD++ RTT++VAHRL+TIR ADTI V+ G V EKG H EL
Sbjct: 1170 EATSALDTSAEKQVQEALDKLMKGRTTILVAHRLSTIRKADTIVVLHKGKVVEKGSHREL 1229
Query: 1233 MRITDGVYASLVALHSS 1249
+ +DG Y L +L +
Sbjct: 1230 VSKSDGFYKKLTSLQEA 1246
Score = 400 bits (1028), Expect = e-111
Identities = 250/630 (39%), Positives = 361/630 (56%), Gaps = 39/630 (6%)
Query: 622 SGEI-VESDIEQGQLDNKKKPKVSIWRL-AKLNKPEIPVILLGAIAAIVNGVVFPIFGFL 679
SG I E+++++ + KK VS+ L + + + ++ LG + ++G P+F
Sbjct: 9 SGNIHAETEVKKEEKKKMKKESVSLMGLFSAADNVDYFLMFLGGLGTCIHGGTLPLFFVF 68
Query: 680 FSAVISMFYK---PPEQQRKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGGKLIERIRS 736
F ++ K P +L V LGLV LV + + G + R+R
Sbjct: 69 FGGMLDSLGKLSTDPNAISSRVSQNALYLVYLGLVNLVSAWIGVACWMQTGERQTARLRI 128
Query: 737 LTFEKIVHQEISWFDDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVGARLSIDAS 796
+ I+ ++I++FD TE +S+ +S DA
Sbjct: 129 NYLKSILAKDITFFD------------------------TEARDSNFI----FHISSDAI 160
Query: 797 TVKSLVGDTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQMKFLKGFS 856
V+ +GD +++ + IAG VI F + W L + L + P+I + G + S
Sbjct: 161 LVQDAIGDKTGHVLRYLCQFIAGFVIGFLSVWQLTLLTLGVVPLIAIAGGGYAIVMSTIS 220
Query: 857 ADAKVMYEEASQVANDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLGLVSGIGFG 916
++ Y +A +VA + +S +RTV +F E K + YS K R GL G+G G
Sbjct: 221 EKSEAAYADAGKVAEEVMSQVRTVYAFVGEEKAVKSYSNSLKKALKLSKRSGLAKGLGVG 280
Query: 917 CSFLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLAPDTNKAKD 976
++ +L+C A +F+ S+LV+HGK + F + + A+ Q +K +
Sbjct: 281 LTYSLLFCAWALLFWYASLLVRHGKTNGAKAFTTILNVIYSGFALGQAVPSLSAISKGRV 340
Query: 977 SAASIFEIIDSKPDIDSSSNA--GVTRETVVGDIELQHVNFNYPTRPDIQIFKDLSLSIP 1034
+AA+IF++I + +++SS G T + VVG IE V+F YP+RP++ +F++LS +I
Sbjct: 341 AAANIFKMIGNN-NLESSERLENGTTLQNVVGKIEFCGVSFAYPSRPNM-VFENLSFTIH 398
Query: 1035 SAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGLVGQEPI 1094
S KT A VG SGSGKST+IS+++RFY+P SG ILLDG D+K +L WLR+QMGLV QEP
Sbjct: 399 SGKTFAFVGPSGSGKSTIISMVQRFYEPRSGEILLDGNDIKNLKLKWLREQMGLVSQEPA 458
Query: 1095 LFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQLSGGQKQ 1154
LF +I +NI GKE A D+II AA AANA SFI +LP+GY+T VGE GTQLSGGQKQ
Sbjct: 459 LFATTIASNILLGKE-KANMDQIIEAAKAANADSFIKSLPNGYNTQVGEGGTQLSGGQKQ 517
Query: 1155 RIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRLTTIRGADT 1214
RIAIAR +L+NPKILLLDEATSALDAESE+IVQ+ALD V RTT+V+AHRL+TIR D
Sbjct: 518 RIAIARAVLRNPKILLLDEATSALDAESEKIVQQALDNVMEKRTTIVIAHRLSTIRNVDK 577
Query: 1215 IAVIKNGAVAEKGRHDELMRITDGVYASLV 1244
I V+++G V E G H EL+ G YA+LV
Sbjct: 578 IVVLRDGQVRETGSHSELIS-RGGDYATLV 606
>At3g28390 P-glycoprotein, putative
Length = 1225
Score = 889 bits (2298), Expect = 0.0
Identities = 507/1262 (40%), Positives = 737/1262 (58%), Gaps = 102/1262 (8%)
Query: 35 LFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSSNPADA--IKQVSKVSL 92
+F AD +D LM +G I AV +G +P++ ++N G S+ D ++ V+K ++
Sbjct: 11 IFMHADGVDWMLMALGLIGAVGDGFITPIIFFICSKLLNNVGGSSFDDETFMQTVAKNAV 70
Query: 93 LFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTE-TNTGEVIGR 151
VY+A S + F++ CW TGERQAA++R YLK +L+QD+ +FD T+T +VI
Sbjct: 71 ALVYVACASWVICFIEGYCWTRTGERQAAKMREKYLKAVLRQDVGYFDLHVTSTSDVITS 130
Query: 152 MSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAVAGAFMSI 211
+S D+++IQ+ + EK+ F S F +++ F+ WRL IV + + + G
Sbjct: 131 VSSDSLVIQDFLSEKLPNFLMNTSAFVASYIVGFLLLWRLTIVGFPFIILLLIPGLMYGR 190
Query: 212 VMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTTMVKQGIV 271
+ ++S + + Y EAG++ +Q + ++RTV +F EKK IEK+++ ++ + ++QG+
Sbjct: 191 ALIRISMKIREEYNEAGSIAEQVISSVRTVYAFGSEKKMIEKFSTALQGSVKLGLRQGLA 250
Query: 272 SGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI----------- 320
G IG I + +G WYGS++V+ G GGTV +VI+ + GG
Sbjct: 251 KGIAIGS-NGITYAIWGFLTWYGSRMVMNHGSKGGTVSSVIVCVTFGGTSLGQSLSNLKY 309
Query: 321 ---------------------------------IKGDIELRDVSFRYPARPDVQIFDGFS 347
+G++E V F YP+RP+ IFD
Sbjct: 310 FSEAFVVGERIMKVINRVPGIDSDNLEGQILEKTRGEVEFNHVKFTYPSRPETPIFDDLC 369
Query: 348 LFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVS 407
L VPSG T ALVG SGSGKSTVISLL+RFYDP AGE+LIDG+ + LQ++W+R Q+GLVS
Sbjct: 370 LRVPSGKTVALVGGSGSGKSTVISLLQRFYDPIAGEILIDGLPINKLQVKWLRSQMGLVS 429
Query: 408 QEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGG 467
QEP+LF TSI+ENI +GKE A+ +E+ A +NA FI + P T G+ G QLSGG
Sbjct: 430 QEPVLFATSIKENILFGKEDASMDEVVEAAKASNAHSFISQFPNSYQTQVGERGVQLSGG 489
Query: 468 QKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLTTIRN 527
QKQRIAIARAI+K+P ILLLDEATSALD+ESER+VQEAL+ + RTT+V+AHRL+TIRN
Sbjct: 490 QKQRIAIARAIIKSPIILLLDEATSALDSESERVVQEALDNASIGRTTIVIAHRLSTIRN 549
Query: 528 ADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEGSRKSEADKLGDNLNID 587
AD+I VV G+I+E G+H L DG Y+ L+RLQ+ DN K D+++++
Sbjct: 550 ADVICVVHNGRIIETGSHEELLEKLDGQYTSLVRLQQVDN-----------KESDHISVE 598
Query: 588 SHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQLDNKKKPKVSIWR 647
A S ++ + + + + S IV D K S R
Sbjct: 599 EGQASSLSK------------DLKYSPKEFIHSTSSNIVRDFPNLSPKDGKSLVP-SFKR 645
Query: 648 LAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKE-SRFWSLLFV 706
L +N+PE L G + A + G V PI+ + +++S+++ Q KE +R + LLFV
Sbjct: 646 LMSMNRPEWKHALYGCLGAALFGAVQPIYSYSSGSMVSVYFLASHDQIKEKTRIYVLLFV 705
Query: 707 GLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQNVCYF 766
GL L T + Q++ F G L +RIR KI+ E++WFD +S
Sbjct: 706 GLALFTFLSNISQHYGFAYMGEYLTKRIRERMLGKILTFEVNWFDKDENS---------- 755
