

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144475.11 - phase: 0 /pseudo
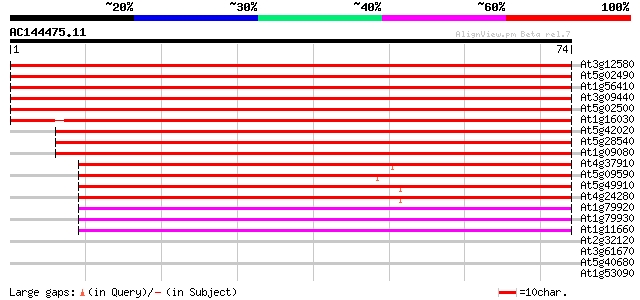
(74 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g12580 putative protein 154 6e-39
At5g02490 dnaK-type molecular chaperone hsc70.1 - like 152 2e-38
At1g56410 hypothetical protein 152 2e-38
At3g09440 heat-shock protein (At-hsc70-3) 152 3e-38
At5g02500 dnaK-type molecular chaperone hsc70.1 151 5e-38
At1g16030 unknown protein 128 6e-31
At5g42020 luminal binding protein (dbj|BAA13948.1) 109 3e-25
At5g28540 luminal binding protein 109 3e-25
At1g09080 putative luminal binding protein 107 8e-25
At4g37910 heat shock protein 70 like protein 69 6e-13
At5g09590 heat shock protein 70 (Hsc70-5) 68 7e-13
At5g49910 heat shock protein 70 (Hsc70-7) 66 3e-12
At4g24280 hsp 70-like protein 66 3e-12
At1g79920 putative heat-shock protein (At1g79920) 52 5e-08
At1g79930 putative heat-shock protein 52 5e-08
At1g11660 heat-shock protein like 47 2e-06
At2g32120 70kD heat shock protein 38 0.001
At3g61670 putative protein 28 0.83
At5g40680 putative protein 25 9.2
At1g53090 phytochrome A supressor spa1, putative 25 9.2
>At3g12580 putative protein
Length = 650
Score = 154 bits (390), Expect = 6e-39
Identities = 74/74 (100%), Positives = 74/74 (100%)
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
Query: 61 QVAMNPTNTVFDAK 74
QVAMNPTNTVFDAK
Sbjct: 61 QVAMNPTNTVFDAK 74
>At5g02490 dnaK-type molecular chaperone hsc70.1 - like
Length = 653
Score = 152 bits (385), Expect = 2e-38
Identities = 73/74 (98%), Positives = 73/74 (98%)
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
Query: 61 QVAMNPTNTVFDAK 74
QVAMNP NTVFDAK
Sbjct: 61 QVAMNPVNTVFDAK 74
>At1g56410 hypothetical protein
Length = 617
Score = 152 bits (385), Expect = 2e-38
Identities = 73/74 (98%), Positives = 73/74 (98%)
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
Query: 61 QVAMNPTNTVFDAK 74
QVAMNP NTVFDAK
Sbjct: 61 QVAMNPVNTVFDAK 74
>At3g09440 heat-shock protein (At-hsc70-3)
Length = 649
Score = 152 bits (384), Expect = 3e-38
Identities = 73/74 (98%), Positives = 73/74 (98%)
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
Query: 61 QVAMNPTNTVFDAK 74
QVAMNP NTVFDAK
Sbjct: 61 QVAMNPINTVFDAK 74
>At5g02500 dnaK-type molecular chaperone hsc70.1
Length = 651
Score = 151 bits (382), Expect = 5e-38
Identities = 72/74 (97%), Positives = 73/74 (98%)
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
M+GKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 1 MSGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
Query: 61 QVAMNPTNTVFDAK 74
QVAMNP NTVFDAK
Sbjct: 61 QVAMNPVNTVFDAK 74
>At1g16030 unknown protein
Length = 646
Score = 128 bits (321), Expect = 6e-31
Identities = 64/74 (86%), Positives = 67/74 (90%), Gaps = 1/74 (1%)
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MA K E AIGIDLGTTYSCVGVW +DRVEII NDQGNRTTPSYVAFTD+ERLIGDAAKN
