

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144474.3 + phase: 0
(449 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
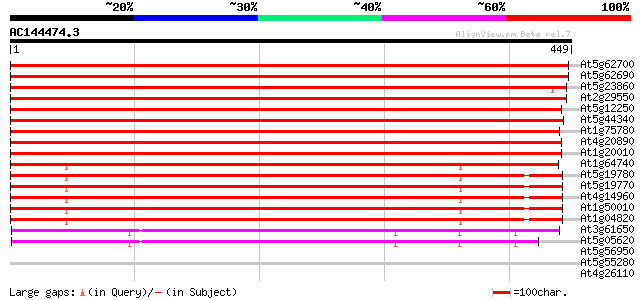
Score E
Sequences producing significant alignments: (bits) Value
At5g62700 tubulin beta-2/beta-3 chain (sp|P29512) 882 0.0
At5g62690 tubulin beta-2/beta-3 chain (sp|P29512) 882 0.0
At5g23860 beta tubulin 874 0.0
At2g29550 tubulin beta-7 chain 868 0.0
At5g12250 tubulin beta-6 chain (sp|P29514) 862 0.0
At5g44340 tubulin beta-4 chain (sp|P24636) 859 0.0
At1g75780 tubulin beta-1 chain 858 0.0
At4g20890 tubulin beta-9 chain 854 0.0
At1g20010 beta tubulin 1, putative 846 0.0
At1g64740 unknown protein 394 e-110
At5g19780 tubulin alpha-5 chain 394 e-110
At5g19770 tubulin alpha-5 chain-like protein 394 e-110
At4g14960 tubulin alpha-6 chain (TUA6) 388 e-108
At1g50010 tubulin alpha-2/alpha-4 chain, putative 385 e-107
At1g04820 tubulin alpha-2/alpha-4 chain 385 e-107
At3g61650 tubulin gamma-1 chain 290 8e-79
At5g05620 tubulin gamma-2 chain (sp|P38558) 290 1e-78
At5g56950 nucleosome assembly protein 38 0.013
At5g55280 cell division protein FtsZ chloroplast homolog precurs... 37 0.022
At4g26110 nucleosome assembly protein I-like protein 35 0.064
>At5g62700 tubulin beta-2/beta-3 chain (sp|P29512)
Length = 450
Score = 882 bits (2280), Expect = 0.0
Identities = 430/449 (95%), Positives = 441/449 (97%), Gaps = 2/449 (0%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQLERINVYYNEASGGRFV 60
MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRY GDSDLQLERINVYYNEAS GRFV
Sbjct: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYTGDSDLQLERINVYYNEASCGRFV 60
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRAVLMDLEPGTMDS+RSGPYGQ FRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV
Sbjct: 61 PRAVLMDLEPGTMDSLRSGPYGQTFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
Query: 121 RKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
RKEAEN DCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV
Sbjct: 121 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
Query: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL
Sbjct: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
Query: 241 RFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKNMM 300
RFPGQLNSDLRKLAVNL+PFPRLHFFM+GFAPLTSRGSQQYR+L+VPE+TQQMWDSKNMM
Sbjct: 241 RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRSLTVPELTQQMWDSKNMM 300
Query: 301 CAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
CAADPRHGRYLTASA+FRGKMSTKEVDEQM+NVQNKNSSYFVEWIPNNVKSTVCDIPPTG
Sbjct: 301 CAADPRHGRYLTASAMFRGKMSTKEVDEQMLNVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
Query: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS
Sbjct: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
Query: 421 EYQQYQDATA-EEDEYEDEEE-DYQQEHD 447
EYQQYQDATA EE +YEDEEE +YQQE +
Sbjct: 421 EYQQYQDATADEEGDYEDEEEGEYQQEEE 449
>At5g62690 tubulin beta-2/beta-3 chain (sp|P29512)
Length = 450
Score = 882 bits (2280), Expect = 0.0
Identities = 430/449 (95%), Positives = 441/449 (97%), Gaps = 2/449 (0%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQLERINVYYNEASGGRFV 60
MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRY GDSDLQLERINVYYNEAS GRFV
Sbjct: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYTGDSDLQLERINVYYNEASCGRFV 60
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRAVLMDLEPGTMDS+RSGPYGQ FRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV
Sbjct: 61 PRAVLMDLEPGTMDSLRSGPYGQTFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
Query: 121 RKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
RKEAEN DCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV
Sbjct: 121 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
Query: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL
Sbjct: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
Query: 241 RFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKNMM 300
