

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144474.7 - phase: 0
(329 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
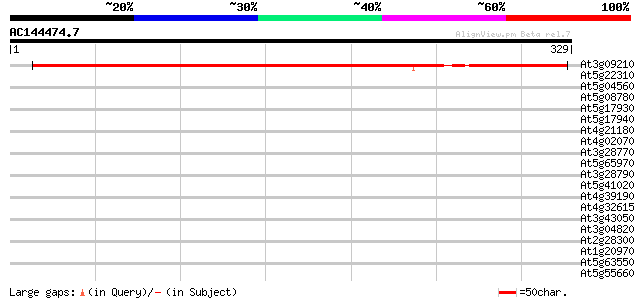
At3g09210 unknown protein 342 2e-94
At5g22310 unknown protein 35 0.043
At5g04560 putative protein 35 0.056
At5g08780 putative protein 33 0.16
At5g17930 unknown protein 33 0.28
At5g17940 putative protein 33 0.28
At4g21180 unknown protein 33 0.28
At4g02070 G/T DNA mismatch repair enzyme 33 0.28
At3g28770 hypothetical protein 33 0.28
At5g65970 Mlo protein-like 32 0.36
At3g28790 unknown protein 32 0.36
At5g41020 putative protein 32 0.47
At4g39190 hypothetical protein 32 0.47
At4g32615 putative protein 32 0.47
At3g43050 putative protein 32 0.47
At3g04820 unknown protein 32 0.47
At2g28300 unknown protein 32 0.47
At1g20970 hypothetical protein 32 0.47
At5g63550 unknown protein 32 0.62
At5g55660 putative protein 32 0.62
>At3g09210 unknown protein
Length = 333
Score = 342 bits (876), Expect = 2e-94
Identities = 177/319 (55%), Positives = 228/319 (70%), Gaps = 11/319 (3%)
Query: 14 PRFYTPSLSTKPPLSISATSSPELLTAKERRRLRNERRESN-ATNWKEEVENKLIQKTKK 72
P YTP T+ ++ LTAKERR+LRNERRES +W+EEVE KLI+K KK
Sbjct: 18 PSIYTPINKTQFSIAACVIERNHQLTAKERRQLRNERRESKLGYSWREEVEEKLIKKPKK 77
Query: 73 VNKSWKDELNLDNLMKLGPQWWGVRVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHE 132
+W +ELNLD L + GPQWW VRVSR++G TA+ LAR+LA+ FP++EF VYAP++
Sbjct: 78 RYATWTEELNLDTLAESGPQWWAVRVSRLRGHETAQILARALARQFPEMEFTVYAPSVQV 137
Query: 133 KKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPR 192
K++LK+GSISVK KP+FPGCIF+RC LNK +HD I++ +GVGGFIGSKVGNTK+QINKPR
Sbjct: 138 KRKLKNGSISVKPKPVFPGCIFIRCILNKEIHDSIRDVDGVGGFIGSKVGNTKRQINKPR 197
Query: 193 PVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKEL----ESDVSKAIVDSKP 248
PV + D+EAIFKQAK QE AD FEE ++ S ++EL SDV + + +SKP
Sbjct: 198 PVDDSDLEAIFKQAKEAQEKADSEFEEADRAEEEASILASQELLALSNSDVIETVAESKP 257
Query: 249 KRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHF 308
KR RK T+ E + KK KL GSTV+++SG+F F G LKKLN KT ATV F
Sbjct: 258 KRAPRKA----TLATETKA--KKKKLAAGSTVRVLSGTFAEFVGNLKKLNRKTAKATVGF 311
Query: 309 TMFGKENIVDLDVSEIVPE 327
T+FGKE +V++D++E+VPE
Sbjct: 312 TLFGKETLVEIDINELVPE 330
>At5g22310 unknown protein
Length = 481
Score = 35.4 bits (80), Expect = 0.043
Identities = 27/115 (23%), Positives = 56/115 (48%), Gaps = 9/115 (7%)
Query: 181 VGNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVS 240
+G+ KK++ K R + + + ++ +V+ + + FE E+K AAV KEL
Sbjct: 309 IGDDKKEMEKEREMMH--IADVLREERVQMKLTEAKFEFEDKYAAVE--RLKKEL----- 359
Query: 241 KAIVDSKPKRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLK 295
+ ++D + +GS + L + + + S + + + + SGS G+ +LK
Sbjct: 360 RRVLDGEEGKGSSEIRRILEVIDGSGSDDDEESDLKSIELNMESGSKWGYVDSLK 414
>At5g04560 putative protein
Length = 1017
