

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144475.7 - phase: 0
(341 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
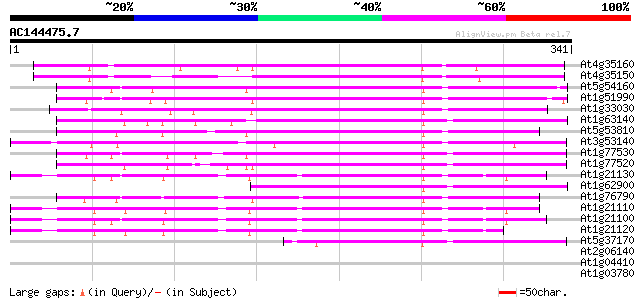
Score E
Sequences producing significant alignments: (bits) Value
At4g35160 O-methyltransferase - like protein 162 2e-40
At4g35150 O-methyltransferase - like protein 160 9e-40
At5g54160 O-methyltransferase 134 5e-32
At1g51990 107 1e-23
At1g33030 putative protein 106 2e-23
At1g63140 caffeic O-methyltransferase, putative 101 5e-22
At5g53810 caffeic acid 3-O-methyltransferase-like protein 101 7e-22
At3g53140 caffeic acid O-methyltransferase - like protein 100 9e-22
At1g77530 putative caffeic acid 3-O-methyltransferase 100 1e-21
At1g77520 caffeic acid 3-O-methyltransferase like protein 100 2e-21
At1g21130 O-methyltransferase like protein 96 2e-20
At1g62900 O-methyltransferase 1, putative 96 4e-20
At1g76790 catechol O-methyltransferase like protein 91 1e-18
At1g21110 O-methyltransferase like protein 91 1e-18
At1g21100 O-methyltransferase, putative 90 2e-18
At1g21120 O-methyltransferase, putative 89 3e-18
At5g37170 caffeic acid O-methyltransferase-like protein 69 5e-12
At2g06140 hypothetical protein 29 3.2
At1g04410 malate dehydrogenase like protein 29 3.2
At1g03780 hypothetical protein 29 3.2
>At4g35160 O-methyltransferase - like protein
Length = 382
Score = 162 bits (411), Expect = 2e-40
Identities = 114/354 (32%), Positives = 181/354 (50%), Gaps = 36/354 (10%)
Query: 15 AQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNH--GKPITLPELVSALRIPEAKAGCVHS 72
A ++ +F F + K A++L IP+ I+NH +P+TL EL SA+ A +
Sbjct: 29 ASLDIWKYVFGFADIAAAKCAIDLKIPEAIENHPSSQPVTLAELSSAV---SASPSHLRR 85
Query: 73 VMRLLAHNKIFAIVKIDDN-KEAYALSPTSK--LLVKGTDHCLTSMVKMVTNPTGVELYY 129
+MR L H IF + D Y +P S+ ++ + L V T P + +
Sbjct: 86 IMRFLVHQGIFKEIPTKDGLATGYVNTPLSRRLMITRRDGKSLAPFVLFETTPEMLAPWL 145
Query: 130 QLTKWTSK----EDLTIFDTT----LWDFMQQNSAYAELFNDAMESDSN-MMRFAMSDCK 180
+L+ S FD +W F Q N +++ N+AM D+ ++ C
Sbjct: 146 RLSSVVSSPVNGSTPPPFDAVHGKDVWSFAQDNPFLSDMINEAMACDARRVVPRVAGACH 205
Query: 181 SVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFES 240
+F+G+T++VDVGGGTG T+ ++ + FP +K FDLP+V+E + + V G+MF+S
Sbjct: 206 GLFDGVTTMVDVGGGTGETMGMLVKEFPWIKGFNFDLPHVIEVAEVLDGVENVEGDMFDS 265
Query: 241 IPQADAILL---------KDCVKILKKCKEAASRQKKGGKVIIIDIVINEK-------QD 284
IP DAI + KDC+KILK CKEA GKV+I++ VI E +
Sbjct: 266 IPACDAIFIKWVLHDWGDKDCIKILKNCKEAV--PPNIGKVLIVESVIGENKKTMIVDER 323
Query: 285 EHEITEVKLCFDITMMANHNS-RERDEKTWKKIITEAGFMSYKIFPIFGFRSLI 337
+ ++ V+L D+ MMA+ ++ +ER K W ++ EAGF Y++ I +SLI
Sbjct: 324 DEKLEHVRLMLDMVMMAHTSTGKERTLKEWDFVLKEAGFARYEVRDIDDVQSLI 377
>At4g35150 O-methyltransferase - like protein
Length = 325
Score = 160 bits (405), Expect = 9e-40
Identities = 111/343 (32%), Positives = 175/343 (50%), Gaps = 57/343 (16%)
Query: 15 AQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNH--GKPITLPELVSALRIPEAKAGCVHS 72
A ++ +F F + K A++L IP+ I+NH +P+TL EL SA+ A +
Sbjct: 15 ASLDIWRYVFGFADIAAAKCAIDLKIPEAIENHPSSQPVTLSELSSAV---SASPSHLRR 71
Query: 73 VMRLLAHNKIFAIVKIDDNKEAYALSPTSKLLVKGTDHCLTSMVKMVTNPTGVELYYQLT 132