RFPGQLNSDLRKLAVNL+PFPRLHFFM+GFAPLTSRGSQQYR+L+VPE+TQQMWDSKNMM
Sbjct: 241 RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRSLTVPELTQQMWDSKNMM 300
Query: 301 CAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
CAADPRHGRYLTASA+FRGKMSTKEVDEQM+NVQNKNSSYFVEWIPNNVKSTVCDIPPTG
Sbjct: 301 CAADPRHGRYLTASAMFRGKMSTKEVDEQMLNVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
Query: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS
Sbjct: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
Query: 421 EYQQYQDATA-EEDEYEDEEE-DYQQEHD 447
EYQQYQDATA EE +YEDEEE +YQQE +
Sbjct: 421 EYQQYQDATADEEGDYEDEEEGEYQQEEE 449
>At5g23860 beta tubulin
Length = 449
Score = 874 bits (2258), Expect = 0.0
Identities = 424/448 (94%), Positives = 439/448 (97%), Gaps = 3/448 (0%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQLERINVYYNEASGGRFV 60
MREILHIQGGQCGNQIGAKFWEVVCAEHGID TGRY G++DLQLER+NVYYNEAS GRFV
Sbjct: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGENDLQLERVNVYYNEASCGRFV 60
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV
Sbjct: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
Query: 121 RKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
RKEAEN DCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV
Sbjct: 121 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
Query: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL
Sbjct: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
Query: 241 RFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKNMM 300
RFPGQLNSDLRKLAVNL+PFPRLHFFM+GFAPLTSRGSQQYRAL+VPE+TQQMWD+KNMM
Sbjct: 241 RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDAKNMM 300
Query: 301 CAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
CAADPRHGRYLTASA+FRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG
Sbjct: 301 CAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
Query: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS
Sbjct: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
Query: 421 EYQQYQDATAEED---EYEDEEEDYQQE 445
EYQQYQDATA+E+ EYE++E + Q+E
Sbjct: 421 EYQQYQDATADEEEGYEYEEDEVEVQEE 448
>At2g29550 tubulin beta-7 chain
Length = 449
Score = 868 bits (2244), Expect = 0.0
Identities = 421/446 (94%), Positives = 437/446 (97%), Gaps = 1/446 (0%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQLERINVYYNEASGGRFV 60
MREILHIQGGQCGNQIG+KFWEVV EHGID TGRYVGDS+LQLER+NVYYNEAS GR+V
Sbjct: 1 MREILHIQGGQCGNQIGSKFWEVVNLEHGIDQTGRYVGDSELQLERVNVYYNEASCGRYV 60
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV
Sbjct: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
Query: 121 RKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
RKEAEN DCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMM+TFSVFPSPKVSDTVV
Sbjct: 121 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMMTFSVFPSPKVSDTVV 180
Query: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKL+TPSFGDLNHLISATMSGVTCCL
Sbjct: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLSTPSFGDLNHLISATMSGVTCCL 240
Query: 241 RFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKNMM 300
RFPGQLNSDLRKLAVNL+PFPRLHFFM+GFAPLTSRGSQQYR L+VPE+TQQMWD+KNMM
Sbjct: 241 RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRNLTVPELTQQMWDAKNMM 300
Query: 301 CAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
CAADPRHGRYLTASA+FRGKMSTKEVDEQM+NVQNKNSSYFVEWIPNNVKSTVCDIPPTG
Sbjct: 301 CAADPRHGRYLTASAMFRGKMSTKEVDEQMLNVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
Query: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS
Sbjct: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
Query: 421 EYQQYQDATA-EEDEYEDEEEDYQQE 445
EYQQYQDATA EE EYE+EE +Y+QE
Sbjct: 421 EYQQYQDATADEEGEYEEEEAEYEQE 446
>At5g12250 tubulin beta-6 chain (sp|P29514)
Length = 449
Score = 862 bits (2227), Expect = 0.0
Identities = 416/442 (94%), Positives = 434/442 (98%), Gaps = 1/442 (0%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQLERINVYYNEASGGRFV 60
MREILHIQGGQCGNQIG+KFWEVVC EHGIDPTGRYVG+SDLQLER+NVYYNEAS GR+V
Sbjct: 1 MREILHIQGGQCGNQIGSKFWEVVCDEHGIDPTGRYVGNSDLQLERVNVYYNEASCGRYV 60
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRA+LMDLEPGTMDSVR+GPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELID+VLDVV
Sbjct: 61 PRAILMDLEPGTMDSVRTGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDAVLDVV 120
Query: 121 RKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
RKEAEN DCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV
Sbjct: 121 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
Query: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL
Sbjct: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
Query: 241 RFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKNMM 300
RFPGQLNSDLRKLAVNL+PFPRLHFFM+GFAPLTSRGSQQYRAL+VPE+TQQMWDSKNMM
Sbjct: 241 RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDSKNMM 300
Query: 301 CAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
CAADPRHGRYLTASA+FRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKS+VCDI P G
Sbjct: 301 CAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSSVCDIAPRG 360
Query: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
L MASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS
Sbjct: 361 LSMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
Query: 421 EYQQYQDATA-EEDEYEDEEED 441
EYQQYQDATA +E EYE++E++
Sbjct: 421 EYQQYQDATADDEGEYEEDEDE 442
>At5g44340 tubulin beta-4 chain (sp|P24636)
Length = 444
Score = 859 bits (2219), Expect = 0.0
Identities = 410/443 (92%), Positives = 434/443 (97%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQLERINVYYNEASGGRFV 60
MREILHIQGGQCGNQIGAKFWEV+C EHGID TG+YVGDS LQLERI+VY+NEASGG++V
Sbjct: 1 MREILHIQGGQCGNQIGAKFWEVICDEHGIDHTGQYVGDSPLQLERIDVYFNEASGGKYV 60
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRAVLMDLEPGTMDS+RSGP+GQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV
Sbjct: 61 PRAVLMDLEPGTMDSLRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
Query: 121 RKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
RKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMM+TFSVFPSPKVSDTVV
Sbjct: 121 RKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMMTFSVFPSPKVSDTVV 180
Query: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKL P+FGDLNHLISATMSGVTCCL
Sbjct: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLANPTFGDLNHLISATMSGVTCCL 240
Query: 241 RFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKNMM 300
RFPGQLNSDLRKLAVNL+PFPRLHFFM+GFAPLTSRGSQQY ALSVPE+TQQMWD+KNMM
Sbjct: 241 RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYSALSVPELTQQMWDAKNMM 300
Query: 301 CAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
CAADPRHGRYLTASA+FRGK+STKEVDEQM+N+QNKNSSYFVEWIPNNVKS+VCDI P G
Sbjct: 301 CAADPRHGRYLTASAVFRGKLSTKEVDEQMMNIQNKNSSYFVEWIPNNVKSSVCDIAPKG 360
Query: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLV+
Sbjct: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVA 420
Query: 421 EYQQYQDATAEEDEYEDEEEDYQ 443
EYQQYQDATA E+EYE+EEE+Y+
Sbjct: 421 EYQQYQDATAGEEEYEEEEEEYE 443
>At1g75780 tubulin beta-1 chain
Length = 447
Score = 858 bits (2216), Expect = 0.0
Identities = 413/442 (93%), Positives = 435/442 (97%), Gaps = 2/442 (0%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDS-DLQLERINVYYNEASGGRF 59
MREILH+QGGQCGNQIG+KFWEV+C EHG+DPTGRY GDS DLQLERINVYYNEASGGR+
Sbjct: 1 MREILHVQGGQCGNQIGSKFWEVICDEHGVDPTGRYNGDSADLQLERINVYYNEASGGRY 60
Query: 60 VPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDV 119
VPRAVLMDLEPGTMDS+RSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELID+VLDV
Sbjct: 61 VPRAVLMDLEPGTMDSIRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDAVLDV 120
Query: 120 VRKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTV 179
VRKEAEN DCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTV
Sbjct: 121 VRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTV 180
Query: 180 VEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCC 239
VEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKL+TPSFGDLNHLISATMSGVTC
Sbjct: 181 VEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLSTPSFGDLNHLISATMSGVTCS 240
Query: 240 LRFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKNM 299
LRFPGQLNSDLRKLAVNL+PFPRLHFFM+GFAPLTSRGSQQY +L+VPE+TQQMWD+KNM
Sbjct: 241 LRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYISLTVPELTQQMWDAKNM 300
Query: 300 MCAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPT 359
MCAADPRHGRYLTASA+FRGKMSTKEVDEQ++NVQNKNSSYFVEWIPNNVKS+VCDIPPT
Sbjct: 301 MCAADPRHGRYLTASAMFRGKMSTKEVDEQILNVQNKNSSYFVEWIPNNVKSSVCDIPPT 360
Query: 360 GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLV 419
G+KMASTF+GNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLV
Sbjct: 361 GIKMASTFVGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLV 420
Query: 420 SEYQQYQDATA-EEDEYEDEEE 440
SEYQQYQDATA EEDEY++EEE
Sbjct: 421 SEYQQYQDATADEEDEYDEEEE 442
>At4g20890 tubulin beta-9 chain
Length = 444
Score = 854 bits (2206), Expect = 0.0
Identities = 408/441 (92%), Positives = 430/441 (96%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQLERINVYYNEASGGRFV 60
MREILHIQGGQCGNQIGAKFWEV+C EHGID TG+ GD+DLQLERINVY+NEASGG++V
Sbjct: 1 MREILHIQGGQCGNQIGAKFWEVICGEHGIDQTGQSCGDTDLQLERINVYFNEASGGKYV 60
Query: 61 PRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
PRAVLMDLEPGTMDS+RSGP+GQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV
Sbjct: 61 PRAVLMDLEPGTMDSLRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDVV 120
Query: 121 RKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTVV 180
RKEAEN DCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMM+TFSVFPSPKVSDTVV
Sbjct: 121 RKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMMTFSVFPSPKVSDTVV 180
Query: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCCL 240
EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKL P+FGDLNHLISATMSGVTCCL
Sbjct: 181 EPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLANPTFGDLNHLISATMSGVTCCL 240
Query: 241 RFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKNMM 300
RFPGQLNSDLRKLAVNL+PFPRLHFFM+GFAPLTSRGSQQY ALSVPE+TQQMWD+KNMM
Sbjct: 241 RFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYSALSVPELTQQMWDAKNMM 300
Query: 301 CAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPTG 360
CAADPRHGRYLTASA+FRGKMSTKEVDEQM+NVQNKNSSYFVEWIPNNVKS+VCDI PTG
Sbjct: 301 CAADPRHGRYLTASAVFRGKMSTKEVDEQMMNVQNKNSSYFVEWIPNNVKSSVCDIAPTG 360
Query: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS 420
LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLV+
Sbjct: 361 LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVA 420
Query: 421 EYQQYQDATAEEDEYEDEEED 441
EYQQYQDAT E+EYE++EE+
Sbjct: 421 EYQQYQDATVGEEEYEEDEEE 441
>At1g20010 beta tubulin 1, putative
Length = 449
Score = 846 bits (2185), Expect = 0.0
Identities = 409/443 (92%), Positives = 432/443 (97%), Gaps = 2/443 (0%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDS-DLQLERINVYYNEASGGRF 59
MREILHIQGGQCGNQIG+KFWEV+C EHGID TGRY GD+ DLQLERINVYYNEASGGR+
Sbjct: 1 MREILHIQGGQCGNQIGSKFWEVICDEHGIDSTGRYSGDTADLQLERINVYYNEASGGRY 60
Query: 60 VPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLDV 119
VPRAVLMDLEPGTMDS+RSGP+GQIFRPDNFVFGQSGAGNNWAKGHYTEGAELID+VLDV
Sbjct: 61 VPRAVLMDLEPGTMDSIRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDAVLDV 120
Query: 120 VRKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTV 179
VRKEAEN DCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTV
Sbjct: 121 VRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDTV 180
Query: 180 VEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTCC 239
VEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKL+TPSFGDLNHLISATMSGVTC
Sbjct: 181 VEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLSTPSFGDLNHLISATMSGVTCS 240
Query: 240 LRFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKNM 299