Query: 767 MTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIAFTA 826
SGA+ +RL+ DA+ V+SLVGD M+L+VQ IS V I
Sbjct: 756 -----------------SGAICSRLAKDANMVRSLVGDRMSLLVQTISAVSITCAIGLVI 798
Query: 827 NWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFCAE 886
+W + +++ + P+I++ Q LK S +A +E+S++A +AVS+IRT+ +F ++
Sbjct: 799 SWRFSIVMMSVQPVIVVCFYTQRVLLKSMSRNAIKGQDESSKLAAEAVSNIRTITAFSSQ 858
Query: 887 SKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTE 946
+++++ GP K R ++GI G S ++ C +A F+ G L+ GK E
Sbjct: 859 ERIINLLKMVQEGPRKDSARQSWLAGIMLGTSQSLITCVSALNFWYGGKLIADGKMMSKE 918
Query: 947 VFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVG 1006
+F T +++ T+ D K D+ AS+F ++D I+ + G + V G
Sbjct: 919 FLEIFLIFASTGRVIAEAGTMTKDLVKGSDAVASVFAVLDRNTTIEPENPDGYVPKKVKG 978
Query: 1007 DIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGR 1066
I +V+F YPTRPD+ IF++ S+ I K+ A+VG SGSGKST+ISL+ERFYDP G
Sbjct: 979 QISFSNVDFAYPTRPDVIIFQNFSIDIEDGKSTAIVGPSGSGKSTIISLIERFYDPLKGI 1038
Query: 1067 ILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGY-GKEGGATEDEIIAAANAAN 1125
+ +DG D+++ L LRQ + LV QEP LF +IR NI Y G E EII AA AAN
Sbjct: 1039 VKIDGRDIRSCHLRSLRQHIALVSQEPTLFAGTIRENIMYGGASNKIDESEIIEAAKAAN 1098
Query: 1126 AHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERI 1185
AH FI++L +GYDT G+RG QLSGGQKQRIAIAR +LKNP +LLLDEATSALD++SE +
Sbjct: 1099 AHDFITSLSNGYDTCCGDRGVQLSGGQKQRIAIARAVLKNPSVLLLDEATSALDSQSESV 1158
Query: 1186 VQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELM-RITDGVYASLV 1244
VQ+AL+R+ V RT+VV+AHRL+TI+ DTIAV++NGAV E G H L+ + G Y SLV
Sbjct: 1159 VQDALERLMVGRTSVVIAHRLSTIQKCDTIAVLENGAVVECGNHSSLLAKGPKGAYFSLV 1218
Query: 1245 AL 1246
+L
Sbjct: 1219 SL 1220
Score = 324 bits (831), Expect = 2e-88
Identities = 206/606 (33%), Positives = 312/606 (50%), Gaps = 50/606 (8%)
Query: 7 VHDNSSSSPTQQHGIRDNKTKQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTL 66
+H SS+ + K VP + + + + G + A G P+ +
Sbjct: 617 IHSTSSNIVRDFPNLSPKDGKSLVPSFKRLMSMNRPEWKHALYGCLGAALFGAVQPIYSY 676
Query: 67 FLGNVINAFGSSNPADAIKQVSKVS-LLFVYLAIGSGIASFLQVTCWMVTGERQAARIRS 125
G++++ + ++ D IK+ +++ LLFV LA+ + +++ Q + GE RIR
Sbjct: 677 SSGSMVSVYFLASH-DQIKEKTRIYVLLFVGLALFTFLSNISQHYGFAYMGEYLTKRIRE 735
Query: 126 LYLKTILQQDIAFFDTETNT-GEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMA 184
L IL ++ +FD + N+ G + R++ D +++ +G+++ Q S +
Sbjct: 736 RMLGKILTFEVNWFDKDENSSGAICSRLAKDANMVRSLVGDRMSLLVQTISAVSITCAIG 795
Query: 185 FIKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASF 244
+ WR +IV+++ P + V +++ MS E+ + + V IRT+ +F
Sbjct: 796 LVISWRFSIVMMSVQPVIVVCFYTQRVLLKSMSRNAIKGQDESSKLAAEAVSNIRTITAF 855
Query: 245 TGEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYN 304
+ +++ I + +Q ++G +G + C L WYG KL+ +
Sbjct: 856 SSQERIINLLKMVQEGPRKDSARQSWLAGIMLGTSQSLITCVSALNFWYGGKLIADGKMM 915
Query: 305 GGTVMTV-IIALMTGGII------------------------------------------ 321
+ + +I TG +I
Sbjct: 916 SKEFLEIFLIFASTGRVIAEAGTMTKDLVKGSDAVASVFAVLDRNTTIEPENPDGYVPKK 975
Query: 322 -KGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPD 380
KG I +V F YP RPDV IF FS+ + G +TA+VG SGSGKST+ISL+ERFYDP
Sbjct: 976 VKGQISFSNVDFAYPTRPDVIIFQNFSIDIEDGKSTAIVGPSGSGKSTIISLIERFYDPL 1035
Query: 381 AGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYG--KEGATDEEITTAIT 438
G V IDG ++++ LR +R+ I LVSQEP LF +IRENI YG + EI A
Sbjct: 1036 KGIVKIDGRDIRSCHLRSLRQHIALVSQEPTLFAGTIRENIMYGGASNKIDESEIIEAAK 1095
Query: 439 LANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAES 498
ANA FI L G DT G G QLSGGQKQRIAIARA+LKNP +LLLDEATSALD++S
Sbjct: 1096 AANAHDFITSLSNGYDTCCGDRGVQLSGGQKQRIAIARAVLKNPSVLLLDEATSALDSQS 1155
Query: 499 ERIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSG-LTMDPDGAYS 557
E +VQ+ALE++++ RT+VV+AHRL+TI+ D IAV++ G +VE G HS L P GAY
Sbjct: 1156 ESVVQDALERLMVGRTSVVIAHRLSTIQKCDTIAVLENGAVVECGNHSSLLAKGPKGAYF 1215
Query: 558 QLIRLQ 563
L+ LQ
Sbjct: 1216 SLVSLQ 1221
>At3g28380 P-glycoprotein, putative
Length = 1240
Score = 882 bits (2280), Expect = 0.0
Identities = 502/1263 (39%), Positives = 733/1263 (57%), Gaps = 101/1263 (7%)
Query: 35 LFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFG--SSNPADAIKQVSKVSL 92
+F AD +D LM +G I AV +G +P++ ++N G SSN ++ +SK +
Sbjct: 23 IFMHADGVDWILMALGLIGAVGDGFITPVVVFIFNTLLNNLGTSSSNNKTFMQTISKNVV 82
Query: 93 LFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTE-TNTGEVIGR 151
+Y+A GS + FL+ CW TGERQAAR+R YL+ +L+QD+ +FD T+T +VI
Sbjct: 83 ALLYVACGSWVICFLEGYCWTRTGERQAARMREKYLRAVLRQDVGYFDLHVTSTSDVITS 142
Query: 152 MSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAVAGAFMSI 211
+S D+++IQ+ + EK+ F AS F ++++FI WRL IV + + V G
Sbjct: 143 ISSDSLVIQDFLSEKLPNFLMNASAFVASYIVSFILMWRLTIVGFPFIILLLVPGLMYGR 202
Query: 212 VMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTTMVKQGIV 271
+ +S + Y EAG++ +Q + ++RTV +F E K I K+++ ++ + ++QG+
Sbjct: 203 ALVSISRKIHEQYNEAGSIAEQAISSVRTVYAFGSENKMIGKFSTALRGSVKLGLRQGLA 262
Query: 272 SGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI----------- 320
G IG + + WYGS+LV+ G GGTV VI + GG+
Sbjct: 263 KGITIGS-NGVTHAIWAFLTWYGSRLVMNHGSKGGTVFVVISCITYGGVSLGQSLSNLKY 321
Query: 321 ---------------------------------IKGDIELRDVSFRYPARPDVQIFDGFS 347
+KG++E V F Y +RP+ IFD
Sbjct: 322 FSEAFVAWERILEVIKRVPDIDSNKKEGQILERMKGEVEFNHVKFTYLSRPETTIFDDLC 381
Query: 348 LFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVS 407
L +P+G T ALVG SGSGKSTVISLL+RFYDP AGE+LIDGV++ LQ+ W+R Q+GLVS