+MR L H +F V PT L G + S M+T G +L
Sbjct: 72 IMRFLVHQGLFKEV------------PTKDGLATGYTNTPLSRRMMITKLHGKDL----- 114
Query: 133 KWTSKEDLTIFDTTLWDFMQQNSAYAELFNDAMESDSN-MMRFAMSDCKSVFEGITSLVD 191
W F Q N +++L N+AM D+ ++ C+ +F+G+ ++VD
Sbjct: 115 ---------------WAFAQDNLCHSQLINEAMACDARRVVPRVAGACQGLFDGVATVVD 159
Query: 192 VGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQADAILL-- 249
VGGGTG T+ I+ + FP +K FDLP+V+E + + V G+MF+SIP +DA+++
Sbjct: 160 VGGGTGETMGILVKEFPWIKGFNFDLPHVIEVAQVLDGVENVEGDMFDSIPASDAVIIKW 219
Query: 250 -------KDCVKILKKCKEAASRQKKGGKVIIIDIVINEKQD-------EHEITEVKLCF 295
KDC+KILK CKEA GKV+I++ VI EK++ + ++ V+L
Sbjct: 220 VLHDWGDKDCIKILKNCKEAV--LPNIGKVLIVECVIGEKKNTMIAEERDDKLEHVRLQL 277
Query: 296 DITMMANHNS-RERDEKTWKKIITEAGFMSYKIFPIFGFRSLI 337
D+ MM + ++ +ER K W ++TEAGF Y++ +SLI
Sbjct: 278 DMVMMVHTSTGKERTLKEWDFVLTEAGFARYEVRDFDDVQSLI 320
>At5g54160 O-methyltransferase
Length = 363
Score = 134 bits (338), Expect = 5e-32
Identities = 101/332 (30%), Positives = 168/332 (50%), Gaps = 26/332 (7%)
Query: 29 PVSIKWAVELGIPDIIQNHGKPITLPELVSAL--RIPEAKAGCVHSVMRLLAHNKIFAI- 85
P+++K A+EL + +I+ +G P++ E+ S L + PEA + ++RLL +
Sbjct: 33 PMALKSALELDLLEIMAKNGSPMSPTEIASKLPTKNPEAPV-MLDRILRLLTSYSVLTCS 91
Query: 86 ---VKIDDNKEAYALSPTSKLLVKGTDHC-LTSMVKMVTNPTGVELYYQLTKWTSKEDLT 141
+ D + Y L P K L K D + ++ M + +E +Y L +
Sbjct: 92 NRKLSGDGVERIYGLGPVCKYLTKNEDGVSIAALCLMNQDKVLMESWYHLKDAILDGGIP 151
Query: 142 I---FDTTLWDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKSVFEGITSLVDVGGGTGN 198
+ + +++ + + ++FN+ M + S + + + FEG+TSLVDVGGG G
Sbjct: 152 FNKAYGMSAFEYHGTDPRFNKVFNNGMSNHSTITMKKILETYKGFEGLTSLVDVGGGIGA 211
Query: 199 TVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQADAILLK-------- 250
T+K+I +P LK I FDLP+V+E + + VGG+MF S+P+ DAI +K
Sbjct: 212 TLKMIVSKYPNLKGINFDLPHVIEDAPSHPGIEHVGGDMFVSVPKGDAIFMKWICHDWSD 271
Query: 251 -DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEITEVKLCFDITMMA-NHNSRER 308
CVK LK C E+ + GKVI+ + ++ E D T+ + D M+A N +ER
Sbjct: 272 EHCVKFLKNCYESL---PEDGKVILAECILPETPDSSLSTKQVVHVDCIMLAHNPGGKER 328
Query: 309 DEKTWKKIITEAGFMSYKIF-PIFGFRSLIEL 339
EK ++ + +GF K+ FG +LIEL
Sbjct: 329 TEKEFEALAKASGFKGIKVVCDAFGV-NLIEL 359
>At1g51990
Length = 363
Score = 107 bits (266), Expect = 1e-23
Identities = 91/339 (26%), Positives = 175/339 (50%), Gaps = 35/339 (10%)
Query: 29 PVSIKWAVELGIPDIIQ------NHGKPITLPELVSALRIPEAKAGCVHSVMRLLAHNKI 82
P +K A EL + +I+ ++ P+ L + +A + P A + ++R L +
Sbjct: 29 PYIVKTARELDLFEIMAKARPLGSYLSPVDLASM-AAPKNPHAPM-MIDRLLRFLVAYSV 86
Query: 83 FA--IVKIDDNKE--AYALSPTSKLLVKGTD-HCLTSMVKMVTNPTGVELYYQLTKWTSK 137
+VK ++ +E AY L K L+K D + V ++ LT+ +
Sbjct: 87 CTCKLVKDEEGRESRAYGLGKVGKKLIKDEDGFSIAPYVLAGCTKAKGGVWSYLTEAIQE 146
Query: 138 EDLTIFDTT----LWDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKSVFEGITSLVDVG 193
+ ++ ++++M++N ++FN++M + ++++ + + FEG++ VDVG
Sbjct: 147 GGASAWERANEALIFEYMKKNENLKKIFNESMTNHTSIVMKKILENYIGFEGVSDFVDVG 206
Query: 194 GGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQADAILLK--- 250
G G+ + I +P +K I FDLP++V+ + + +GG+MF+ IP+ + IL+K
