

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144477.14 + phase: 1 /pseudo
(190 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
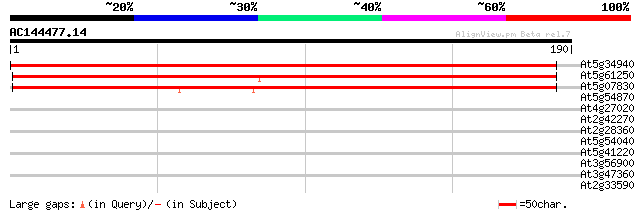
Score E
Sequences producing significant alignments: (bits) Value
At5g34940 putative protein 250 4e-67
At5g61250 unknown protein 210 3e-55
At5g07830 unknown protein 207 4e-54
At5g54870 unknown protein 28 2.2
At4g27020 unknown protein 28 3.8
At2g42270 ATP-dependent RNA helicase like protein 27 5.0
At2g28360 hypothetical protein 27 5.0
At5g54040 CHP-rich zinc finger protein-like 27 8.5
At5g41220 glutathione transferase-like 27 8.5
At3g56900 unknown protein 27 8.5
At3g47360 putative protein 27 8.5
At2g33590 putative cinnamoyl-CoA reductase 27 8.5
>At5g34940 putative protein
Length = 536
Score = 250 bits (638), Expect = 4e-67
Identities = 114/185 (61%), Positives = 143/185 (76%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKP 60
A+SFIR+T NYTI GWELGNELCG G+G + QYA D LRNIV VY+ V P
Sbjct: 180 AESFIRFTAENNYTIDGWELGNELCGSGVGARVGANQYAIDTINLRNIVNRVYKNVSPMP 239
Query: 61 LIIAPGGFFDANWFKKFLNRSEKLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGTF 120
L+I PGGFF+ +WF ++LN++E + T HIY+LGPGVD+H+ EKIL+P+YLD A +F
Sbjct: 240 LVIGPGGFFEVDWFTEYLNKAENSLNATTRHIYDLGPGVDEHLIEKILNPSYLDQEAKSF 299
Query: 121 SSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCRQ 180
SLKN+++ SST A AWVGE+GGA+NSG +LVS+AFV SFWY DQLGM++ Y TKTYCRQ
Sbjct: 300 RSLKNIIKNSSTKAVAWVGESGGAYNSGRNLVSNAFVYSFWYLDQLGMASLYDTKTYCRQ 359
Query: 181 TLIGG 185
+LIGG
Sbjct: 360 SLIGG 364
>At5g61250 unknown protein
Length = 539
Score = 210 bits (535), Expect = 3e-55
Identities = 95/185 (51%), Positives = 126/185 (67%), Gaps = 1/185 (0%)
Query: 2 KSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKPL 61
+ F+ YTVSK Y I WE GNEL G GI S+S Y D +L+N+++ VY+ KPL
Sbjct: 177 QDFMNYTVSKGYAIDSWEFGNELSGSGIWASVSVELYGKDLIVLKNVIKNVYKNSRTKPL 236
Query: 62 IIAPGGFFDANWFKKFLNRSEK-LADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGTF 120
++APGGFF+ W+ + L S + DV+THHIYNLGPG D + KILDP YL G++ F
Sbjct: 237 VVAPGGFFEEQWYSELLRLSGPGVLDVLTHHIYNLGPGNDPKLVNKILDPNYLSGISELF 296
Query: 121 SSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCRQ 180
+++ +Q A AWVGEAGGA+NSG VS+ F+NSFWY DQLG+S+ ++TK YCRQ
Sbjct: 297 ANVNQTIQEHGPWAAAWVGEAGGAFNSGGRQVSETFINSFWYLDQLGISSKHNTKVYCRQ 356
Query: 181 TLIGG 185
L+GG
Sbjct: 357 ALVGG 361
>At5g07830 unknown protein
Length = 543
Score = 207 bits (526), Expect = 4e-54
Identities = 94/186 (50%), Positives = 127/186 (67%), Gaps = 2/186 (1%)
Query: 2 KSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREV-VQKP 60
+ F+ YTVSK Y I WE GNEL G G+G S+S Y D +L++++ +VY+ + KP
Sbjct: 180 QDFLNYTVSKGYVIDSWEFGNELSGSGVGASVSAELYGKDLIVLKDVINKVYKNSWLHKP 239
Query: 61 LIIAPGGFFDANWFKKFLNRS-EKLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGT 119
+++APGGF++ W+ K L S + DVVTHHIYNLG G D + +KI+DP+YL V+ T
Sbjct: 240 ILVAPGGFYEQQWYTKLLEISGPSVVDVVTHHIYNLGSGNDPALVKKIMDPSYLSQVSKT 299
Query: 120 FSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCR 179
F + +Q A WVGE+GGA+NSG VSD F++SFWY DQLGMSA ++TK YCR
Sbjct: 300 FKDVNQTIQEHGPWASPWVGESGGAYNSGGRHVSDTFIDSFWYLDQLGMSARHNTKVYCR 359
Query: 180 QTLIGG 185
QTL+GG
Sbjct: 360 QTLVGG 365
>At5g54870 unknown protein
Length = 531
Score = 28.5 bits (62), Expect = 2.2
Identities = 15/50 (30%), Positives = 25/50 (50%), Gaps = 1/50 (2%)