LRFPGQLNSDLRKLAVNL+PFPRLHFFM+GFAPLTSRGSQQY +L+VPE+TQQMWDSKNM
Sbjct: 241 LRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYISLTVPELTQQMWDSKNM 300
Query: 300 MCAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPPT 359
MCAADPRHGRYLTASAIFRG+MSTKEVDEQ++N+QNKNSSYFVEWIPNNVKS+VCDIPP
Sbjct: 301 MCAADPRHGRYLTASAIFRGQMSTKEVDEQILNIQNKNSSYFVEWIPNNVKSSVCDIPPK 360
Query: 360 GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLV 419
GLKMA+TF+GNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLV
Sbjct: 361 GLKMAATFVGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLV 420
Query: 420 SEYQQYQDATA-EEDEYEDEEED 441
+EYQQYQDATA EE EY+ EEE+
Sbjct: 421 AEYQQYQDATADEEGEYDVEEEE 443
>At1g64740 unknown protein
Length = 450
Score = 394 bits (1013), Expect = e-110
Identities = 187/449 (41%), Positives = 287/449 (63%), Gaps = 10/449 (2%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQL--ERINVYYNEASGGR 58
MREI+ I GQ G Q+G WE+ C EHGI P G DS + + N +++E S G+
Sbjct: 1 MREIISIHIGQAGIQVGNSCWELYCLEHGIQPDGTMPSDSTVGACHDAFNTFFSETSSGQ 60
Query: 59 FVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
VPRAV +DLEP +D VR+G Y Q+F P+ + G+ A NN+A+GHYT G E++D+ L+
Sbjct: 61 HVPRAVFLDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTVGREIVDTCLE 120
Query: 119 VVRKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
+RK A+N LQGF V +++GGGTGSG+G+LL+ ++ ++ + L F+++PSP+VS
Sbjct: 121 RLRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDFGKKSKLGFTIYPSPQVSTA 180
Query: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN LIS T+S +T
Sbjct: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYSNLNRLISQTISSLTT 240
Query: 239 CLRFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKN 298
LRF G +N D+ + NLVP+PR+HF + +AP+ S + SVPEIT +++ N
Sbjct: 241 SLRFDGAINVDITEFQTNLVPYPRIHFMLSSYAPVISSAKAYHEQFSVPEITTSVFEPSN 300
Query: 299 MMCAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPP 358
MM DPRHG+Y+ ++RG + K+V+ + ++ K + FV+W P K + PP
Sbjct: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNTAVAAIKAKRTIQFVDWCPTGFKCGINYQPP 360
Query: 359 T--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
+ ++ A I N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 361 SVVPGGDLAKVQRAVCMISNNTAVAEVFSRIDHKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
Query: 411 AESNMNDLVSEYQQYQDATAEEDEYEDEE 439
A ++ L +Y++ AE+D+ E +E
Sbjct: 421 AREDLAALEKDYEEVGGEGAEDDDEEGDE 449
>At5g19780 tubulin alpha-5 chain
Length = 450
Score = 394 bits (1012), Expect = e-110
Identities = 193/453 (42%), Positives = 288/453 (62%), Gaps = 14/453 (3%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQL--ERINVYYNEASGGR 58
MREI+ I GQ G Q+G WE+ C EHGI P G D+ + + + N +++E G+
Sbjct: 1 MREIISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTTVGVAHDAFNTFFSETGAGK 60
Query: 59 FVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
VPRAV +DLEP +D VR+G Y Q+F P+ + G+ A NN+A+GHYT G E++D LD
Sbjct: 61 HVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTVGKEIVDLCLD 120
Query: 119 VVRKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
VRK A+N LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+++PSP+VS
Sbjct: 121 RVRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
Query: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN LIS +S +T
Sbjct: 181 VVEPYNSVLSTHSLLEHTDVAVLLDNEAIYDICRRSLDIERPTYTNLNRLISQIISSLTT 240
Query: 239 CLRFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKN 298
LRF G +N D+ + NLVP+PR+HF + +AP+ S + LSVPEIT +++ +
Sbjct: 241 SLRFDGAINVDITEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQLSVPEITNAVFEPAS 300
Query: 299 MMCAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPP 358
MM DPRHG+Y+ ++RG + K+V+ + ++ K + FV+W P K + PP
Sbjct: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVGTIKTKRTVQFVDWCPTGFKCGINYQPP 360
Query: 359 T--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
T ++ A I N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 361 TVVPGGDLAKVQRAVCMISNNTAVAEVFSRIDHKFDLMYAKRAFVHWYVGEGMEEGEFSE 420