Sbjct: 382 LKIPAGKTVALVGGSGSGKSTVISLLQRFYDPIAGEILIDGVSIDKLQVNWLRSQMGLVS 441
Query: 408 QEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGG 467
QEP+LF TSI ENI +GKE A+ +E+ A +NA FI + P G T G+ G Q+SGG
Sbjct: 442 QEPVLFATSITENILFGKEDASLDEVVEAAKASNAHTFISQFPLGYKTQVGERGVQMSGG 501
Query: 468 QKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLTTIRN 527
QKQRIAIARAI+K+PKILLLDEATSALD+ESER+VQE+L+ + RTT+V+AHRL+TIRN
Sbjct: 502 QKQRIAIARAIIKSPKILLLDEATSALDSESERVVQESLDNASIGRTTIVIAHRLSTIRN 561
Query: 528 ADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEGSRKSEADKLGDNLNID 587
AD+I V+ G+IVE G+H L DG Y+ L+ LQ+ +NE N+NI+
Sbjct: 562 ADVICVIHNGQIVETGSHEELLKRIDGQYTSLVSLQQMENEE------------SNVNIN 609
Query: 588 SHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQLDNKKKPKV-SIW 646
+ S SQ +S+ S + +S I N +P V S
Sbjct: 610 VSVTKDQVMSLSKDFKYSQHNSIGSTSSSIVTNVSDLI----------PNDNQPLVPSFT 659
Query: 647 RLAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKE-SRFWSLLF 705
RL +N+PE L G ++A + GV+ P+ + +VIS+F+ Q KE +R + LLF
Sbjct: 660 RLMVMNRPEWKHALYGCLSAALVGVLQPVSAYSAGSVISVFFLTSHDQIKEKTRIYVLLF 719
Query: 706 VGLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQNVCY 765
VGL + + ++ Q++ F G L +RIR KI+ E++WFD +S
Sbjct: 720 VGLAIFSFLVNISQHYGFAYMGEYLTKRIREQMLSKILTFEVNWFDIDDNS--------- 770
Query: 766 FMTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIAFT 825
SGA+ +RL+ DA+ V+S+VGD M+L+VQ IS VI +I
Sbjct: 771 ------------------SGAICSRLAKDANVVRSMVGDRMSLLVQTISAVIIACIIGLV 812
Query: 826 ANWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFCA 885
W LA +++ + P+I++ Q LK S A +E+S++A +AVS+IRT+ +F +
Sbjct: 813 IAWRLAIVMISVQPLIVVCFYTQRVLLKSLSEKASKAQDESSKLAAEAVSNIRTITAFSS 872
Query: 886 ESKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFT 945
+ +++ + K GP ++ V ++GI G S ++ CT+A F+ G L+ GK
Sbjct: 873 QERIIKLLKKVQEGPRRESVHRSWLAGIVLGTSRSLITCTSALNFWYGGRLIADGKIVSK 932
Query: 946 EVFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVV 1005
F +F T ++ T+ D + D+ S+F ++D I+ + G E +
Sbjct: 933 AFFEIFLIFVTTGRVIADAGTMTTDLARGLDAVGSVFAVLDRCTTIEPKNPDGYVAEKIK 992
Query: 1006 GDIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSG 1065
G I +V+F YPTRPD+ IF++ S+ I K+ A+VG SGSGKST+I L+ERFYDP G
Sbjct: 993 GQITFLNVDFAYPTRPDVVIFENFSIEIDEGKSTAIVGTSGSGKSTIIGLIERFYDPLKG 1052
Query: 1066 RILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGY-GKEGGATEDEIIAAANAA 1124
+ +DG D++++ L LR+ + LV QEP+LF +IR NI Y G E EII AA AA
Sbjct: 1053 TVKIDGRDIRSYHLRSLRKYISLVSQEPMLFAGTIRENIMYGGTSDKIDESEIIEAAKAA 1112
Query: 1125 NAHSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESER 1184
NAH FI++L +GYDT+ G++G QLSGGQKQRIAIAR +LKNP +LLLDEATSALD++SER
Sbjct: 1113 NAHDFITSLSNGYDTNCGDKGVQLSGGQKQRIAIARAVLKNPSVLLLDEATSALDSKSER 1172
Query: 1185 IVQEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELM-RITDGVYASL 1243
+VQ+AL+RV V RT++++AHRL+TI+ D I V+ G + E G H L+ + G Y SL
Sbjct: 1173 VVQDALERVMVGRTSIMIAHRLSTIQNCDMIVVLGKGKIVESGTHSSLLEKGPTGTYFSL 1232
Query: 1244 VAL 1246
+
Sbjct: 1233 AGI 1235
Score = 343 bits (880), Expect = 4e-94
Identities = 213/611 (34%), Positives = 325/611 (52%), Gaps = 50/611 (8%)
Query: 2 AENPNVHDNSSSSPTQQHGIRDNKTKQKVPFYMLFNFADHLDVTLMIIGTISAVANGLAS 61
+++ ++ SSS T + N + VP + + + + G +SA G+
Sbjct: 627 SQHNSIGSTSSSIVTNVSDLIPNDNQPLVPSFTRLMVMNRPEWKHALYGCLSAALVGVLQ 686
Query: 62 PLMTLFLGNVINAFGSSNPADAIKQVSKVS-LLFVYLAIGSGIASFLQVTCWMVTGERQA 120
P+ G+VI+ F ++ D IK+ +++ LLFV LAI S + + Q + GE
Sbjct: 687 PVSAYSAGSVISVFFLTSH-DQIKEKTRIYVLLFVGLAIFSFLVNISQHYGFAYMGEYLT 745
Query: 121 ARIRSLYLKTILQQDIAFFDTETNT-GEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCG 179
RIR L IL ++ +FD + N+ G + R++ D +++ +G+++ Q S
Sbjct: 746 KRIREQMLSKILTFEVNWFDIDDNSSGAICSRLAKDANVVRSMVGDRMSLLVQTISAVII 805
Query: 180 GFVMAFIKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIR 239
++ + WRLAIV+++ P + V +++ +S + A E+ + + V IR
Sbjct: 806 ACIIGLVIAWRLAIVMISVQPLIVVCFYTQRVLLKSLSEKASKAQDESSKLAAEAVSNIR 865
Query: 240 TVASFTGEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVI 299
T+ +F+ +++ I+ + V + ++G +G + CT L WYG +L+
Sbjct: 866 TITAFSSQERIIKLLKKVQEGPRRESVHRSWLAGIVLGTSRSLITCTSALNFWYGGRLIA 925
Query: 300 EKGYNGGTVMTVIIALMTGG---------------------------------------- 319
+ + + +T G
Sbjct: 926 DGKIVSKAFFEIFLIFVTTGRVIADAGTMTTDLARGLDAVGSVFAVLDRCTTIEPKNPDG 985
Query: 320 ----IIKGDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLER 375
IKG I +V F YP RPDV IF+ FS+ + G +TA+VG SGSGKST+I L+ER
Sbjct: 986 YVAEKIKGQITFLNVDFAYPTRPDVVIFENFSIEIDEGKSTAIVGTSGSGKSTIIGLIER 1045
Query: 376 FYDPDAGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYG--KEGATDEEI 433
FYDP G V IDG ++++ LR +R+ I LVSQEP+LF +IRENI YG + + EI
Sbjct: 1046 FYDPLKGTVKIDGRDIRSYHLRSLRKYISLVSQEPMLFAGTIRENIMYGGTSDKIDESEI 1105
Query: 434 TTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSA 493
A ANA FI L G DT G G QLSGGQKQRIAIARA+LKNP +LLLDEATSA
Sbjct: 1106 IEAAKAANAHDFITSLSNGYDTNCGDKGVQLSGGQKQRIAIARAVLKNPSVLLLDEATSA 1165
Query: 494 LDAESERIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSG-LTMDP 552
LD++SER+VQ+ALE++++ RT++++AHRL+TI+N D+I V+ +GKIVE GTHS L P
Sbjct: 1166 LDSKSERVVQDALERVMVGRTSIMIAHRLSTIQNCDMIVVLGKGKIVESGTHSSLLEKGP 1225
Query: 553 DGAYSQLIRLQ 563
G Y L +Q
Sbjct: 1226 TGTYFSLAGIQ 1236
>At3g28360 P-glycoprotein like protein
Length = 1158
Score = 858 bits (2218), Expect = 0.0
Identities = 489/1201 (40%), Positives = 705/1201 (57%), Gaps = 100/1201 (8%)