Score = 35.0 bits (79), Expect = 0.056
Identities = 34/135 (25%), Positives = 53/135 (39%), Gaps = 20/135 (14%)
Query: 175 GFIGSKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAA--------- 225
GF+ V T +I P P ++ M++I + V NA +A E+ +
Sbjct: 213 GFLEQIVTTTGHEI--PEPKSDKSMQSIMDSSAV---NATEATEQNDGSRQDVLEFDLNK 267
Query: 226 VNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISG 285
P+K + K +V+ KPKR RK + + E +K K T V+
Sbjct: 268 TPQQKPSKRKRKFMPKVVVEGKPKRKPRKPAELPKVVVEGKPKRKPRKAATQEKVKSKE- 326
Query: 286 SFLGFAGTLKKLNSK 300
G+ KK N K
Sbjct: 327 -----TGSAKKKNLK 336
>At5g08780 putative protein
Length = 463
Score = 33.5 bits (75), Expect = 0.16
Identities = 24/80 (30%), Positives = 37/80 (46%), Gaps = 8/80 (10%)
Query: 202 IFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPK-------RGSRK 254
++K A+ Q + + ++ S +P ELE +SK + PK GS K
Sbjct: 362 LWKAAQKLQSQLSEIIDSCDETVVPYSVSPGCELEG-ISKGLCKEDPKMKTPRGNNGSEK 420
Query: 255 TSNQLTITEEASSAKKKPKL 274
S QL E S+KKKP++
Sbjct: 421 PSTQLEQKEREKSSKKKPQV 440
>At5g17930 unknown protein
Length = 707
Score = 32.7 bits (73), Expect = 0.28
Identities = 23/96 (23%), Positives = 45/96 (45%), Gaps = 11/96 (11%)
Query: 163 LHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEE- 221
L+D + V +GS++G+++K+ K R + D E + + A + E+ + F +EE
Sbjct: 136 LNDLFEGLPSVLDSMGSELGDSRKKRKKKRSEEKQDHEDVDELANEDLEHEESEFSDEES 195
Query: 222 ----------KKAAVNSGNPNKELESDVSKAIVDSK 247
K+ + ++ELESD+ D +
Sbjct: 196 EEEPVGKRDRKRHKKKKKSVDEELESDLMNITDDGE 231
>At5g17940 putative protein
Length = 427
Score = 32.7 bits (73), Expect = 0.28
Identities = 23/96 (23%), Positives = 45/96 (45%), Gaps = 11/96 (11%)
Query: 163 LHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEE- 221
L+D + V +GS++G+++K+ K R + D E + + A + E+ + F +EE
Sbjct: 156 LNDLFEGLPSVLDSMGSELGDSRKKRKKKRSEEKQDHEDVDELANEDLEHEESEFSDEES 215
Query: 222 ----------KKAAVNSGNPNKELESDVSKAIVDSK 247
K+ + ++ELESD+ D +
Sbjct: 216 EEEPVGKRDRKRHKKKKKSVDEELESDLMNITDDGE 251
>At4g21180 unknown protein
Length = 643
Score = 32.7 bits (73), Expect = 0.28
Identities = 22/69 (31%), Positives = 38/69 (54%), Gaps = 8/69 (11%)
Query: 199 MEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQ 258
+E + + VE ENA++ EEE+ + ++ ES+ S+ D KRGS+K N+
Sbjct: 580 VEVLKRTRDVEGENAEEGLEEEDDEI------EEEDYESEYSEDEEDK--KRGSKKKVNK 631
Query: 259 LTITEEASS 267
+ +EE+ S
Sbjct: 632 ESSSEESGS 640
>At4g02070 G/T DNA mismatch repair enzyme
Length = 1324
Score = 32.7 bits (73), Expect = 0.28
Identities = 24/99 (24%), Positives = 45/99 (45%), Gaps = 2/99 (2%)
Query: 196 EDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPK--RGSR 253
EDD+E + + E+E ++ EE K V+ + K S+V+K+ + K K G+
Sbjct: 239 EDDVELVDENEMDEEELVEEKDEETSKVNRVSKTDSRKRKTSEVTKSGGEKKSKTDTGTI 298
Query: 254 KTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAG 292
+ ++ E A + ++V G ++ G L G