Sbjct: 207 GSLGSNLAQILSKYPHIKGINFDLPHIVKEAPQIHGVEHIGGDMFDEIPRGEVILMKWIL 266
Query: 251 ------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEI-TEVKLCFDITMMA-N 302
CV+ILK CK+A + G++I+I++++ + E ++ T+ L D+TMM+
Sbjct: 267 HDWNDEKCVEILKNCKKAL---PETGRIIVIEMIVPREVSETDLATKNSLSADLTMMSLT 323
Query: 303 HNSRERDEKTWKKIITEAGFMSYKIFPIFGFRS--LIEL 339
+ER +K ++ + EAGF KI I+G S +IEL
Sbjct: 324 SGGKERTKKEFEDLAKEAGFKLPKI--IYGAYSYWIIEL 360
>At1g33030 putative protein
Length = 352
Score = 106 bits (265), Expect = 2e-23
Identities = 88/325 (27%), Positives = 159/325 (48%), Gaps = 26/325 (8%)
Query: 25 SFMRPVSIKWAVELGIPDIIQNHGKPITLPELVSALRIPEAK---AGCVHSVMRLLAHNK 81
S + P+ +K A++LG+ DI+ G P + ++ S L K + V+ ++R LA
Sbjct: 16 SSVLPMVLKTAIDLGLFDILAESG-PSSASQIFSLLSNETKKHHDSSLVNRILRFLASYS 74
Query: 82 IFAIVKIDDNKEAYA---LSPTSKLLVKGTDH--CLTSMVKMVTNPTGVELYYQLTKWTS 136
I ++ E +A L+P +K K + L MV + + +++Y L
Sbjct: 75 ILTCSVSTEHGEPFAIYGLAPVAKYFTKNQNGGGSLAPMVNLFQDKVVTDMWYNLKDSVL 134
Query: 137 KEDLTIFDT---TLWDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKSVFEGITSLVDVG 193
+ L +T + + + +S + E+F +M+ + + + F+G+ SLVDVG
Sbjct: 135 EGGLPFNNTHGSSAVELVGSDSRFREVFQSSMKGFNEVFIEEFLKNYNGFDGVKSLVDVG 194
Query: 194 GGTGNTV-KIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQADAILLK-- 250
GG G+ + +IIS+ +K I FDLP V+ + + V G+MF + P+ +AI +K
Sbjct: 195 GGDGSLLSRIISKHTHIIKAINFDLPTVINTSLPSPGIEHVAGDMFTNTPKGEAIFMKWM 254
Query: 251 -------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEITEVKLCFDITMM-AN 302
CVKIL C ++ GKVI++D+VI E + + F++ MM N
Sbjct: 255 LHSWDDDHCVKILSNCYQSL---PSNGKVIVVDMVIPEFPGDTLLDRSLFQFELFMMNMN 311
Query: 303 HNSRERDEKTWKKIITEAGFMSYKI 327
+ +ER +K ++ + AGF + ++
Sbjct: 312 PSGKERTKKEFEILARLAGFSNVQV 336
>At1g63140 caffeic O-methyltransferase, putative
Length = 381
Score = 101 bits (252), Expect = 5e-22
Identities = 91/345 (26%), Positives = 162/345 (46%), Gaps = 42/345 (12%)
Query: 29 PVSIKWAVELGIPDIIQNHGKPITLPELVSALRIPEAKAG-----CVHSVMRLLAHNKI- 82
P+ +K A+ELG+ D+I + + L AL +P + ++ LLA + I
Sbjct: 43 PMVLKTALELGVIDMITSVDDGVWLSPSEIALGLPTKPTNPEAPVLLDRMLVLLASHSIL 102
Query: 83 -FAIVKIDDN------KEAYALSPTSKLLVKGTDHC--LTSMVKMVTNPTGVELYYQLTK 133
+ V+ DN + YA P + D L ++ ++ ++ + L
Sbjct: 103 KYRTVETGDNIGSRKTERVYAAEPVCTFFLNRGDGLGSLATLFMVLQGEVCMKPWEHLKD 162
Query: 134 --------WTSKEDLTIFDTTLWDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKSVFEG 185
+TS + F+ + N +AE+FN AM S ++ + + FE
Sbjct: 163 MILEGKDAFTSAHGMRFFE-----LIGSNEQFAEMFNRAMSEASTLIMKKVLEVYKGFED 217
Query: 186 ITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQAD 245
+ +LVDVGGG G + ++ +P +K I FDL +V+ N + V G+MF+ IP+ D
Sbjct: 218 VNTLVDVGGGIGTIIGQVTSKYPHIKGINFDLASVLAHAPFNKGVEHVSGDMFKEIPKGD 277
Query: 246 AILLK---------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEI-TEVKLCF 295
AI +K DCVKILK ++ + GKVII+++V E+ ++I + +
Sbjct: 278 AIFMKWILHDWTDEDCVKILKNYWKSLPEK---GKVIIVEVVTPEEPKINDISSNIVFGM 334
Query: 296 DITMMA-NHNSRERDEKTWKKIITEAGFMSYKIFPIFGFRSLIEL 339
D+ M+A + +ER ++ + +++GF+ +I S+IEL
Sbjct: 335 DMLMLAVSSGGKERSLSQFETLASDSGFLRCEIICHAFSYSVIEL 379
>At5g53810 caffeic acid 3-O-methyltransferase-like protein