Query: 95 VYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTE-TNTGEVIGRMS 153
+Y+A S + FL+ CW TGERQAA++R YL+ +L+QD+ +FD T+T ++I +S
Sbjct: 2 LYVACASWVICFLEGYCWTRTGERQAAKMRERYLRAVLRQDVGYFDLHVTSTSDIITSVS 61
Query: 154 GDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAVAGAFMSIVM 213
D+++IQ+ + EK+ AS F G +++ F+ WRL IV + + + G +
Sbjct: 62 SDSLVIQDFLSEKLPNILMNASAFVGSYIVGFMLLWRLTIVGFPFIILLLIPGLMYGRAL 121
Query: 214 AKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTTMVKQGIVSG 273
+S + + Y EAG++ +Q + ++RTV +F EKK IEK++ ++ + ++QG+ G
Sbjct: 122 IGISRKIREEYNEAGSIAEQAISSVRTVYAFVSEKKMIEKFSDALQGSVKLGLRQGLAKG 181
Query: 274 FGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI------------- 320
IG I + +G WYGS++V+ GY GGTV TV + + GG
Sbjct: 182 IAIGS-NGIVYAIWGFLTWYGSRMVMNYGYKGGTVSTVTVCVTFGGTALGQALSNLKYFS 240
Query: 321 -------------------------------IKGDIELRDVSFRYPARPDVQIFDGFSLF 349
I+G++E +V +YP+RP+ IFD L
Sbjct: 241 EAFVAGERIQKMIKRVPDIDSDNLNGHILETIRGEVEFNNVKCKYPSRPETLIFDDLCLK 300
Query: 350 VPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVSQE 409
+PSG T ALVG SGSGKSTVISLL+RFYDP+ G++LID V++ N+Q++W+R Q+G+VSQE
Sbjct: 301 IPSGKTVALVGGSGSGKSTVISLLQRFYDPNEGDILIDSVSINNMQVKWLRSQMGMVSQE 360
Query: 410 PILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGGQK 469
P LF TSI+ENI +GKE A+ +E+ A +NA FI + P G T G+ G +SGGQK
Sbjct: 361 PSLFATSIKENILFGKEDASFDEVVEAAKASNAHNFISQFPHGYQTQVGERGVHMSGGQK 420
Query: 470 QRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLTTIRNAD 529
QRIAIARA++K+P ILLLDEATSALD ESER+VQEAL+ + RTT+V+AHRL+TIRNAD
Sbjct: 421 QRIAIARALIKSPIILLLDEATSALDLESERVVQEALDNASVGRTTIVIAHRLSTIRNAD 480
Query: 530 IIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEGSRKSEADKLGDNLNIDSH 589
II V+ G IVE G+H L M+ DG Y+ L+RLQ+ NE + D+
Sbjct: 481 IICVLHNGCIVETGSHDKL-MEIDGKYTSLVRLQQMKNEE---------------SCDNT 524
Query: 590 MAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQLDNKKKPKV-SIWRL 648
G R S +R+ + HS +S IV + + KKP V S RL
Sbjct: 525 SVGVKEGRVSSLRNDLDYNPRDLAHS-----MSSSIVTN--LSDSIPQDKKPLVPSFKRL 577
Query: 649 AKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKE-SRFWSLLFVG 707
+N+PE L G ++A + G V PI+ + +IS+F+ +Q KE +R + LLF G
Sbjct: 578 MAMNRPEWKHALCGCLSASLGGAVQPIYAYSSGLMISVFFLTNHEQIKENTRIYVLLFFG 637
Query: 708 LGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQNVCYFM 767
L L T Q + F G L +RIR KI+ E++WFD+
Sbjct: 638 LALFTFFTSISQQYSFSYMGEYLTKRIREQMLSKILTFEVNWFDE--------------- 682
Query: 768 TK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIAFTAN 827
E + SGA+ +RL+ DA+ V+SLVG+ M+L+VQ ISTV+ I
Sbjct: 683 ------------EENSSGAICSRLAKDANVVRSLVGERMSLLVQTISTVMVACTIGLVIA 730
Query: 828 WILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFCAES 887
W +++ + P+I++ +Q LK S A + +E+S++A +AVS+IRT+ +F ++
Sbjct: 731 WRFTIVMISVQPVIIVCYYIQRVLLKNMSKKAIIAQDESSKLAAEAVSNIRTITTFSSQE 790
Query: 888 KVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTEV 947
++M + + GP ++ R ++GI G + ++ CT+A F+ G L+ GK
Sbjct: 791 RIMKLLERVQEGPRRESARQSWLAGIMLGTTQSLITCTSALNFWYGGKLIADGKMVSKAF 850
Query: 948 FRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVGD 1007
F +F T A+++ T+ D K +S S+F ++D + I+ + G E + G
Sbjct: 851 FELFLIFKTTGRAIAEAGTMTTDLAKGSNSVDSVFTVLDRRTTIEPENPDGYILEKIKGQ 910
Query: 1008 IELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRI 1067
I +V+F YPTRP++ IF + S+ I K+ A+VG S SGKSTVI L+ERFYDP G +
Sbjct: 911 ITFLNVDFAYPTRPNMVIFNNFSIEIHEGKSTAIVGPSRSGKSTVIGLIERFYDPLQGIV 970
Query: 1068 LLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGG-ATEDEIIAAANAANA 1126
+DG D++++ L LRQ M LV QEP LF +IR NI YG+ E EII A ANA
Sbjct: 971 KIDGRDIRSYHLRSLRQHMSLVSQEPTLFAGTIRENIMYGRASNKIDESEIIEAGKTANA 1030
Query: 1127 HSFISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIV 1186
H FI++L DGYDT G+RG QLSGGQKQRIAIART+LKNP ILLLDEATSALD++SER+V
Sbjct: 1031 HEFITSLSDGYDTYCGDRGVQLSGGQKQRIAIARTILKNPSILLLDEATSALDSQSERVV 1090
Query: 1187 QEALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELM-RITDGVYASLVA 1245
Q+AL+ V V +T+VV+AHRL+TI+ DTIAV+ G V E G H L+ + G Y SLV+
Sbjct: 1091 QDALEHVMVGKTSVVIAHRLSTIQNCDTIAVLDKGKVVESGTHASLLAKGPTGSYFSLVS 1150
Query: 1246 L 1246
L
Sbjct: 1151 L 1151
Score = 335 bits (858), Expect = 1e-91
Identities = 217/604 (35%), Positives = 312/604 (50%), Gaps = 48/604 (7%)
Query: 8 HDNSSSSPTQQHGIRDNKTKQKVPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLF 67
H SSS T K VP + + + + G +SA G P+
Sbjct: 549 HSMSSSIVTNLSDSIPQDKKPLVPSFKRLMAMNRPEWKHALCGCLSASLGGAVQPIYAYS 608
Query: 68 LGNVINAFGSSNPADAIKQVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLY 127
G +I+ F +N + LLF LA+ + S Q + GE RIR
Sbjct: 609 SGLMISVFFLTNHEQIKENTRIYVLLFFGLALFTFFTSISQQYSFSYMGEYLTKRIREQM 668
Query: 128 LKTILQQDIAFFDTETNT-GEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFI 186
L IL ++ +FD E N+ G + R++ D +++ +GE++ Q S + +
Sbjct: 669 LSKILTFEVNWFDEEENSSGAICSRLAKDANVVRSLVGERMSLLVQTISTVMVACTIGLV 728
Query: 187 KGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTG 246
WR IV+++ P + V +++ MS + IA E+ + + V IRT+ +F+
Sbjct: 729 IAWRFTIVMISVQPVIIVCYYIQRVLLKNMSKKAIIAQDESSKLAAEAVSNIRTITTFSS 788
Query: 247 EKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLV-------- 298
+++ ++ + +Q ++G +G + CT L WYG KL+
Sbjct: 789 QERIMKLLERVQEGPRRESARQSWLAGIMLGTTQSLITCTSALNFWYGGKLIADGKMVSK 848
Query: 299 -------------------------IEKGYNG-GTVMTVIIALMT-------GGI---IK 322
+ KG N +V TV+ T G I IK