Sbjct: 299 LKGFKASVVEPAKKIGQADRVVKGLEDNVLDGDALARFG 337
>At3g28770 hypothetical protein
Length = 2081
Score = 32.7 bits (73), Expect = 0.28
Identities = 27/107 (25%), Positives = 50/107 (46%), Gaps = 4/107 (3%)
Query: 168 KEYEGVGGFIGSKVGNTKKQINKPRPVAEDDMEAIFKQ--AKVEQENADKAFEEEEKKAA 225
K+ E + K + K+ +N ED+ + K +K+++EN D E++E + +
Sbjct: 947 KKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNK-EKKESEDS 1005
Query: 226 VNSGNPNKELESDVSKAIVDS-KPKRGSRKTSNQLTITEEASSAKKK 271
+ KE E SK ++ K K+ S+ + +EE S K+K
Sbjct: 1006 ASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEK 1052
Score = 28.5 bits (62), Expect = 5.2
Identities = 23/95 (24%), Positives = 46/95 (48%), Gaps = 2/95 (2%)
Query: 179 SKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESD 238
S+ +KK+ + R + E K+ K E EN K+ ++E+KK ++ + KE +
Sbjct: 1043 SEERKSKKEKEESRDLKAKKKEEETKEKK-ESEN-HKSKKKEDKKEHEDNKSMKKEEDKK 1100
Query: 239 VSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKPK 273
K +SK ++ + + ++ S+ KK+ K
Sbjct: 1101 EKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDK 1135
Score = 27.7 bits (60), Expect = 9.0
Identities = 24/98 (24%), Positives = 46/98 (46%), Gaps = 7/98 (7%)
Query: 180 KVGNTKKQIN----KPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKK-AAVNSGNPNKE 234
++ ++K Q N K + ++D + K+ K +E K EE+ KK +V KE
Sbjct: 1167 EIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKE 1226
Query: 235 LESDVSKAIVDSK--PKRGSRKTSNQLTITEEASSAKK 270
+ + +K D K K+ K + + ++EA + +K
Sbjct: 1227 TKKEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQK 1264
Score = 27.7 bits (60), Expect = 9.0
Identities = 19/70 (27%), Positives = 32/70 (45%), Gaps = 2/70 (2%)
Query: 204 KQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQLTITE 263
K+ K E +N++ +EE+KK VN N K+ E + + K N+
Sbjct: 945 KKKKKESKNSNMKKKEEDKKEYVN--NELKKQEDNKKETTKSENSKLKEENKDNKEKKES 1002
Query: 264 EASSAKKKPK 273
E S++K + K
Sbjct: 1003 EDSASKNREK 1012
>At5g65970 Mlo protein-like
Length = 569
Score = 32.3 bits (72), Expect = 0.36
Identities = 21/96 (21%), Positives = 44/96 (44%), Gaps = 4/96 (4%)
Query: 234 ELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKPKLVTG--STVQIISGSFLGFA 291
++ S++ KA+ D + + +K +T+ ++ A+K P G TV + SF
Sbjct: 435 QMGSNMKKAVFDEQMAKALKKW--HMTVKKKKGKARKPPTETLGVSDTVSTSTSSFHASG 492
Query: 292 GTLKKLNSKTKMATVHFTMFGKENIVDLDVSEIVPE 327
TL + + + + F +++ DL+ + PE
Sbjct: 493 ATLLRSKTTGHSTASYMSNFEDQSMSDLEAEPLSPE 528
>At3g28790 unknown protein
Length = 608
Score = 32.3 bits (72), Expect = 0.36
Identities = 62/285 (21%), Positives = 109/285 (37%), Gaps = 40/285 (14%)
Query: 12 PPPRFYTPSLSTK--PPLSISATSSPEL-LTAKERRRLRNERRESNATNWKEEVENKLIQ 68
P P TPS T P S A S+P T+++ + ++ESN+ + E + +
Sbjct: 291 PTPSTPTPSTPTPSTPTPSTPAPSTPAAGKTSEKGSESASMKKESNSKSESESAASGSVS 350