Length = 378
Score = 101 bits (251), Expect = 7e-22
Identities = 91/328 (27%), Positives = 154/328 (46%), Gaps = 41/328 (12%)
Query: 29 PVSIKWAVELGIPDIIQN-HGKPITLPELVSALRIP------EAKAGCVHSVMRLLAHNK 81
P+ +K A+ELG+ D I G + L ALR+P EA A + L++H+
Sbjct: 39 PMVLKAALELGVIDTITTVGGGDLWLSPSEIALRLPTKPCNLEAPALLDRMLRFLVSHSV 98
Query: 82 IFAIVKIDDN------KEAYALSPTSKLLVKGTDHCLTSMVKMVTNPTGVELYYQLTK-W 134
+ I++N + YA P K L+ +D S + ++L K W
Sbjct: 99 LKCRTVIEENGQTGKVERVYAAEPVCKYLLNKSDDVSGSFASLFM----LDLSDVFIKTW 154
Query: 135 TSKEDLTI---------FDTTLWDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKSVFEG 185
T ED+ + L++++Q + + ++FN AM S M+ + F+
Sbjct: 155 THLEDVILEGRDAFSSAHGMKLFEYIQADERFGKVFNRAMLESSTMVTEKVLKFYEGFKD 214
Query: 186 ITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQAD 245
+ +LVDVGGG GNT+ +I+ +P L I FDL V+ ++ V G+MF IP+ D
Sbjct: 215 VKTLVDVGGGLGNTLGLITSKYPHLIGINFDLAPVLANAHSYPGVNHVAGDMFIKIPKGD 274
Query: 246 AILLK---------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEI-TEVKLCF 295
AI +K CV ILK C ++ ++ GK+II+++V + +I + +
Sbjct: 275 AIFMKWILHDWTDEQCVAILKNCWKSL---EENGKLIIVEMVTPVEAKSGDICSNIVFGM 331
Query: 296 DITMMAN-HNSRERDEKTWKKIITEAGF 322
D+TM+ +ERD ++ + +GF
Sbjct: 332 DMTMLTQCSGGKERDLYEFENLAYASGF 359
>At3g53140 caffeic acid O-methyltransferase - like protein
Length = 359
Score = 100 bits (250), Expect = 9e-22
Identities = 97/366 (26%), Positives = 167/366 (45%), Gaps = 38/366 (10%)
Query: 1 MDSENEYKASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHG--KPITLPELVS 58
M++E+ + +A L N I P+S+ AV LGI D I N G P++ E++
Sbjct: 1 MENESSESRNRARLAIMELANMISV---PMSLNAAVRLGIADAIWNGGANSPLSAAEILP 57
Query: 59 ALRIPE-----AKAGCVHSVMRLLAHNKIFAIVKIDDNKEAYALSPTSKLLVKGTDHCLT 113
L +P + ++R+L +F+ + + Y+L+ K LV +
Sbjct: 58 RLHLPSHTTIGGDPENLQRILRMLTSYGVFSEHLVGSIERKYSLTDVGKTLVTDSGGLSY 117
Query: 114 SMVKMVTNPTGVELYYQLTKWTSKEDLTIFDTTLWDFMQQNSAYAE---------LFNDA 164
+ + + + + L E +T + +AYA+ L A
Sbjct: 118 AAYVLQHHQEALMRAWPLVHTAVVEP----ETEPYVKANGEAAYAQYGKSEEMNGLMQKA 173
Query: 165 MESDSNMMRFAMSDCKSVFEGITSLVDVGGGTGNTVKIISEAFPTLK-CIVFDLPNVVEG 223
M S A+ D F+ + LVDVGG G+ +++I + FP ++ I FDLP VV
Sbjct: 174 MSGVSVPFMKAILDGYDGFKSVDILVDVGGSAGDCLRMILQQFPNVREGINFDLPEVVAK 233
Query: 224 LTGNNYLSFVGGNMFESIPQADAILLK---------DCVKILKKCKEAASRQKKGGKVII 274
++ VGG+MF+S+P ADAI +K +C +I+K C A GGK+I
Sbjct: 234 APNIPGVTHVGGDMFQSVPSADAIFMKWVLTTWTDEECKQIMKNCYNAL---PVGGKLIA 290
Query: 275 IDIVINEKQDEHEITEVKLCFDITMMANHNS--RERDEKTWKKIITEAGFMSYKIFPIFG 332
+ V+ ++ DE T L DI +M + + + R E+ + ++ AGF +++ F I
Sbjct: 291 CEPVLPKETDESHRTRALLEGDIFVMTIYRTKGKHRTEEEFIELGLSAGFPTFRPFYIDY 350
Query: 333 FRSLIE 338
F +++E
Sbjct: 351 FYTILE 356
>At1g77530 putative caffeic acid 3-O-methyltransferase
Length = 381
Score = 100 bits (248), Expect = 1e-21
Identities = 87/346 (25%), Positives = 165/346 (47%), Gaps = 46/346 (13%)
Query: 29 PVSIKWAVELGIPDIIQ--NHGKPITLPELVSAL----RIPEAKAGCVHSVMRLLAHNKI 82
P+ +K A+ELG+ D I ++G ++ E+ +L PEA + ++RLL + I
Sbjct: 43 PMVLKAALELGVIDTIAAASNGTWLSPSEIAVSLPNKPTNPEAPV-LLDRMLRLLVSHSI 101
Query: 83 FAIVKIDDNKEA--------YALSPTSKLLVKGTDHC--LTSMVKMVTNPTGVELYYQLT 132