Sbjct: 1 MATKSE-KAIGIDLGTTYSCVGVWMNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKN 59
Query: 61 QVAMNPTNTVFDAK 74
QVA+NP NTVFDAK
Sbjct: 60 QVALNPQNTVFDAK 73
>At5g42020 luminal binding protein (dbj|BAA13948.1)
Length = 668
Score = 109 bits (272), Expect = 3e-25
Identities = 52/68 (76%), Positives = 58/68 (84%)
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+V FTDSERLIG+AAKNQ A+NP
Sbjct: 35 GSVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAVNP 94
Query: 67 TNTVFDAK 74
TVFD K
Sbjct: 95 ERTVFDVK 102
>At5g28540 luminal binding protein
Length = 669
Score = 109 bits (272), Expect = 3e-25
Identities = 52/68 (76%), Positives = 58/68 (84%)
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+V FTDSERLIG+AAKNQ A+NP
Sbjct: 35 GSVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAVNP 94
Query: 67 TNTVFDAK 74
TVFD K
Sbjct: 95 ERTVFDVK 102
>At1g09080 putative luminal binding protein
Length = 678
Score = 107 bits (268), Expect = 8e-25
Identities = 51/68 (75%), Positives = 57/68 (83%)
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+ + VEIIANDQGNR TPS+VAFTD+ERLIG+AAKNQ A NP
Sbjct: 50 GTVIGIDLGTTYSCVGVYHNKHVEIIANDQGNRITPSWVAFTDTERLIGEAAKNQAAKNP 109
Query: 67 TNTVFDAK 74
T+FD K
Sbjct: 110 ERTIFDPK 117
>At4g37910 heat shock protein 70 like protein
Length = 682
Score = 68.6 bits (166), Expect = 6e-13
Identities = 35/66 (53%), Positives = 42/66 (63%), Gaps = 1/66 (1%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD-SERLIGDAAKNQVAMNPTN 68
IGIDLGTT SCV V + +I N +G+RTTPS VA E L+G AK Q NPTN
Sbjct: 55 IGIDLGTTNSCVSVMEGKTARVIENAEGSRTTPSVVAMNQKGELLVGTPAKRQAVTNPTN 114
Query: 69 TVFDAK 74
T+F +K
Sbjct: 115 TIFGSK 120
>At5g09590 heat shock protein 70 (Hsc70-5)
Length = 682
Score = 68.2 bits (165), Expect = 7e-13
Identities = 37/66 (56%), Positives = 42/66 (63%), Gaps = 1/66 (1%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAF-TDSERLIGDAAKNQVAMNPTN 68
IGIDLGTT SCV V + ++I N +G RTTPS VAF T E L+G AK Q NPTN
Sbjct: 60 IGIDLGTTNSCVAVMEGKNPKVIENAEGARTTPSVVAFNTKGELLVGTPAKRQAVTNPTN 119
Query: 69 TVFDAK 74
TV K
Sbjct: 120 TVSGTK 125
>At5g49910 heat shock protein 70 (Hsc70-7)
Length = 718
Score = 66.2 bits (160), Expect = 3e-12
Identities = 33/66 (50%), Positives = 42/66 (63%), Gaps = 1/66 (1%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIGDAAKNQVAMNPTN 68
+GIDLGTT S V + + I+ N +G RTTPS VA+T S +RL+G AK Q +NP N
Sbjct: 81 VGIDLGTTNSAVAAMEGGKPTIVTNAEGQRTTPSVVAYTKSKDRLVGQIAKRQAVVNPEN 140
Query: 69 TVFDAK 74
T F K
Sbjct: 141 TFFSVK 146
>At4g24280 hsp 70-like protein
Length = 718
Score = 66.2 bits (160), Expect = 3e-12
Identities = 33/66 (50%), Positives = 42/66 (63%), Gaps = 1/66 (1%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIGDAAKNQVAMNPTN 68
+GIDLGTT S V + + I+ N +G RTTPS VA+T S +RL+G AK Q +NP N
Sbjct: 81 VGIDLGTTNSAVAAMEGGKPTIVTNAEGQRTTPSVVAYTKSGDRLVGQIAKRQAVVNPEN 140