Query: 69 KTKKVNK-----SWKDELNLDNLMKLG-------PQWWGVRVSRVKGQYTAEALARSLAK 116
KTK+ NK ++KD + G P + KG +A A A + A
Sbjct: 351 KTKETNKGSSGDTYKDTTGTSSGSPSGSPSGSPTPSTSTDGKASSKGSASASAGASASAS 410
Query: 117 FFPDIEFKVYAPAIHEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEG---- 172
+ A + ++ KS S S + + E+N + + K+Y G
Sbjct: 411 AGASASAEESAASQKKESNSKSSSSSSSTTSVKEVETQTSSEVNSFISNLEKKYTGNSEL 470
Query: 173 ----------VGGFIGSKVGNTKKQINKPRPVAEDDMEAIF----KQAKVEQENADKAFE 218
+ N K+ + R A EA+ + +K E+ A
Sbjct: 471 KVFFEKLKTSMSASAKLSTSNAKELVTGMRSAASKIAEAMMFVSSRFSKSEETKTSMASC 530
Query: 219 EEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQLTITE 263
++E ++ KEL+ D++ IV K +++T + TIT+
Sbjct: 531 QQEVMQSL------KELQ-DINSQIVSGKTVTSTQQTELKQTITK 568
>At5g41020 putative protein
Length = 588
Score = 32.0 bits (71), Expect = 0.47
Identities = 26/96 (27%), Positives = 42/96 (43%), Gaps = 11/96 (11%)
Query: 185 KKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIV 244
K++ NK +P + D+E I N D + +KK + + E E + +
Sbjct: 185 KRKNNKKKPSVDSDVEDI---------NLDST-NDGKKKRKKKKQSEDSETEENGLNSTK 234
Query: 245 DSKPKRGSRKTSNQLTITE-EASSAKKKPKLVTGST 279
D+K +R +K Q ++E E S K L T ST
Sbjct: 235 DAKKRRKKKKKKKQSEVSEAEEKSDKSDEDLTTPST 270
Score = 28.5 bits (62), Expect = 5.2
Identities = 24/92 (26%), Positives = 39/92 (42%), Gaps = 3/92 (3%)
Query: 185 KKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIV 244
KK+ KP + + ++A+ K++K + E E+ S +KE + D
Sbjct: 75 KKKSKKPIRIDSEAVDAVKKKSKKRSKETKADSEAEDDGVEKKSKEKSKETKVDSEAHDG 134
Query: 245 DSKPKRGSRKTSNQLTITEEASSA---KKKPK 273
+ K+ S+K S I SS KKK K
Sbjct: 135 VKRKKKKSKKESGGDVIENTESSKVSDKKKGK 166
>At4g39190 hypothetical protein
Length = 277
Score = 32.0 bits (71), Expect = 0.47
Identities = 33/129 (25%), Positives = 51/129 (38%), Gaps = 41/129 (31%)
Query: 156 RCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQE---- 211
+C+ N D++ E GGF ++D+M + KQ++ E E
Sbjct: 97 KCKSNDLYEDFVLESSRRGGF------------------SQDEMRSGEKQSEAENEAKQS 138
Query: 212 -NADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSAKK 270
+KA E EEK++ ES V K++ + K KR I+E+ K
Sbjct: 139 ITENKAKENEEKQSIT---------ESRVKKSVTEKKTKR---------IISEKKVKQSK 180
Query: 271 KPKLVTGST 279
KL ST
Sbjct: 181 PEKLTKQST 189
>At4g32615 putative protein
Length = 315
Score = 32.0 bits (71), Expect = 0.47
Identities = 34/119 (28%), Positives = 51/119 (42%), Gaps = 10/119 (8%)
Query: 186 KQINKPRPVAEDDMEAIFKQAKV--EQENADKAFEEEEKKAAVNSGNPNKELESDVSKAI 243
K+ K + +AE +EA+ V + EN + +++ +K VN KE + SKA
Sbjct: 163 KKERKKKELAE--LEALLADFGVATKDENGQQDSQDKGEKKEVNDEGEKKENTTGESKA- 219
Query: 244 VDSKPKRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTK 302
SK K +K Q + E S K + S Q S S + LKK+ S K
Sbjct: 220 --SKKK---KKKDKQKELKESQSEVKSNSDAASESAEQEESSSSIDVKERLKKIASMKK 273
>At3g43050 putative protein
Length = 608
Score = 32.0 bits (71), Expect = 0.47