Sbjct: 849 AFFELFLIFKTTGRAIAEAGTMTTDLAKGSNSVDSVFTVLDRRTTIEPENPDGYILEKIK 908
Query: 323 GDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAG 382
G I +V F YP RP++ IF+ FS+ + G +TA+VG S SGKSTVI L+ERFYDP G
Sbjct: 909 GQITFLNVDFAYPTRPNMVIFNNFSIEIHEGKSTAIVGPSRSGKSTVIGLIERFYDPLQG 968
Query: 383 EVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEG--ATDEEITTAITLA 440
V IDG ++++ LR +R+ + LVSQEP LF +IRENI YG+ + EI A A
Sbjct: 969 IVKIDGRDIRSYHLRSLRQHMSLVSQEPTLFAGTIRENIMYGRASNKIDESEIIEAGKTA 1028
Query: 441 NAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESER 500
NA +FI L G DT G G QLSGGQKQRIAIAR ILKNP ILLLDEATSALD++SER
Sbjct: 1029 NAHEFITSLSDGYDTYCGDRGVQLSGGQKQRIAIARTILKNPSILLLDEATSALDSQSER 1088
Query: 501 IVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSG-LTMDPDGAYSQL 559
+VQ+ALE +++ +T+VV+AHRL+TI+N D IAV+ +GK+VE GTH+ L P G+Y L
Sbjct: 1089 VVQDALEHVMVGKTSVVIAHRLSTIQNCDTIAVLDKGKVVESGTHASLLAKGPTGSYFSL 1148
Query: 560 IRLQ 563
+ LQ
Sbjct: 1149 VSLQ 1152
>At3g28415 putative protein
Length = 1098
Score = 732 bits (1889), Expect = 0.0
Identities = 420/1144 (36%), Positives = 641/1144 (55%), Gaps = 104/1144 (9%)
Query: 35 LFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSSNPADA--IKQVSKVSL 92
+F A+ +D+ LM +G I AV +G +P++ G ++N G S+ D + + K ++
Sbjct: 10 IFMHANSVDLVLMGLGLIGAVGDGFITPIIFFITGLLLNDIGDSSFGDKTFMHAIMKNAV 69
Query: 93 LFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTE-TNTGEVIGR 151
+Y+A S + F+ GERQA+R+R YL+ +L+QD+ +FD T+T +VI
Sbjct: 70 ALLYVAGASLVICFV--------GERQASRMREKYLRAVLRQDVGYFDLHVTSTSDVITS 121
Query: 152 MSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAVAGAFMSI 211
+S DT++IQ+ + EK+ F AS F +++ FI WRL IV + + G
Sbjct: 122 VSSDTLVIQDVLSEKLPNFLMSASAFVASYIVGFIMLWRLTIVGFPFFILLLIPGLMCGR 181
Query: 212 VMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTTMVKQGIV 271
+ +S + + Y EAG++ +Q + +RTV +F E+K I K+++ ++ + ++QGI
Sbjct: 182 ALINISRKIREEYNEAGSIAEQAISLVRTVYAFGSERKMISKFSAALEGSVKLGLRQGIA 241
Query: 272 SGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI----------- 320
G IG + + +G WYGS++V+ G GGT+ VII + GG
Sbjct: 242 KGIAIGS-NGVTYAIWGFMTWYGSRMVMYHGAKGGTIFAVIICITYGGTSLGRGLSNLKY 300
Query: 321 ---------------------------------IKGDIELRDVSFRYPARPDVQIFDGFS 347
IKG+++ + V F Y +RP+ IFD
Sbjct: 301 FSEAVVAGERIIEVIKRVPDIDSDNPRGQVLENIKGEVQFKHVKFMYSSRPETPIFDDLC 360
Query: 348 LFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVS 407
L +PSG + ALVG SGSGKSTVISLL+RFYDP GE+LIDGV++K LQ++W+R Q+GLVS
Sbjct: 361 LRIPSGKSVALVGGSGSGKSTVISLLQRFYDPIVGEILIDGVSIKKLQVKWLRSQMGLVS 420
Query: 408 QEPILFTTSIRENIAYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGG 467
QEP LF TSI ENI +GKE A+ +E+ A +NA FI + P G T G+ G Q+SGG
Sbjct: 421 QEPALFATSIEENILFGKEDASFDEVVEAAKSSNAHDFISQFPLGYKTQVGERGVQMSGG 480
Query: 468 QKQRIAIARAILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLTTIRN 527
QKQRI+IARAI+K+P +LLLDEATSALD+ESER+VQEAL+ + RTT+V+AHRL+TIRN
Sbjct: 481 QKQRISIARAIIKSPTLLLLDEATSALDSESERVVQEALDNATIGRTTIVIAHRLSTIRN 540
Query: 528 ADIIAVVQQGKIVERGTHSGLTMDPDGAYSQLIRLQEGDNEAEGSRKSEADKLGDNLNID 587
D+I V + G+IVE G+H L + DG Y+ L+RLQ +NE S + + G N +
Sbjct: 541 VDVICVFKNGQIVETGSHEELMENVDGQYTSLVRLQIMENEESNDNVSVSMREGQFSNFN 600
Query: 588 SHMAGSSTQRTSFVRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQLDNKKKPKVSIWR 647
+ SS S+ TSS+ L+G I K K S R
Sbjct: 601 KDVKYSSRLSIQSRSSLFATSSID-------TNLAGSI------------PKDKKPSFKR 641
Query: 648 LAKLNKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKE-SRFWSLLFV 706
L +NKPE L G ++A++ G + PI+ + +++S+++ + KE +R + LLFV
Sbjct: 642 LMAMNKPEWKHALYGCLSAVLYGALHPIYAYASGSMVSVYFLTSHDEMKEKTRIYVLLFV 701
Query: 707 GLGLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQNVCYF 766
GL ++ +I +Q + F G L +RIR K++ E+SWFD+ +S
Sbjct: 702 GLAVLCFLISIIQQYSFAYMGEYLTKRIRENILSKLLTFEVSWFDEDENS---------- 751
Query: 767 MTK**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIAFTA 826
SG++ +RL+ DA+ V+SLVG+ ++L+VQ IS V +
Sbjct: 752 -----------------SGSICSRLAKDANVVRSLVGERVSLLVQTISAVSVACTLGLAI 794
Query: 827 NWILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFCAE 886
+W L+ +++ + P+++ Q LK S A +E+S++A +AVS+IRT+ +F ++
Sbjct: 795 SWKLSIVMIAIQPVVVGCFYTQRIVLKSISKKAIKAQDESSKLAAEAVSNIRTITAFSSQ 854
Query: 887 SKVMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTE 946
+++ + GP ++ +R ++GI S ++ CT+A ++ G+ L+ GK T
Sbjct: 855 ERILKLLKMVQEGPQRENIRQSWLAGIVLATSRSLMTCTSALNYWYGARLIIDGKITSKA 914
Query: 947 VFRVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVG 1006
F +F T ++ + D K D+ S+F ++D +I+ G + + G
Sbjct: 915 FFELFILFVSTGRVIADAGAMTMDLAKGSDAVGSVFAVLDRYTNIEPEKPDGFVPQNIKG 974
Query: 1007 DIELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGR 1066
I+ +V+F YPTRPD+ IFK+ S+ I K+ A+VG SGSGKST+I L+ERFYDP G
Sbjct: 975 QIKFVNVDFAYPTRPDVIIFKNFSIDIDEGKSTAIVGPSGSGKSTIIGLIERFYDPLKGI 1034
Query: 1067 ILLDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGY-GKEGGATEDEIIAAANAAN 1125
+ +DG D++++ L LRQ +GLV QEPILF +IR NI Y G E EII AA AAN
Sbjct: 1035 VKIDGRDIRSYHLRSLRQHIGLVSQEPILFAGTIRENIMYGGASDKIDESEIIEAAKAAN 1094
Query: 1126 AHSF 1129
AH F
Sbjct: 1095 AHDF 1098
Score = 381 bits (978), Expect = e-105
Identities = 224/598 (37%), Positives = 332/598 (55%), Gaps = 40/598 (6%)
Query: 652 NKPEIPVILLGAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKESRFWSLLFVGL--- 708
N ++ ++ LG I A+ +G + PI F+ +++ +S F F+
Sbjct: 15 NSVDLVLMGLGLIGAVGDGFITPIIFFITGLLLNDI--------GDSSFGDKTFMHAIMK 66