++ + YA P K +K +D L+S++ ++ + L
Sbjct: 102 LKCCMVESRENGQTGKIERVYAAEPICKYFLKDSDGSGSLSSLLLLLHSQV------ILK 155
Query: 133 KWTSKEDLTI---------FDTTLWDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKSVF 183
WT+ +D+ + D L++++ + +++LF+ AM S M+ + + F
Sbjct: 156 TWTNLKDVILEGKDAFSSAHDMRLFEYISSDDQFSKLFHRAMSESSTMVMKKVLEEYRGF 215
Query: 184 EGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQ 243
E + +LVDVGGG G + +I+ +P +K + FDL V+ + V G+MF +P+
Sbjct: 216 EDVNTLVDVGGGIGTILGLITSKYPHIKGVNFDLAQVLTQAPFYPGVKHVSGDMFIEVPK 275
Query: 244 ADAILLK---------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEIT-EVKL 293
DAI +K DC+KILK C ++ + GKVII++++ + ++ + L
Sbjct: 276 GDAIFMKWILHDWGDEDCIKILKNCWKSLPEK---GKVIIVEMITPMEPKPNDFSCNTVL 332
Query: 294 CFDITMMAN-HNSRERDEKTWKKIITEAGFMSYKIFPIFGFRSLIE 338
D+ M+ +ER ++ + +GF+ +I + S+IE
Sbjct: 333 GMDLLMLTQCSGGKERSLSQFENLAFASGFLLCEIICLSYSYSVIE 378
>At1g77520 caffeic acid 3-O-methyltransferase like protein
Length = 381
Score = 99.8 bits (247), Expect = 2e-21
Identities = 86/345 (24%), Positives = 157/345 (44%), Gaps = 44/345 (12%)
Query: 29 PVSIKWAVELGIPDIIQNHGKPITLPELVSALRIPEAKAG-----CVHSVMRLLAHNKIF 83
P+ +K A ELG+ D I G L A +P + ++ LL + I
Sbjct: 43 PMVLKAAFELGVIDTIAAAGNDTWLSPCEIACSLPTKPTNPEAPVLLDRMLSLLVSHSIL 102
Query: 84 AIVKIDDNKEA--------YALSPTSKLLVKGTDHCLTSMVKMVTNPTGVELYYQL--TK 133
I+ + YA P K ++ +D S+V P + L+ Q+
Sbjct: 103 KCRMIETGENGRTGKIERVYAAEPVCKYFLRDSDGT-GSLV-----PLFMLLHTQVFFKT 156
Query: 134 WTSKEDLTI-----FDTT----LWDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKSVFE 184
WT+ +D+ + F++ +++++ + +AELFN AM S M+ + D FE
Sbjct: 157 WTNLKDVILEGRDAFNSAHGMKIFEYINSDQPFAELFNRAMSEPSTMIMKKVLDVYRGFE 216
Query: 185 GITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQA 244
+ +LVDVGGG G + +++ +P +K + FDL V+ + V G+MF +P+
Sbjct: 217 DVNTLVDVGGGNGTVLGLVTSKYPHIKGVNFDLAQVLTQAPFYPGVEHVSGDMFVEVPKG 276
Query: 245 DAILLK---------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHEI-TEVKLC 294
DA+ +K DC+KILK C ++ + GK+II++ V ++ ++ +
Sbjct: 277 DAVFMKWILHDWGDEDCIKILKNCWKSLPEK---GKIIIVEFVTPKEPKGGDLSSNTVFA 333
Query: 295 FDITMMAN-HNSRERDEKTWKKIITEAGFMSYKIFPIFGFRSLIE 338
D+ M+ +ER ++ + +GF+ +I + S+IE
Sbjct: 334 MDLLMLTQCSGGKERSLSQFENLAFASGFLRCEIICLAYSYSVIE 378
>At1g21130 O-methyltransferase like protein
Length = 373
Score = 96.3 bits (238), Expect = 2e-20
Identities = 89/359 (24%), Positives = 162/359 (44%), Gaps = 52/359 (14%)
Query: 1 MDSENEYKASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKP----ITLPEL 56
+D +NE + +A A + P+ +K A+ELG+ D + ++ E+
Sbjct: 19 IDDDNELGLMAVRLANAAAF--------PMVLKAALELGVFDTLYAEASRSDSFLSPSEI 70
Query: 57 VSAL----RIPEAKAGCVHSVMRLLAHNKIFAIVKIDDNK--EAYALSPTSKLLVKGTDH 110
S L R PEA + ++RLLA + K+ + K Y P + +K
Sbjct: 71 ASKLPTTPRNPEAPV-LLDRMLRLLASYSVVKCGKVSEGKGERVYRAEPICRFFLKDNIQ 129
Query: 111 CLTSMV-KMVTNPTGVELYYQLTKWTSKEDLTIFD----------TTLWDFMQQNSAYAE 159
+ S+ +++ N V L W +D+ + L+D+M + +++
Sbjct: 130 DIGSLASQVIVNFDSVFL----NTWAQLKDVVLEGGDAFGRAHGGMKLFDYMGTDERFSK 185
Query: 160 LFNDAMESDSNMMRFAMSDCKSVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPN 219