Identities = 17/49 (34%), Positives = 29/49 (58%), Gaps = 1/49 (2%)
Query: 36 ELLTAKERRRLRNERRESNATNWKEE-VENKLIQKTKKVNKSWKDELNL 83
E + +E+ + E RE A +E+ ++N+ I K+VNK W+D +NL
Sbjct: 168 EAVACEEQEEMACEEREEMACVEREDGMDNQGIGVCKRVNKEWEDGVNL 216
>At3g04820 unknown protein
Length = 699
Score = 32.0 bits (71), Expect = 0.47
Identities = 33/131 (25%), Positives = 58/131 (44%), Gaps = 20/131 (15%)
Query: 136 LKSGSISVKSKPL-FPGC--IFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKP- 191
L++G+ S L PG I+ ++ + HD K+Y I S G ++ KP
Sbjct: 508 LEAGTYSTSDIVLPLPGSRVIYPSNDIAEIYHDLAKKYCFREFSITSMTGGYRRVFQKPI 567
Query: 192 -------------RPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVN---SGNPNKEL 235
+P+AE D++ I VE+ + + E+E K+ N SG N +
Sbjct: 568 DFEWELLTYTDSNKPLAETDLDRIPMNKPVEKVGSTEEIEDESMKSDTNPHDSGETNLKD 627
Query: 236 ESDVSKAIVDS 246
++D ++A D+
Sbjct: 628 QTDSNEAEKDT 638
>At2g28300 unknown protein
Length = 2218
Score = 32.0 bits (71), Expect = 0.47
Identities = 31/112 (27%), Positives = 52/112 (45%), Gaps = 27/112 (24%)
Query: 188 INKPRPVAEDDMEAIFKQ---------AKVEQENADKA-------FEEEEKKAAVNSGNP 231
I+K + +D E++F Q A + E+ + A E EEK+ + S P
Sbjct: 831 ISKSADIEQDPEESVFVQGVGRPKVGTADTQMEDTNDAKLLVGCSVESEEKEKTLQSLIP 890
Query: 232 --NKELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQ 281
+ + E D +++ D +PK GS T Q+ T+EA KL+ G +V+
Sbjct: 891 GDDADTEQDPEESVSDQRPKVGSAYT--QMEDTDEA-------KLLMGCSVE 933
>At1g20970 hypothetical protein
Length = 1498
Score = 32.0 bits (71), Expect = 0.47
Identities = 34/131 (25%), Positives = 52/131 (38%), Gaps = 9/131 (6%)
Query: 159 LNKPLHDYIKEYEGVGGFIGSKVGN---TKKQINKPRPVAEDDMEAIFKQAKVEQENADK 215
L L ++ E V S+VG+ TK + +P E D E VE DK
Sbjct: 158 LESGLEGKVESKEEVEQLHDSEVGSKDLTKNNVEEPEVEIESDSET-----DVEGHQGDK 212
Query: 216 AFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKPKLV 275
E +EK + + +L +V K VDS R S S +++ TE + +
Sbjct: 213 -IEAQEKSDRDLDVSQDLKLNENVEKHPVDSDEVRESELVSAKVSPTEPSDGGMDLGQPT 271
Query: 276 TGSTVQIISGS 286
+ I+GS
Sbjct: 272 VTDPAETINGS 282
Score = 27.7 bits (60), Expect = 9.0
Identities = 25/112 (22%), Positives = 44/112 (38%), Gaps = 11/112 (9%)
Query: 171 EGVGGFIGSKVGNTKKQINKP-----------RPVAEDDMEAIFKQAKVEQENADKAFEE 219
E V F GSKV N K++ KP + V D +I + +E +K+
Sbjct: 1144 EKVVKFEGSKVENNGKEVAKPTEQKSQTTKSKKAVKPDQPPSIVTELVSGKEEIEKSATP 1203
Query: 220 EEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSAKKK 271
EE++ + + ++ + K K + ++ +EA KKK
Sbjct: 1204 EEEEPPKLTKEEEELIKKEEEKRKQKEAAKMKEQHRLEEIAKAKEAMERKKK 1255
>At5g63550 unknown protein
Length = 530
Score = 31.6 bits (70), Expect = 0.62
Identities = 41/157 (26%), Positives = 68/157 (43%), Gaps = 32/157 (20%)
Query: 187 QINKPRPVAEDDMEAIFKQAK-VEQENADK---AFEEEEKKAAVNSGNPN------KELE 236
++N P A+++++ + K+ K VE+E D EEEKK G K+ E
Sbjct: 12 EVNSP---AKEEIDVVPKEEKEVEKEKVDSPRIGEAEEEKKEDEEEGEAKEGELGEKDKE 68