Query: 709 GLVTLVILPLQNFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQNVCYFMT 768
V L+ + + G + R+R ++ Q++ +FD
Sbjct: 67 NAVALLYVAGASLVICFVGERQASRMREKYLRAVLRQDVGYFD----------------- 109
Query: 769 K**KNYETERNESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIAFTANW 828
+ +S S + + +S D ++ ++ + + + + S +A ++ F W
Sbjct: 110 ---------LHVTSTSDVITS-VSSDTLVIQDVLSEKLPNFLMSASAFVASYIVGFIMLW 159
Query: 829 ILAFIVLVLTPMILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFCAESK 888
L + ++L+ G++ + L S + Y EA +A A+S +RTV +F +E K
Sbjct: 160 RLTIVGFPFFILLLIPGLMCGRALINISRKIREEYNEAGSIAEQAISLVRTVYAFGSERK 219
Query: 889 VMDMYSKKCLGPAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTEVF 948
++ +S G K G+R G+ GI G + V Y F+ + GS +V + A +F
Sbjct: 220 MISKFSAALEGSVKLGLRQGIAKGIAIGSNG-VTYAIWGFMTWYGSRMVMYHGAKGGTIF 278
Query: 949 RVFFALTMTAIAVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVGDI 1008
V +T ++ + + ++A + I E+I PDIDS + G E + G++
Sbjct: 279 AVIICITYGGTSLGRGLSNLKYFSEAVVAGERIIEVIKRVPDIDSDNPRGQVLENIKGEV 338
Query: 1009 ELQHVNFNYPTRPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRIL 1068
+ +HV F Y +RP+ IF DL L IPS K++ALVG SGSGKSTVISLL+RFYDP G IL
Sbjct: 339 QFKHVKFMYSSRPETPIFDDLCLRIPSGKSVALVGGSGSGKSTVISLLQRFYDPIVGEIL 398
Query: 1069 LDGVDLKTFRLSWLRQQMGLVGQEPILFNESIRANIGYGKEGGATEDEIIAAANAANAHS 1128
+DGV +K ++ WLR QMGLV QEP LF SI NI +GKE A+ DE++ AA ++NAH
Sbjct: 399 IDGVSIKKLQVKWLRSQMGLVSQEPALFATSIEENILFGKED-ASFDEVVEAAKSSNAHD 457
Query: 1129 FISNLPDGYDTSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIVQE 1188
FIS P GY T VGERG Q+SGGQKQRI+IAR ++K+P +LLLDEATSALD+ESER+VQE
Sbjct: 458 FISQFPLGYKTQVGERGVQMSGGQKQRISIARAIIKSPTLLLLDEATSALDSESERVVQE 517
Query: 1189 ALDRVSVNRTTVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELMRITDGVYASLVAL 1246
ALD ++ RTT+V+AHRL+TIR D I V KNG + E G H+ELM DG Y SLV L
Sbjct: 518 ALDNATIGRTTIVIAHRLSTIRNVDVICVFKNGQIVETGSHEELMENVDGQYTSLVRL 575
>At3g55320 P-glycoprotein - like
Length = 1408
Score = 695 bits (1794), Expect = 0.0
Identities = 450/1360 (33%), Positives = 694/1360 (50%), Gaps = 175/1360 (12%)
Query: 30 VPFYMLFNFADHLDVTLMIIGTISAVANGLASPLMTLFLGNVINAFGSSNPADAIK---- 85
VPF LF AD D LMI+G+++A A+G A + + +++ SN + +
Sbjct: 71 VPFSQLFACADRFDWVLMIVGSVAAAAHGTALIVYLHYFAKIVDVLAFSNDSSQQRSEHQ 130
Query: 86 --QVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTET 143
++ ++SL VY+A G I+ +++V+CW++TGERQ A IRS Y++ +L QD++FFDT
Sbjct: 131 FDRLVQLSLTIVYIAGGVFISGWIEVSCWILTGERQTAVIRSKYVQVLLNQDMSFFDTYG 190
Query: 144 NTGEVIGRMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVA 203
N G+++ ++ D +LIQ A+ EKVG + + F G V+ F+ W +A++ LA P +
Sbjct: 191 NNGDIVSQVLSDVLLIQSALSEKVGNYIHNMATFISGLVIGFVNCWEIALITLATGPFIV 250
Query: 204 VAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYT 263
AG +I + +++ Q AYAEA + +Q + IRT+ +FT E A Y + ++
Sbjct: 251 AAGGISNIFLHRLAENIQDAYAEAAGIAEQAISYIRTLYAFTNETLAKYSYATSLQATLR 310
Query: 264 TMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGYNGGTVMTVIIALMTGGI--- 320
+ +V G G+G +A C+ L +W G V NGG ++ + A++ G+
Sbjct: 311 YGILISLVQGLGLGFTYGLAICSCALQLWIGRFFVHNGRANGGEIIAALFAVILSGLGLN 370
Query: 321 ---------------------------------------IKGDIELRDVSFRYPARPDVQ 341
++G+IE R+V F Y +RP++
Sbjct: 371 QAATNFYSFDQGRIAAYRLFEMITRSSSVANQEGAVLASVQGNIEFRNVYFSYLSRPEIP 430
Query: 342 IFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIRE 401
I GF L VP+ ALVG++GSGKS++I L+ERFYDP GEVL+DG N+KNL+L W+R
Sbjct: 431 ILSGFYLTVPAKKAVALVGRNGSGKSSIIPLMERFYDPTLGEVLLDGENIKNLKLEWLRS 490
Query: 402 QIGLVSQEPILFTTSI----------------------------------------RENI 421
QIGLV+QEP L + SI R +
Sbjct: 491 QIGLVTQEPALLSLSIRENIAYGRDATLDQIEEAAKNAHAHTFISSLEKGYETQVGRAGL 550
Query: 422 AYGKEGATDEEITTAITLANAKKFIDKLPQGLDTMAG---QNGTQLSGGQKQRIAIAR-- 476
A +E I A+ L +D++ GLD A Q L + I IAR
Sbjct: 551 AMTEEQKIKLSIARAVLLNPTILLLDEVTGGLDFEAERIVQEALDLLMLGRSTIIIARRL 610
Query: 477 AILKNPKILLLDEATSALD--AESERIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVV 534
+++KN + + E ++ E I L +LK R +RN AV
Sbjct: 611 SLIKNADYIAVMEEGQLVEMGTHDELINLGGLYAELLKCEEATKLPRRMPVRNYKESAVF 670
Query: 535 QQGKIVERGTHSGLTMDPDGAY-----------SQLIRLQEGDNEAEGSRKSEA---DKL 580
+ VER + +G + + S + R QE + E S K+ + +K
Sbjct: 671 E----VERDSSAGCGVQEPSSPKMIKSPSLQRGSGVFRPQELCFDTEESPKAHSPASEKT 726
Query: 581 G-DNLNIDSHMAGSSTQRT-SF-----------VRSISQTSSVSH--------------- 612
G D +++D + +R SF V+ Q S+ S
Sbjct: 727 GEDGMSLDCADKEPTIKRQDSFEMRLPHLPKVDVQCPQQKSNGSEPESPVSPLLTSDPKN 786
Query: 613 --RHSQSL-RGLSGEIVESDIEQGQLDNKKKPKVSIWRLAKLNKPEIPVILLGAIAAIVN 669
HSQ+ R LS + D + K S WRLA+L+ PE +LG++ A +
Sbjct: 787 ERSHSQTFSRPLSSPDDTKANGKASKDAQHKESPSFWRLAQLSFPEWLYAVLGSLGAAIF 846
Query: 670 GVVFPIFGFLFSAVISMFYKPPEQQ-RKESRFWSLLFVGLGLVTLVILPLQNFFFGIAGG 728
G P+ ++ + V++ +YK R+E W L+ +G+VT+V LQ+F+FGI G
Sbjct: 847 GSFNPLLAYVIALVVTEYYKSKGGHLREEVDKWCLIIACMGIVTVVANFLQHFYFGIMGE 906
Query: 729 KLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGAVG 788
K+ ER+R + F ++ E+ WFDD E + +
Sbjct: 907 KMTERVRRMMFSAMLRNEVGWFDD---------------------------EENSPDTLS 939
Query: 789 ARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGIVQ 848
RL+ DA+ V++ + +++ +Q+ VI L+I W LA + L P++ + I Q
Sbjct: 940 MRLANDATFVRAAFSNRLSIFIQDSFAVIVALLIGLLLGWRLALVALATLPILTLSAIAQ 999
Query: 849 MKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVRLG 908
+L GFS + M+ +AS V DAV +I TV +FCA +KVM++Y + +Q G