LFN + + + + + F+G+ LVDVGGG GNT+ +++ +P +K I FDL
Sbjct: 186 LFNQTGFTIAVVKKAL--EVYQGFKGVNVLVDVGGGVGNTLGVVASKYPNIKGINFDLTC 243
Query: 220 VVEGLTGNNYLSFVGGNMFESIPQADAILLK---------DCVKILKKCKEAASRQKKGG 270
+ + V G+MF +P DA++LK DCVKILK C ++ + G
Sbjct: 244 ALAQAPSYPGVEHVAGDMFVDVPTGDAMILKRILHDWTDEDCVKILKNCWKSL---PESG 300
Query: 271 KVIIIDIVINEKQDEHEITEVKLCFDITMM---ANHNSRERDEKTWKKIITEAGFMSYK 326
KV++I++V ++ + +I + FD+ M+ +ER ++ + +GF K
Sbjct: 301 KVVVIELVTPDEAENGDI-NANIAFDMDMLMFTQCSGGKERSRAEFEALAAASGFTHCK 358
>At1g62900 O-methyltransferase 1, putative
Length = 205
Score = 95.5 bits (236), Expect = 4e-20
Identities = 62/204 (30%), Positives = 110/204 (53%), Gaps = 14/204 (6%)
Query: 147 LWDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKSVFEGITSLVDVGGGTGNTVKIISEA 206
+++ + N +AE+FN M S ++ + + FE + +LVDVGGG G + ++
Sbjct: 3 VFELIGSNEQFAEMFNRTMSEASTLIMKKVLEVYKGFEDVNTLVDVGGGIGTIIGQVTSK 62
Query: 207 FPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQADAILLK---------DCVKILK 257
+P +K I FDL +V+ N + V G+MF+ IP+ DAI +K DCVKILK
Sbjct: 63 YPHIKGINFDLASVLAHAPFNKGVEHVSGDMFKEIPKGDAIFMKWILHDWTDEDCVKILK 122
Query: 258 KCKEAASRQKKGGKVIIIDIVINEKQDEHEI-TEVKLCFDITMMA-NHNSRERDEKTWKK 315
++ + GKVII+++V E+ ++I + + D+ M+A + +ER ++
Sbjct: 123 NYWKSLPEK---GKVIIVEVVTPEEPKINDISSNIVFGMDMLMLAVSSGGKERSLSQFET 179
Query: 316 IITEAGFMSYKIFPIFGFRSLIEL 339
+ +++GF+ +I S+IEL
Sbjct: 180 LASDSGFLRCEIICHAFSYSVIEL 203
>At1g76790 catechol O-methyltransferase like protein
Length = 367
Score = 90.5 bits (223), Expect = 1e-18
Identities = 86/328 (26%), Positives = 145/328 (43%), Gaps = 44/328 (13%)
Query: 29 PVSIKWAVELGIPDII------QNHGKPITLPELVSALRIP------EAKAGCVHSVMRL 76
P+ K A+ELG+ D + G L A+R+P EA A + ++RL
Sbjct: 29 PMVFKAAIELGVIDTLYLAARDDVTGSSSFLTPSEIAIRLPTKPSNPEAPA-LLDRILRL 87
Query: 77 LAHNKIFAIVKIDDNKEAYALSPTSKLLVK-GTDHCLTSMVKMVTNPTGVELYYQLTKWT 135
LA + ID N+ Y P + +K D L ++ + L W
Sbjct: 88 LASYSMVKCQIIDGNR-VYKAEPICRYFLKDNVDEELGTLASQLIVTLDTVF---LNTWG 143
Query: 136 SKEDLTIFDTT----------LWDFMQQNSAYAELFNDAMESDSNMMRFAMSDCKSVFEG 185
+++ + L+D++ ++ ++LFN S + + + S FEG
Sbjct: 144 ELKNVVLEGGVAFGRANGGLKLFDYISKDERLSKLFNRTGFSVAVLKKILQ--VYSGFEG 201
Query: 186 ITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQAD 245
+ LVDVGGG G+T+ ++ +P +K I FDL + + V G+MF +P+ D
Sbjct: 202 VNVLVDVGGGVGDTLGFVTSKYPNIKGINFDLTCALTQAPSYPNVEHVAGDMFVDVPKGD 261
Query: 246 AILLK---------DCVKILKKCKEAASRQKKGGKVIIIDIVINEKQDEHE-ITEVKLCF 295
AILLK DC KILK C +A + GKVI++++V ++ D + I+ +
Sbjct: 262 AILLKRILHDWTDEDCEKILKNCWKAL---PENGKVIVMEVVTPDEADNRDVISNIAFDM 318
Query: 296 DITMMAN-HNSRERDEKTWKKIITEAGF 322
D+ M+ +ER + + +GF
Sbjct: 319 DLLMLTQLSGGKERSRAEYVAMAANSGF 346
>At1g21110 O-methyltransferase like protein
Length = 373
Score = 90.5 bits (223), Expect = 1e-18
Identities = 84/354 (23%), Positives = 158/354 (43%), Gaps = 50/354 (14%)
Query: 1 MDSENEYKASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKP----ITLPEL 56
+D +NE + +A A + P+ +K ++ELG+ D + ++ E+
Sbjct: 19 VDDDNELGLMAVRLANAAAF--------PMVLKASLELGVFDTLYAEASRTDSFLSPSEI 70
Query: 57 VSALRIPEAKAGC---VHSVMRLLAHNKIFAIVKIDDNKE--AYALSPTSKLLVKGTDHC 111