Query: 116 VAGTFSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQ 165
V+G ++ V R S K WVGE+G + G +++ +W F+Q
Sbjct: 250 VSGAYAGHSAVALRDSE-GKLWVGESGNENDKGEDVIAILPWEEWWAFEQ 298
>At4g27020 unknown protein
Length = 523
Score = 27.7 bits (60), Expect = 3.8
Identities = 15/50 (30%), Positives = 24/50 (48%), Gaps = 1/50 (2%)
Query: 116 VAGTFSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQ 165
V+G ++ V R S K WVGE+G G +++ +W F+Q
Sbjct: 242 VSGAYAGHTAVCLRDSE-GKLWVGESGNENEKGEDVIAILPWEEWWEFEQ 290
>At2g42270 ATP-dependent RNA helicase like protein
Length = 2172
Score = 27.3 bits (59), Expect = 5.0
Identities = 22/87 (25%), Positives = 38/87 (43%), Gaps = 4/87 (4%)
Query: 13 YTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKPLIIAPGGFFD-- 70
Y + ELG +C +G + Y + L VQ + R V+Q L + P +D
Sbjct: 1179 YDLSSQELGELICNPKMGRPLHKYIHQFPKLKLAAHVQPISRSVLQVELTVTPDFHWDDK 1238
Query: 71 ANWFKK--FLNRSEKLADVVTHHIYNL 95
AN + + ++ + + + HH Y L
Sbjct: 1239 ANKYVEPFWIIVEDNDGEKILHHEYFL 1265
>At2g28360 hypothetical protein
Length = 826
Score = 27.3 bits (59), Expect = 5.0
Identities = 22/69 (31%), Positives = 31/69 (44%), Gaps = 11/69 (15%)
Query: 98 GVDDHITEKILDPTYLDGVAGTFSSLK-NVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAF 156
GV+ TEK +D +G+ G LK N++Q+ A NSG +DA
Sbjct: 762 GVEPEGTEKAMDQALKEGIVGEAGPLKRNIVQKVPENENQ-------AENSGVTEFNDA- 813
Query: 157 VNSFWYFDQ 165
+FW DQ
Sbjct: 814 --NFWRVDQ 820
>At5g54040 CHP-rich zinc finger protein-like
Length = 596
Score = 26.6 bits (57), Expect = 8.5
Identities = 9/24 (37%), Positives = 14/24 (57%)
Query: 18 WELGNELCGKGIGISISPYQYAND 41
W +LC K ++SPY++A D
Sbjct: 562 WYCSKDLCPKEFNFAVSPYRFALD 585
>At5g41220 glutathione transferase-like
Length = 590
Score = 26.6 bits (57), Expect = 8.5
Identities = 10/39 (25%), Positives = 20/39 (50%)
Query: 72 NWFKKFLNRSEKLADVVTHHIYNLGPGVDDHITEKILDP 110
+W+ L++ ++ V+ H NL PG ++ +L P
Sbjct: 84 HWYPTDLSKRARIHSVLDWHHTNLRPGAAGYVLNSVLGP 122
>At3g56900 unknown protein
Length = 447
Score = 26.6 bits (57), Expect = 8.5
Identities = 10/32 (31%), Positives = 19/32 (59%)
Query: 122 SLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVS 153
SLK +LQ + T + + G +W+ G H+++
Sbjct: 84 SLKQILQPTDVTLLSEIDLQGVSWHQGKHIIA 115
>At3g47360 putative protein
Length = 309
Score = 26.6 bits (57), Expect = 8.5
Identities = 22/78 (28%), Positives = 34/78 (43%), Gaps = 10/78 (12%)
Query: 28 GIGISISPYQYANDATILR---------NIVQEVYREVVQKPLIIAPGGFFDANWFKKFL 78
GIG ++ Y+YA L IV E R++ +II PG + KKF+
Sbjct: 58 GIGEHVA-YEYAKKGAYLALVARRRDRLEIVAETSRQLGSGNVIIIPGDVSNVEDCKKFI 116
Query: 79 NRSEKLADVVTHHIYNLG 96
+ + + + H I N G
Sbjct: 117 DETIRHFGKLDHLINNAG 134
>At2g33590 putative cinnamoyl-CoA reductase
Length = 321
Score = 26.6 bits (57), Expect = 8.5
Identities = 11/30 (36%), Positives = 19/30 (62%)
Query: 152 VSDAFVNSFWYFDQLGMSATYSTKTYCRQT 181
V+ AF+N W +Q+ A +S + YC++T
Sbjct: 129 VAAAFMNPMWSKNQVLDEACWSDQEYCKKT 158
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,412,076
Number of Sequences: 26719
Number of extensions: 179670
Number of successful extensions: 394
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 383
Number of HSP's gapped (non-prelim): 12
length of query: 190
length of database: 11,318,596
effective HSP length: 94
effective length of query: 96
effective length of database: 8,807,010
effective search space: 845472960
effective search space used: 845472960
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC144477.14