Query: 69 TVFDAK 74
T F K
Sbjct: 141 TFFSVK 146
>At1g79920 putative heat-shock protein (At1g79920)
Length = 736
Score = 52.0 bits (123), Expect = 5e-08
Identities = 23/65 (35%), Positives = 34/65 (51%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPTNT 69
+G D G V V + ++++ ND+ NR TP+ V F D +R IG A MNP N+
Sbjct: 4 VGFDFGNENCLVAVARQRGIDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPKNS 63
Query: 70 VFDAK 74
+ K
Sbjct: 64 ISQIK 68
>At1g79930 putative heat-shock protein
Length = 831
Score = 52.0 bits (123), Expect = 5e-08
Identities = 23/65 (35%), Positives = 34/65 (51%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPTNT 69
+G D G V V + ++++ ND+ NR TP+ V F D +R IG A MNP N+
Sbjct: 4 VGFDFGNENCLVAVARQRGIDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPKNS 63
Query: 70 VFDAK 74
+ K
Sbjct: 64 ISQIK 68
>At1g11660 heat-shock protein like
Length = 773
Score = 46.6 bits (109), Expect = 2e-06
Identities = 19/65 (29%), Positives = 36/65 (55%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPTNT 69
+G D+G + V + ++++ ND+ NR P+ V+F + +R +G AA M+P +T
Sbjct: 4 VGFDVGNENCVIAVAKQRGIDVLLNDESNRENPAMVSFGEKQRFMGAAAAASATMHPKST 63
Query: 70 VFDAK 74
+ K
Sbjct: 64 ISQLK 68
>At2g32120 70kD heat shock protein
Length = 563
Score = 37.7 bits (86), Expect = 0.001
Identities = 15/41 (36%), Positives = 24/41 (57%)
Query: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD 49
A+GID+GT+ + VW +V I+ N + + S+V F D
Sbjct: 30 ALGIDIGTSQCSIAVWNGSQVHILRNTRNQKLIKSFVTFKD 70
>At3g61670 putative protein
Length = 790
Score = 28.1 bits (61), Expect = 0.83
Identities = 15/46 (32%), Positives = 23/46 (49%)
Query: 27 DRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPTNTVFD 72
D + I ND+GN++ S +ERL+ A K + P N +D
Sbjct: 635 DLTKSIQNDEGNKSNVSINGHPLTERLLRKAEKQAGVIQPGNYWYD 680
>At5g40680 putative protein
Length = 415
Score = 24.6 bits (52), Expect = 9.2
Identities = 12/24 (50%), Positives = 13/24 (54%)
Query: 43 SYVAFTDSERLIGDAAKNQVAMNP 66
SY TDS +LI D K MNP
Sbjct: 279 SYDELTDSWKLIPDMLKGMTFMNP 302
>At1g53090 phytochrome A supressor spa1, putative
Length = 794
Score = 24.6 bits (52), Expect = 9.2
Identities = 10/28 (35%), Positives = 15/28 (52%)
Query: 37 GNRTTPSYVAFTDSERLIGDAAKNQVAM 64
G+ T SYV F DS L+ + N + +
Sbjct: 660 GHHKTVSYVRFVDSSTLVSSSTDNTLKL 687
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,729,119
Number of Sequences: 26719
Number of extensions: 57966
Number of successful extensions: 88
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 64
Number of HSP's gapped (non-prelim): 20
length of query: 74
length of database: 11,318,596
effective HSP length: 50
effective length of query: 24
effective length of database: 9,982,646
effective search space: 239583504
effective search space used: 239583504
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 52 (24.6 bits)
Medicago: description of AC144475.11