Sbjct: 1000 KLWLAGFSKGIQEMHRKASLVLEDAVRNIYTVVAFCAGNKVMELYRMQLQRILRQSYLHG 1059
Query: 909 LVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTTLA 968
+ G FG S +L+ NA + + ++ V G + + + A+ + LA
Sbjct: 1060 MAIGFAFGFSQFLLFACNALLLWCTALSVNRGYMKLSTAITEYMVFSFATFALVEPFGLA 1119
Query: 969 PDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIFKD 1028
P K + S S+FEI+D P I+ N+ + V G IEL++V+F YPTRP+I + +
Sbjct: 1120 PYILKRRKSLISVFEIVDRVPTIEPDDNSALKPPNVYGSIELKNVDFCYPTRPEILVLSN 1179
Query: 1029 LSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQMGL 1088
SL I +T+A+VG SGSGKST+ISL+ER+YDP +G++LLDG DLK + L WLR MGL
Sbjct: 1180 FSLKISGGQTVAVVGVSGSGKSTIISLVERYYDPVAGQVLLDGRDLKLYNLRWLRSHMGL 1239
Query: 1089 VGQEPILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGTQL 1148
V QEPI+F+ +IR NI Y + A+E E+ AA ANAH FIS+LP GYDT +G RG +L
Sbjct: 1240 VQQEPIIFSTTIRENIIYARH-NASEAEMKEAARIANAHHFISSLPHGYDTHIGMRGVEL 1298
Query: 1149 SGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSV-NRTTVVVAHRLT 1207
+ GQKQRIAIAR +LKN I+L+DEA+S++++ES R+VQEALD + + N+TT+++AHR
Sbjct: 1299 TPGQKQRIAIARVVLKNAPIILIDEASSSIESESSRVVQEALDTLIMGNKTTILIAHRAA 1358
Query: 1208 TIRGADTIAVIKNGAVAEKGRHDELMRITDGVYASLVALH 1247
+R D I V+ G + E+G HD L +G+Y L+ H
Sbjct: 1359 MMRHVDNIVVLNGGRIVEEGTHDSL-AAKNGLYVRLMQPH 1397
Score = 347 bits (891), Expect = 2e-95
Identities = 211/638 (33%), Positives = 352/638 (55%), Gaps = 49/638 (7%)
Query: 620 GLSGEIVESDIEQGQLDNKKKPKVSI---WRLAKLNKPEIPVILLGAIAAIVNGVVFPIF 676
G + + E+D E D + P ++ A ++ + ++++G++AA +G ++
Sbjct: 46 GTAAALAEADEEMDDQDELEPPPAAVPFSQLFACADRFDWVLMIVGSVAAAAHGTALIVY 105
Query: 677 GFLFSAVISM--FYKPPEQQRKESRFWSLLFVGLGLVTLVILPLQNFFFG--------IA 726
F+ ++ + F QQR E +F L+ + L T+V + F G +
Sbjct: 106 LHYFAKIVDVLAFSNDSSQQRSEHQFDRLVQLSL---TIVYIAGGVFISGWIEVSCWILT 162
Query: 727 GGKLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQNVCYFMTK**KNYETERNESSCSGA 786
G + IRS + +++Q++S+FD ++ G
Sbjct: 163 GERQTAVIRSKYVQVLLNQDMSFFDTYGNN----------------------------GD 194
Query: 787 VGARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTPMILMQGI 846
+ +++ D ++S + + + + N++T I+GLVI F W +A I L P I+ G
Sbjct: 195 IVSQVLSDVLLIQSALSEKVGNYIHNMATFISGLVIGFVNCWEIALITLATGPFIVAAGG 254
Query: 847 VQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFCAESKVMDMYSKKCLGPAKQGVR 906
+ FL + + + Y EA+ +A A+S IRT+ +F E+ Y+ + G+
Sbjct: 255 ISNIFLHRLAENIQDAYAEAAGIAEQAISYIRTLYAFTNETLAKYSYATSLQATLRYGIL 314
Query: 907 LGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAIAVSQTTT 966
+ LV G+G G ++ + C+ A +IG V +G+A E+ FA+ ++ + ++Q T
Sbjct: 315 ISLVQGLGLGFTYGLAICSCALQLWIGRFFVHNGRANGGEIIAALFAVILSGLGLNQAAT 374
Query: 967 LAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPTRPDIQIF 1026
++ + +A +FE+I + ++ G +V G+IE ++V F+Y +RP+I I
Sbjct: 375 NFYSFDQGRIAAYRLFEMITRSSSV--ANQEGAVLASVQGNIEFRNVYFSYLSRPEIPIL 432
Query: 1027 KDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRLSWLRQQM 1086
L++P+ K +ALVG +GSGKS++I L+ERFYDP G +LLDG ++K +L WLR Q+
Sbjct: 433 SGFYLTVPAKKAVALVGRNGSGKSSIIPLMERFYDPTLGEVLLDGENIKNLKLEWLRSQI 492
Query: 1087 GLVGQEPILFNESIRANIGYGKEGGATEDEIIAAANAANAHSFISNLPDGYDTSVGERGT 1146
GLV QEP L + SIR NI YG++ AT D+I AA A+AH+FIS+L GY+T VG G
Sbjct: 493 GLVTQEPALLSLSIRENIAYGRD--ATLDQIEEAAKNAHAHTFISSLEKGYETQVGRAGL 550
Query: 1147 QLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRTTVVVAHRL 1206
++ QK +++IAR +L NP ILLLDE T LD E+ERIVQEALD + + R+T+++A RL
Sbjct: 551 AMTEEQKIKLSIARAVLLNPTILLLDEVTGGLDFEAERIVQEALDLLMLGRSTIIIARRL 610
Query: 1207 TTIRGADTIAVIKNGAVAEKGRHDELMRITDGVYASLV 1244
+ I+ AD IAV++ G + E G HDEL+ + G+YA L+
Sbjct: 611 SLIKNADYIAVMEEGQLVEMGTHDELINL-GGLYAELL 647
Score = 330 bits (847), Expect = 2e-90
Identities = 199/602 (33%), Positives = 324/602 (53%), Gaps = 56/602 (9%)
Query: 11 SSSSPTQQHG--IRDNKTKQKVPFYML--FNFADHLDVTLMIIGTISAVANGLASPLMTL 66
SS T+ +G +D + K+ F+ L +F + L ++G++ A G +PL+
Sbjct: 799 SSPDDTKANGKASKDAQHKESPSFWRLAQLSFPEWL---YAVLGSLGAAIFGSFNPLLAY 855
Query: 67 FLGNVINAFGSSNPADAIKQVSKVSLLFVYLAIGSGIASFLQVTCWMVTGERQAARIRSL 126
+ V+ + S ++V K L+ + I + +A+FLQ + + GE+ R+R +
Sbjct: 856 VIALVVTEYYKSKGGHLREEVDKWCLIIACMGIVTVVANFLQHFYFGIMGEKMTERVRRM 915
Query: 127 YLKTILQQDIAFFDTETNTGEVIG-RMSGDTILIQEAMGEKVGKFFQLASNFCGGFVMAF 185
+L+ ++ +FD E N+ + + R++ D ++ A ++ F Q + ++
Sbjct: 916 MFSAMLRNEVGWFDDEENSPDTLSMRLANDATFVRAAFSNRLSIFIQDSFAVIVALLIGL 975
Query: 186 IKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYAEAGNVVDQTVGAIRTVASFT 245
+ GWRLA+V LA +P + ++ + +A S Q + +A V++ V I TV +F
Sbjct: 976 LLGWRLALVALATLPILTLSAIAQKLWLAGFSKGIQEMHRKASLVLEDAVRNIYTVVAFC 1035
Query: 246 GEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFCTYGLAMWYGSKLVIEKGY-N 304
K +E Y +++ G+ GF G F+ F L +W + L + +GY
Sbjct: 1036 AGNKVMELYRMQLQRILRQSYLHGMAIGFAFGFSQFLLFACNALLLWC-TALSVNRGYMK 1094
Query: 305 GGTVMT--VIIALMTGGIIK---------------------------------------- 322
T +T ++ + T +++
Sbjct: 1095 LSTAITEYMVFSFATFALVEPFGLAPYILKRRKSLISVFEIVDRVPTIEPDDNSALKPPN 1154
Query: 323 --GDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPD 380
G IEL++V F YP RP++ + FSL + G T A+VG SGSGKST+ISL+ER+YDP
Sbjct: 1155 VYGSIELKNVDFCYPTRPEILVLSNFSLKISGGQTVAVVGVSGSGKSTIISLVERYYDPV 1214
Query: 381 AGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIRENIAYGKEGATDEEITTAITLA 440
AG+VL+DG +LK LRW+R +GLV QEPI+F+T+IRENI Y + A++ E+ A +A
Sbjct: 1215 AGQVLLDGRDLKLYNLRWLRSHMGLVQQEPIIFSTTIRENIIYARHNASEAEMKEAARIA 1274