Query: 237 SDV-SKAIVDSKPKRGSRKTSNQLTIT---EEASSAKKKPKLVTGST---------VQII 283
DV S+ + + GS+K+S + T+T E + +KK + + ST V I
Sbjct: 69 DDVESEEEEEEEEGSGSKKSSEKETVTPTSERPTRERKKVERFSLSTPMRAPPSKSVSIE 128
Query: 284 SG------SFLGFAGTLKKLNSKTKMATVHFTMFGKE 314
G A L K + + +H +FGK+
Sbjct: 129 KGRGTPLREIPNVAHKLSKRKADDNLMLLHTILFGKK 165
>At5g55660 putative protein
Length = 759
Score = 31.6 bits (70), Expect = 0.62
Identities = 20/92 (21%), Positives = 43/92 (46%), Gaps = 2/92 (2%)
Query: 180 KVGNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDV 239
K N + + VAE ++E ++K E E+ ++ E+E++ S + ++ + D+
Sbjct: 216 KEANKEDDVEADTKVAEPEVEDKKTESKDENEDKEEEKEDEKEDEKEESNDDKEDKKEDI 275
Query: 240 SKAIVDSKPKRGSRKTSNQLTITEEASSAKKK 271
K+ + + K + KT + EE + K
Sbjct: 276 KKS--NKRGKGKTEKTRGKTKSDEEKKDIEPK 305
Score = 31.2 bits (69), Expect = 0.81
Identities = 23/84 (27%), Positives = 36/84 (42%), Gaps = 7/84 (8%)
Query: 177 IGSKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEE--KKAAVNSGNPNKE 234
+G G+ + K V EDD E K+ +N + EEEE K V N +
Sbjct: 169 VGGDKGDDVDEAEKVENVDEDDKEEALKE-----KNEAELAEEEETNKGEEVKEANKEDD 223
Query: 235 LESDVSKAIVDSKPKRGSRKTSNQ 258
+E+D A + + K+ K N+
Sbjct: 224 VEADTKVAEPEVEDKKTESKDENE 247
Score = 28.9 bits (63), Expect = 4.0
Identities = 26/97 (26%), Positives = 43/97 (43%), Gaps = 18/97 (18%)
Query: 192 RPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKE-------------LESD 238
+ VA D E+ ++ K + E +K E EE++ +G P+K +ES+
Sbjct: 501 KSVAHSDDES--EEEKEDDEEEEKEQEVEEEEEENENGIPDKSEDEAPQLSESEENVESE 558
Query: 239 VSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKPKLV 275
K KRGSR +S++ E A ++ K V
Sbjct: 559 EESEEETKKKKRGSRTSSDK---KESAGKSRSKKTAV 592
Score = 28.1 bits (61), Expect = 6.9
Identities = 28/98 (28%), Positives = 38/98 (38%), Gaps = 16/98 (16%)
Query: 182 GNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSK 241
G K+ NK EDD+EA K A+ E E +KK N +KE E + K
Sbjct: 212 GEEVKEANK-----EDDVEADTKVAEPEVE---------DKKTESKDENEDKEEEKEDEK 257
Query: 242 AIVDSKPKRGSRKTSNQLTITEEASSAKKKPKLVTGST 279
D K + K + I + K K + G T
Sbjct: 258 E--DEKEESNDDKEDKKEDIKKSNKRGKGKTEKTRGKT 293
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.129 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,531,274
Number of Sequences: 26719
Number of extensions: 330712
Number of successful extensions: 1953
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 73
Number of HSP's that attempted gapping in prelim test: 1810
Number of HSP's gapped (non-prelim): 185
length of query: 329
length of database: 11,318,596
effective HSP length: 100
effective length of query: 229
effective length of database: 8,646,696
effective search space: 1980093384
effective search space used: 1980093384
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144474.7