Query: 411 AESNMNDLVSEYQQYQDATAE-EDEYEDEEEDY 442
A DL + + Y++ AE D+ EDE EDY
Sbjct: 421 AR---EDLAALEKDYEEVGAEGGDDEEDEGEDY 450
>At5g19770 tubulin alpha-5 chain-like protein
Length = 450
Score = 394 bits (1012), Expect = e-110
Identities = 193/453 (42%), Positives = 288/453 (62%), Gaps = 14/453 (3%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQL--ERINVYYNEASGGR 58
MREI+ I GQ G Q+G WE+ C EHGI P G D+ + + + N +++E G+
Sbjct: 1 MREIISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTTVGVAHDAFNTFFSETGAGK 60
Query: 59 FVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
VPRAV +DLEP +D VR+G Y Q+F P+ + G+ A NN+A+GHYT G E++D LD
Sbjct: 61 HVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTVGKEIVDLCLD 120
Query: 119 VVRKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
VRK A+N LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+++PSP+VS
Sbjct: 121 RVRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
Query: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN LIS +S +T
Sbjct: 181 VVEPYNSVLSTHSLLEHTDVAVLLDNEAIYDICRRSLDIERPTYTNLNRLISQIISSLTT 240
Query: 239 CLRFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKN 298
LRF G +N D+ + NLVP+PR+HF + +AP+ S + LSVPEIT +++ +
Sbjct: 241 SLRFDGAINVDITEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQLSVPEITNAVFEPAS 300
Query: 299 MMCAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPP 358
MM DPRHG+Y+ ++RG + K+V+ + ++ K + FV+W P K + PP
Sbjct: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVGTIKTKRTVQFVDWCPTGFKCGINYQPP 360
Query: 359 T--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
T ++ A I N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 361 TVVPGGDLAKVQRAVCMISNNTAVAEVFSRIDHKFDLMYAKRAFVHWYVGEGMEEGEFSE 420
Query: 411 AESNMNDLVSEYQQYQDATAE-EDEYEDEEEDY 442
A DL + + Y++ AE D+ EDE EDY
Sbjct: 421 AR---EDLAALEKDYEEVGAEGGDDEEDEGEDY 450
>At4g14960 tubulin alpha-6 chain (TUA6)
Length = 450
Score = 388 bits (996), Expect = e-108
Identities = 192/453 (42%), Positives = 286/453 (62%), Gaps = 14/453 (3%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQL--ERINVYYNEASGGR 58
MRE + I GQ G Q+G WE+ C EHGI P G+ GD + + N +++E G+
Sbjct: 1 MRECISIHIGQAGIQVGNACWELYCLEHGIQPDGQMPGDKTVGGGDDAFNTFFSETGAGK 60
Query: 59 FVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
VPRAV +DLEP +D VR+G Y Q+F P+ + G+ A NN+A+GHYT G E++D LD
Sbjct: 61 HVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTIGKEIVDLCLD 120
Query: 119 VVRKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
+RK A+N LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+V+PSP+VS +
Sbjct: 121 RIRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTVYPSPQVSTS 180
Query: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN L+S +S +T
Sbjct: 181 VVEPYNSVLSTHSLLEHTDVSILLDNEAIYDICRRSLNIERPTYTNLNRLVSQVISSLTA 240
Query: 239 CLRFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKN 298
LRF G LN D+ + NLVP+PR+HF + +AP+ S + LSV EIT ++ +
Sbjct: 241 SLRFDGALNVDVTEFQTNLVPYPRIHFMLSSYAPVISAEKAFHEQLSVAEITNSAFEPAS 300
Query: 299 MMCAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPP 358
MM DPRHG+Y+ ++RG + K+V+ + ++ K + FV+W P K + PP
Sbjct: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVGTIKTKRTIQFVDWCPTGFKCGINYQPP 360
Query: 359 T--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
T ++ A I NSTS+ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 361 TVVPGGDLAKVQRAVCMISNSTSVAEVFSRIDHKFDLMYAKRAFVHWYVGEGMEEGEFSE 420
Query: 411 AESNMNDLVSEYQQYQDATAE-EDEYEDEEEDY 442
A DL + + Y++ AE D+ +DE E+Y
Sbjct: 421 AR---EDLAALEKDYEEVGAEGGDDEDDEGEEY 450
>At1g50010 tubulin alpha-2/alpha-4 chain, putative
Length = 450
Score = 385 bits (990), Expect = e-107
Identities = 191/453 (42%), Positives = 285/453 (62%), Gaps = 14/453 (3%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQL--ERINVYYNEASGGR 58
MRE + I GQ G Q+G WE+ C EHGI P G+ D + + N +++E G+
Sbjct: 1 MRECISIHIGQAGIQVGNACWELYCLEHGIQPDGQMPSDKTVGGGDDAFNTFFSETGAGK 60