S L G + ++RLLA + K+ KE Y P + +K
Sbjct: 71 ASKLPTTPRNPGAPVLLDRMLRLLASYSMVKCEKVSVGKEQRVYRAEPICRFFLKNNIQD 130
Query: 112 LTSMV-KMVTNPTGVELYYQLTKWTSKEDLTIFD----------TTLWDFMQQNSAYAEL 160
+ S+ +++ N V L W +D+ + L+D+M + +++L
Sbjct: 131 IGSLASQVIVNFDSVFL----NTWAQLKDVVLEGGDAFGRAHGGMKLFDYMGTDERFSKL 186
Query: 161 FNDAMESDSNMMRFAMSDCKSVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNV 220
FN + + + + + F+G+ LVDVGGG GNT+ +++ +P +K I FDL
Sbjct: 187 FNQTGFTIAVVKKAL--EVYQGFKGVNVLVDVGGGVGNTLGVVTSKYPNIKGINFDLTCA 244
Query: 221 VEGLTGNNYLSFVGGNMFESIPQADAILLK---------DCVKILKKCKEAASRQKKGGK 271
+ + V G+MF +P +A++LK DCVKILK C ++ + GK
Sbjct: 245 LAQAPTYPGVEHVAGDMFVDVPTGNAMILKRILHDWTDEDCVKILKNCWKSL---PQNGK 301
Query: 272 VIIIDIVINEKQDEHEITEVKLCFDITMM---ANHNSRERDEKTWKKIITEAGF 322
V++I++V ++ + +I + FD+ M+ +ER ++ + +GF
Sbjct: 302 VVVIELVTPDEAENGDI-NANIAFDMDMLMFTQCSGGKERSRAEFEALAAASGF 354
>At1g21100 O-methyltransferase, putative
Length = 373
Score = 89.7 bits (221), Expect = 2e-18
Identities = 89/359 (24%), Positives = 160/359 (43%), Gaps = 52/359 (14%)
Query: 1 MDSENEYKASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKP----ITLPEL 56
+D +NE + +A A + P+ +K A+ELG+ D + ++ E+
Sbjct: 19 VDDDNELGLMAVRLANAAAF--------PMVLKAALELGVFDTLYAAASRTDSFLSPYEI 70
Query: 57 VSAL----RIPEAKAGCVHSVMRLLAHNKIFAIVKIDDNK--EAYALSPTSKLLVKGTDH 110
S L R PEA + ++RLLA + K K Y P + +K
Sbjct: 71 ASKLPTTPRNPEAPV-LLDRMLRLLASYSMVKCGKALSGKGERVYRAEPICRFFLKDNIQ 129
Query: 111 CLTSMV-KMVTNPTGVELYYQLTKWTSKEDLTIFD----------TTLWDFMQQNSAYAE 159
+ S+ +++ N V L W +D+ + L+D+M + +++
Sbjct: 130 DIGSLASQVIVNFDSVFL----NTWAQLKDVVLEGGDAFGRAHGGMKLFDYMGTDERFSK 185
Query: 160 LFNDAMESDSNMMRFAMSDCKSVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPN 219
LFN + + + + + F+G+ LVDVGGG GNT+ +++ +P +K I FDL
Sbjct: 186 LFNQTGFTIAVVKKAL--EVYEGFKGVKVLVDVGGGVGNTLGVVTSKYPNIKGINFDLTC 243
Query: 220 VVEGLTGNNYLSFVGGNMFESIPQADAILLK---------DCVKILKKCKEAASRQKKGG 270
+ + V G+MF +P DA++LK DCVKILK C ++ + G
Sbjct: 244 ALAQAPSYPGVEHVAGDMFVDVPTGDAMILKRILHDWTDEDCVKILKNCWKSL---PENG 300
Query: 271 KVIIIDIVINEKQDEHEITEVKLCFDITMM---ANHNSRERDEKTWKKIITEAGFMSYK 326
KV++I++V ++ + +I + FD+ M+ +ER ++ + +GF K
Sbjct: 301 KVVVIELVTPDEAENGDI-NANIAFDMDMLMFTQCSGGKERSRAEFEALAAASGFTHCK 358
>At1g21120 O-methyltransferase, putative
Length = 373
Score = 89.4 bits (220), Expect = 3e-18
Identities = 80/329 (24%), Positives = 148/329 (44%), Gaps = 47/329 (14%)
Query: 1 MDSENEYKASELFVAQAHLYNQIFSFMRPVSIKWAVELGIPDIIQNHGKP----ITLPEL 56
+D +NE + +A A + P+ +K ++ELG+ D + ++ E+
Sbjct: 19 VDDDNELGLMAVRLANAAAF--------PMVLKASLELGVFDTLYAEASRTDSFLSPSEI 70
Query: 57 VSALRIPEAKAGC---VHSVMRLLAHNKIFAIVKIDDNK--EAYALSPTSKLLVKGTDHC 111
S L G + ++RLLA + K+ K Y P + +K
Sbjct: 71 ASKLPTTPRNPGAPVLLDRMLRLLASYSMVKCEKVSVGKGERVYRAEPICRFFLKNNIQD 130
Query: 112 LTSMV-KMVTNPTGVELYYQLTKWTSKEDLTIFD----------TTLWDFMQQNSAYAEL 160
+ S+ +++ N V L W +D+ + L+D+M + +++L
Sbjct: 131 IGSLASQVIVNFDSVFL----NTWAQLKDVVLEGGDAFGRAHGGMKLFDYMGTDERFSKL 186
Query: 161 FNDAMESDSNMMRFAMSDCKSVFEGITSLVDVGGGTGNTVKIISEAFPTLKCIVFDLPNV 220
FN + + + + + F+G+ LVDVGGG GNT+ +++ +P +K I FDL