Query: 441 NAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESER 500
NA FI LP G DT G G +L+ GQKQRIAIAR +LKN I+L+DEA+S++++ES R
Sbjct: 1275 NAHHFISSLPHGYDTHIGMRGVELTPGQKQRIAIARVVLKNAPIILIDEASSSIESESSR 1334
Query: 501 IVQEALEKIIL-KRTTVVVAHRLTTIRNADIIAVVQQGKIVERGTHSGLTMDPDGAYSQL 559
+VQEAL+ +I+ +TT+++AHR +R+ D I V+ G+IVE GTH L +G Y +L
Sbjct: 1335 VVQEALDTLIMGNKTTILIAHRAAMMRHVDNIVVLNGGRIVEEGTHDSLAA-KNGLYVRL 1393
Query: 560 IR 561
++
Sbjct: 1394 MQ 1395
>At3g28344 P-glycoprotein, 5' partial
Length = 626
Score = 431 bits (1108), Expect = e-120
Identities = 251/647 (38%), Positives = 374/647 (57%), Gaps = 36/647 (5%)
Query: 601 VRSISQTSSVSHRHSQSLRGLSGEIVESDIEQGQLDNKKKPKVSIWRLAKLNKPEIPVIL 660
+R+ S+ S++S S S ++G S I+ DNK + S RL +N PE L
Sbjct: 13 IRNSSRVSTLSR--SSSANSVTGP---STIKNLSEDNKPQLP-SFKRLLAMNLPEWKQAL 66
Query: 661 LGAIAAIVNGVVFPIFGFLFSAVISMFYKPPEQQRKE-SRFWSLLFVGLGLVTLVILPLQ 719
G I+A + G + P + + +++S+++ + KE +R ++L FVGL +++ +I Q
Sbjct: 67 YGCISATLFGAIQPAYAYSLGSMVSVYFLTSHDEIKEKTRIYALSFVGLAVLSFLINISQ 126
Query: 720 NFFFGIAGGKLIERIRSLTFEKIVHQEISWFDDPSHSRYVIQNVCYFMTK**KNYETERN 779
++ F G L +RIR K++ E+ WFD R+
Sbjct: 127 HYNFAYMGEYLTKRIRERMLSKVLTFEVGWFD--------------------------RD 160
Query: 780 ESSCSGAVGARLSIDASTVKSLVGDTMALIVQNISTVIAGLVIAFTANWILAFIVLVLTP 839
E+S SGA+ + V+SLVGD MAL+VQ +S V + W LA +++ + P
Sbjct: 161 ENS-SGAICSMFFFLNDKVRSLVGDRMALVVQTVSAVTIAFTMGLVIAWRLALVMIAVQP 219
Query: 840 MILMQGIVQMKFLKGFSADAKVMYEEASQVANDAVSSIRTVASFCAESKVMDMYSKKCLG 899
+I++ + LK S A +E+S++A +AVS++RT+ +F ++ ++M M K
Sbjct: 220 VIIVCFYTRRVLLKSMSKKAIKAQDESSKLAAEAVSNVRTITAFSSQERIMKMLEKAQES 279
Query: 900 PAKQGVRLGLVSGIGFGCSFLVLYCTNAFIFYIGSVLVQHGKATFTEVFRVFFALTMTAI 959
P ++ +R +G G S + CT A F+ G L+Q G T +F F L T
Sbjct: 280 PRRESIRQSWFAGFGLAMSQSLTSCTWALDFWYGGRLIQDGYITAKALFETFMILVSTGR 339
Query: 960 AVSQTTTLAPDTNKAKDSAASIFEIIDSKPDIDSSSNAGVTRETVVGDIELQHVNFNYPT 1019
++ ++ D K D+ S+F ++D ID G E + G +E V+F+YPT
Sbjct: 340 VIADAGSMTTDLAKGSDAVGSVFAVLDRYTSIDPEDPDGYETERITGQVEFLDVDFSYPT 399
Query: 1020 RPDIQIFKDLSLSIPSAKTIALVGESGSGKSTVISLLERFYDPNSGRILLDGVDLKTFRL 1079
RPD+ IFK+ S+ I K+ A+VG SGSGKST+I L+ERFYDP G + +DG D++++ L
Sbjct: 400 RPDVIIFKNFSIKIEEGKSTAIVGPSGSGKSTIIGLIERFYDPLKGIVKIDGRDIRSYHL 459
Query: 1080 SWLRQQMGLVGQEPILFNESIRANIGYGKEGGATED-EIIAAANAANAHSFISNLPDGYD 1138
LR+ + LV QEP LF +IR NI YG ++ EII AA AANAH FI++L +GYD
Sbjct: 460 RSLRRHIALVSQEPTLFAGTIRENIIYGGVSDKIDEAEIIEAAKAANAHDFITSLTEGYD 519
Query: 1139 TSVGERGTQLSGGQKQRIAIARTMLKNPKILLLDEATSALDAESERIVQEALDRVSVNRT 1198
T G+RG QLSGGQKQRIAIAR +LKNP +LLLDEATSALD++SER+VQ+AL+RV V RT
Sbjct: 520 TYCGDRGVQLSGGQKQRIAIARAVLKNPSVLLLDEATSALDSQSERVVQDALERVMVGRT 579
Query: 1199 TVVVAHRLTTIRGADTIAVIKNGAVAEKGRHDELM-RITDGVYASLV 1244
+VV+AHRL+TI+ D IAV+ G + E+G H L+ + G+Y SLV
Sbjct: 580 SVVIAHRLSTIQNCDAIAVLDKGKLVERGTHSSLLSKGPTGIYFSLV 626
Score = 345 bits (885), Expect = 9e-95
Identities = 212/565 (37%), Positives = 311/565 (54%), Gaps = 56/565 (9%)
Query: 48 IIGTISAVANGLASPLMTLFLGNVINAFGSSNPADAIKQVSKV-SLLFVYLAIGSGIASF 106
+ G ISA G P LG++++ + ++ D IK+ +++ +L FV LA+ S + +
Sbjct: 66 LYGCISATLFGAIQPAYAYSLGSMVSVYFLTSH-DEIKEKTRIYALSFVGLAVLSFLINI 124
Query: 107 LQVTCWMVTGERQAARIRSLYLKTILQQDIAFFDTETNT-GEVIGRMSGDTILIQEAMGE 165
Q + GE RIR L +L ++ +FD + N+ G + ++ +G+
Sbjct: 125 SQHYNFAYMGEYLTKRIRERMLSKVLTFEVGWFDRDENSSGAICSMFFFLNDKVRSLVGD 184
Query: 166 KVGKFFQLASNFCGGFVMAFIKGWRLAIVLLACVPCVAVAGAFMSIVMAKMSSRGQIAYA 225
++ Q S F M + WRLA+V++A P + V +++ MS + A
Sbjct: 185 RMALVVQTVSAVTIAFTMGLVIAWRLALVMIAVQPVIIVCFYTRRVLLKSMSKKAIKAQD 244
Query: 226 EAGNVVDQTVGAIRTVASFTGEKKAIEKYNSKIKIAYTTMVKQGIVSGFGIGMLTFIAFC 285
E+ + + V +RT+ +F+ +++ ++ + ++Q +GFG+ M + C
Sbjct: 245 ESSKLAAEAVSNVRTITAFSSQERIMKMLEKAQESPRRESIRQSWFAGFGLAMSQSLTSC 304
Query: 286 TYGLAMWYGSKLVIEKGYNGGTVM--TVIIALMTGGIIK--------------------- 322
T+ L WYG +L I+ GY + T +I + TG +I
Sbjct: 305 TWALDFWYGGRL-IQDGYITAKALFETFMILVSTGRVIADAGSMTTDLAKGSDAVGSVFA 363
Query: 323 ----------------------GDIELRDVSFRYPARPDVQIFDGFSLFVPSGTTTALVG 360
G +E DV F YP RPDV IF FS+ + G +TA+VG
Sbjct: 364 VLDRYTSIDPEDPDGYETERITGQVEFLDVDFSYPTRPDVIIFKNFSIKIEEGKSTAIVG 423
Query: 361 QSGSGKSTVISLLERFYDPDAGEVLIDGVNLKNLQLRWIREQIGLVSQEPILFTTSIREN 420
SGSGKST+I L+ERFYDP G V IDG ++++ LR +R I LVSQEP LF +IREN
Sbjct: 424 PSGSGKSTIIGLIERFYDPLKGIVKIDGRDIRSYHLRSLRRHIALVSQEPTLFAGTIREN 483
Query: 421 IAYGKEGATDE----EITTAITLANAKKFIDKLPQGLDTMAGQNGTQLSGGQKQRIAIAR 476
I YG G +D+ EI A ANA FI L +G DT G G QLSGGQKQRIAIAR
Sbjct: 484 IIYG--GVSDKIDEAEIIEAAKAANAHDFITSLTEGYDTYCGDRGVQLSGGQKQRIAIAR 541
Query: 477 AILKNPKILLLDEATSALDAESERIVQEALEKIILKRTTVVVAHRLTTIRNADIIAVVQQ 536
A+LKNP +LLLDEATSALD++SER+VQ+ALE++++ RT+VV+AHRL+TI+N D IAV+ +
Sbjct: 542 AVLKNPSVLLLDEATSALDSQSERVVQDALERVMVGRTSVVIAHRLSTIQNCDAIAVLDK 601
Query: 537 GKIVERGTHSG-LTMDPDGAYSQLI 560
GK+VERGTHS L+ P G Y L+
Sbjct: 602 GKLVERGTHSSLLSKGPTGIYFSLV 626
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,004,593
Number of Sequences: 26719
Number of extensions: 1026362
Number of successful extensions: 4148
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 117
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 3202
Number of HSP's gapped (non-prelim): 508
length of query: 1250
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1139
effective length of database: 8,352,787
effective search space: 9513824393
effective search space used: 9513824393
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144478.12