Query: 59 FVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
VPRAV +DLEP +D VR+G Y Q+F P+ + G+ A NN+A+GHYT G E++D LD
Sbjct: 61 HVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTIGKEIVDLCLD 120
Query: 119 VVRKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
+RK A+N LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+V+PSP+VS +
Sbjct: 121 RIRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTVYPSPQVSTS 180
Query: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN L+S +S +T
Sbjct: 181 VVEPYNSVLSTHSLLEHTDVSILLDNEAIYDICRRSLSIERPTYTNLNRLVSQVISSLTA 240
Query: 239 CLRFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKN 298
LRF G LN D+ + NLVP+PR+HF + +AP+ S + LSV EIT ++ +
Sbjct: 241 SLRFDGALNVDVTEFQTNLVPYPRIHFMLSSYAPVISAEKAFHEQLSVAEITNSAFEPAS 300
Query: 299 MMCAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPP 358
MM DPRHG+Y+ ++RG + K+V+ + ++ K + FV+W P K + PP
Sbjct: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVGTIKTKRTIQFVDWCPTGFKCGINYQPP 360
Query: 359 T--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
T ++ A I NSTS+ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 361 TVVPGGDLAKVQRAVCMISNSTSVAEVFSRIDHKFDLMYAKRAFVHWYVGEGMEEGEFSE 420
Query: 411 AESNMNDLVSEYQQYQDATAE-EDEYEDEEEDY 442
A DL + + Y++ AE D+ +DE E+Y
Sbjct: 421 AR---EDLAALEKDYEEVGAEGGDDEDDEGEEY 450
>At1g04820 tubulin alpha-2/alpha-4 chain
Length = 450
Score = 385 bits (990), Expect = e-107
Identities = 191/453 (42%), Positives = 285/453 (62%), Gaps = 14/453 (3%)
Query: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQL--ERINVYYNEASGGR 58
MRE + I GQ G Q+G WE+ C EHGI P G+ D + + N +++E G+
Sbjct: 1 MRECISIHIGQAGIQVGNACWELYCLEHGIQPDGQMPSDKTVGGGDDAFNTFFSETGAGK 60
Query: 59 FVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
VPRAV +DLEP +D VR+G Y Q+F P+ + G+ A NN+A+GHYT G E++D LD
Sbjct: 61 HVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTIGKEIVDLCLD 120
Query: 119 VVRKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
+RK A+N LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+V+PSP+VS +
Sbjct: 121 RIRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTVYPSPQVSTS 180
Query: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN L+S +S +T
Sbjct: 181 VVEPYNSVLSTHSLLEHTDVSILLDNEAIYDICRRSLSIERPTYTNLNRLVSQVISSLTA 240
Query: 239 CLRFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLTSRGSQQYRALSVPEITQQMWDSKN 298
LRF G LN D+ + NLVP+PR+HF + +AP+ S + LSV EIT ++ +
Sbjct: 241 SLRFDGALNVDVTEFQTNLVPYPRIHFMLSSYAPVISAEKAFHEQLSVAEITNSAFEPAS 300
Query: 299 MMCAADPRHGRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPP 358
MM DPRHG+Y+ ++RG + K+V+ + ++ K + FV+W P K + PP
Sbjct: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVGTIKTKRTIQFVDWCPTGFKCGINYQPP 360
Query: 359 T--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
T ++ A I NSTS+ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 361 TVVPGGDLAKVQRAVCMISNSTSVAEVFSRIDHKFDLMYAKRAFVHWYVGEGMEEGEFSE 420
Query: 411 AESNMNDLVSEYQQYQDATAE-EDEYEDEEEDY 442
A DL + + Y++ AE D+ +DE E+Y
Sbjct: 421 AR---EDLAALEKDYEEVGAEGGDDEDDEGEEY 450
>At3g61650 tubulin gamma-1 chain
Length = 474
Score = 290 bits (743), Expect = 8e-79
Identities = 158/455 (34%), Positives = 264/455 (57%), Gaps = 17/455 (3%)
Query: 2 REILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQLERINVYYNEASGGRFVP 61
REI+ +Q GQCGNQIG +FW+ +C EHGI G + +R +V++ +A ++P
Sbjct: 3 REIITLQVGQCGNQIGMEFWKQLCLEHGISKDGILEDFATQGGDRKDVFFYQADDQHYIP 62
Query: 62 RAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQ--SGAGNNWAKGHYTEGAELIDSVLDV 119
RA+L+DLEP ++ +++G Y ++ +N GAGNNWA G Y +G + + ++D+
Sbjct: 63 RALLIDLEPRVINGIQNGDYRNLYNHENIFVADHGGGAGNNWASG-YHQGKGVEEEIMDM 121
Query: 120 VRKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSP-KVSDT 178
+ +EA+ SD L+GF +CHS+ GGTGSGMG+ L+ + + Y +++ T+SVFP+ + SD
Sbjct: 122 IDREADGSDSLEGFVLCHSIAGGTGSGMGSYLLETLNDRYSKKLVQTYSVFPNQMETSDV 181