Sbjct: 187 FNQTGFTIAVVKKAL--EVYQGFKGVNVLVDVGGGVGNTLGVVTSKYPNIKGINFDLTCA 244
Query: 221 VEGLTGNNYLSFVGGNMFESIPQADAILLK---------DCVKILKKCKEAASRQKKGGK 271
+ + V G+MF +P DA++LK DCVKILK C ++ + GK
Sbjct: 245 LAQAPSYPGVEHVAGDMFVDVPTGDAMILKRILHDWTDEDCVKILKNCWKSL---PENGK 301
Query: 272 VIIIDIVINEKQDEHEITEVKLCFDITMM 300
V++I++V ++ + +I + FD+ M+
Sbjct: 302 VVVIELVTPDEAENGDI-NANIAFDMDML 329
>At5g37170 caffeic acid O-methyltransferase-like protein
Length = 186
Score = 68.6 bits (166), Expect = 5e-12
Identities = 56/187 (29%), Positives = 96/187 (50%), Gaps = 20/187 (10%)
Query: 167 SDSNMMRFAMSDCKSVFEG---ITSLVDVGGGTGNTVK-IISEAFPTLKCIVFDLPNVVE 222
SDS+ M M+ V++G + +LVD+GGG G + +IS +P +K I FDL V+
Sbjct: 2 SDSSTM--IMTKILEVYKGLKDVNTLVDIGGGLGTILNLVISSKYPQIKGINFDLAAVLA 59
Query: 223 GLTGNNYLSFVGGNMFESIPQADAILL---------KDCVKILKKCKEAASRQKKGGKVI 273
+ V G+MF +P+ DAI + KDCVKIL C ++ + GKVI
Sbjct: 60 TAPSYPGVEHVPGDMFIDVPKGDAIFMRRILRDWNDKDCVKILTNCWKSLPEK---GKVI 116
Query: 274 IIDIVI-NEKQDEHEITEVKLCFDITMMANHN-SRERDEKTWKKIITEAGFMSYKIFPIF 331
I+D+V +E + + ++V D+ M+ + + R ++ + + +GF ++ +
Sbjct: 117 IVDMVAPSEPKSDDIFSKVVFGTDMLMLTQCSCGKVRSFAQFEALASASGFHKCEVSGLA 176
Query: 332 GFRSLIE 338
S+IE
Sbjct: 177 YTYSVIE 183
>At2g06140 hypothetical protein
Length = 633
Score = 29.3 bits (64), Expect = 3.2
Identities = 24/87 (27%), Positives = 41/87 (46%), Gaps = 13/87 (14%)
Query: 239 ESIPQADAILLKDCVK----ILKKCKEAASRQKKGG-------KVIIIDIVINEKQDEHE 287
+ P+A A+ ++ K +++K K+ S +KG +V I + + +K D+ E
Sbjct: 167 DGFPKAQALYEEETKKEIRFLVEKGKDLVSYSEKGNPDPIKSLRVSIPLVSLQDKVDQEE 226
Query: 288 ITEVK--LCFDITMMANHNSRERDEKT 312
E + FD MA HN RE +T
Sbjct: 227 GLEKEETASFDEDSMAEHNPREEGSET 253
>At1g04410 malate dehydrogenase like protein
Length = 332
Score = 29.3 bits (64), Expect = 3.2
Identities = 28/86 (32%), Positives = 42/86 (48%), Gaps = 15/86 (17%)
Query: 197 GNTVKIISEAFPTLKCIVFDLPNVVEGLTGNNYLSFVGGNMFESIPQADAILLKDC---- 252
G +++I AFP LK +V + VEG TG N VGG P+ + + KD
Sbjct: 53 GVKMELIDAAFPLLKGVVATT-DAVEGCTGVNVAVMVGG-----FPRKEGMERKDVMSKN 106
Query: 253 VKILKKCKEAASRQKKGG---KVIII 275
V I K +AA+ +K KV+++
Sbjct: 107 VSIYK--SQAAALEKHAAPNCKVLVV 130
>At1g03780 hypothetical protein
Length = 753
Score = 29.3 bits (64), Expect = 3.2
Identities = 21/80 (26%), Positives = 33/80 (41%), Gaps = 8/80 (10%)
Query: 64 EAKAGCVHSVMRLLAHNKIFAIVKIDDNKEAYALSPTSKLLVKGTDHCLTSMVKMVTNPT 123
E + S L+ N+ K+DD K L+P L+ T + L + T
Sbjct: 210 ETNLNNIASTTNLIQENQAIKRQKLDDGKSRQILNPKPATLLHKTRNGLVN--------T 261
Query: 124 GVELYYQLTKWTSKEDLTIF 143
G L +TK T KE+ ++
Sbjct: 262 GFNLCPSVTKHTPKENRKVY 281
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,520,632
Number of Sequences: 26719
Number of extensions: 313899
Number of successful extensions: 814
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 760
Number of HSP's gapped (non-prelim): 33
length of query: 341
length of database: 11,318,596
effective HSP length: 100
effective length of query: 241
effective length of database: 8,646,696
effective search space: 2083853736
effective search space used: 2083853736
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144475.7