Query: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
VV+PYN+ L++ +L NAD +VLDN AL I L LT P+F N L+S MS T
Sbjct: 182 VVQPYNSLLTLKRLTLNADCVVVLDNTALGRIAVERLHLTNPTFAQTNSLVSTVMSASTT 241
Query: 239 CLRFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLT-SRGSQQYRALSVPEITQQMWDSK 297
LR+PG +N+DL L +L+P PR HF M G+ PLT R + R +V ++ +++ +K
Sbjct: 242 TLRYPGYMNNDLVGLLASLIPTPRCHFLMTGYTPLTVERQANVIRKTTVLDVMRRLLQTK 301
Query: 298 NMMCAADPRH-----GRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKST 352
N+M ++ R+ +Y++ I +G++ +V E + ++ + F+EW P +++
Sbjct: 302 NIMVSSYARNKEASQAKYISILNIIQGEVDPTQVHESLQRIRERKLVNFIEWGPASIQVA 361
Query: 353 VCDIPP---TGLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGM----DE 405
+ P T +++ + + TSI+ +F + Q+ + +++AFL Y M D
Sbjct: 362 LSKKSPYVQTAHRVSGLMLASHTSIRHLFSKCLSQYDKLRKKQAFLDNYRKFPMFADNDL 421
Query: 406 MEFTEAESNMNDLVSEYQQYQDATAEEDEYEDEEE 440
EF E+ + LV EY+ + + ED E+
Sbjct: 422 SEFDESRDIIESLVDEYKACESPDYIKWGMEDPEQ 456
>At5g05620 tubulin gamma-2 chain (sp|P38558)
Length = 474
Score = 290 bits (742), Expect = 1e-78
Identities = 155/438 (35%), Positives = 258/438 (58%), Gaps = 17/438 (3%)
Query: 2 REILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYVGDSDLQLERINVYYNEASGGRFVP 61
REI+ +Q GQCGNQIG +FW+ +C EHGI G + +R +V++ +A ++P
Sbjct: 3 REIITLQVGQCGNQIGMEFWKQLCLEHGISKDGILEDFATQGGDRKDVFFYQADDQHYIP 62
Query: 62 RAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQ--SGAGNNWAKGHYTEGAELIDSVLDV 119
RA+L+DLEP ++ +++G Y ++ +N GAGNNWA G Y +G + + ++D+
Sbjct: 63 RALLIDLEPRVINGIQNGEYRNLYNHENIFLSDHGGGAGNNWASG-YHQGKGVEEEIMDM 121
Query: 120 VRKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSP-KVSDT 178
+ +EA+ SD L+GF +CHS+ GGTGSGMG+ L+ + + Y +++ T+SVFP+ + SD
Sbjct: 122 IDREADGSDSLEGFVLCHSIAGGTGSGMGSYLLETLNDRYSKKLVQTYSVFPNQMETSDV 181
Query: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
VV+PYN+ L++ +L NAD +VLDN AL I L LT P+F N L+S MS T
Sbjct: 182 VVQPYNSLLTLKRLTLNADCVVVLDNTALNRIAVERLHLTNPTFAQTNSLVSTVMSASTT 241
Query: 239 CLRFPGQLNSDLRKLAVNLVPFPRLHFFMLGFAPLT-SRGSQQYRALSVPEITQQMWDSK 297
LR+PG +N+DL L +L+P PR HF M G+ PLT R + R +V ++ +++ +K
Sbjct: 242 TLRYPGYMNNDLVGLLASLIPTPRCHFLMTGYTPLTVERQANVIRKTTVLDVMRRLLQTK 301
Query: 298 NMMCAADPRH-----GRYLTASAIFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKST 352
N+M ++ R+ +Y++ I +G++ +V E + ++ + F++W P +++
Sbjct: 302 NIMVSSYARNKEASQAKYISILNIIQGEVDPTQVHESLQRIRERKLVNFIDWGPASIQVA 361
Query: 353 VCDIPP---TGLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGM----DE 405
+ P T +++ + + TSI+ +F R Q+ + +++AFL Y M D
Sbjct: 362 LSKKSPYVQTSHRVSGLMLASHTSIRHLFSRCLSQYDKLRKKQAFLDNYRKFPMFADNDL 421
Query: 406 MEFTEAESNMNDLVSEYQ 423
EF E+ + LV EY+
Sbjct: 422 SEFDESRDIIESLVDEYK 439
>At5g56950 nucleosome assembly protein
Length = 374
Score = 37.7 bits (86), Expect = 0.013
Identities = 21/52 (40%), Positives = 31/52 (59%), Gaps = 5/52 (9%)
Query: 397 WYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATAEEDEYEDEEEDYQQEHDE 448
W+TGE ++ EF E +++ D + E + EEDE EDEEED + E +E
Sbjct: 293 WFTGEAIEGEEF-EIDNDDEDDIDE----DEDEDEEDEDEDEEEDDEDEEEE 339
>At5g55280 cell division protein FtsZ chloroplast homolog precursor
(sp|Q42545)
Length = 433
Score = 37.0 bits (84), Expect = 0.022
Identities = 35/143 (24%), Positives = 61/143 (42%), Gaps = 12/143 (8%)
Query: 109 GAELIDSVLDVVRKEAENSDCLQGFQVCHSLGGGTGSGMGTLL--ISKIREEYPDRMMLT 166
G + + D + + SD + + +GGGTGSG ++ ISK D LT
Sbjct: 140 GEQAAEESKDAIANALKGSDLVF---ITAGMGGGTGSGAAPVVAQISK------DAGYLT 190
Query: 167 FSVFPSPKVSDTVVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLN 226
V P + A ++ +L +N D +V+ N+ L DI L +F +
Sbjct: 191 VGVVTYPFSFEGRKRSLQALEAIEKLQKNVDTLIVIPNDRLLDIADEQTPL-QDAFLLAD 249
Query: 227 HLISATMSGVTCCLRFPGQLNSD 249
++ + G++ + PG +N D
Sbjct: 250 DVLRQGVQGISDIITIPGLVNVD 272
>At4g26110 nucleosome assembly protein I-like protein
Length = 372
Score = 35.4 bits (80), Expect = 0.064
Identities = 17/48 (35%), Positives = 30/48 (62%), Gaps = 4/48 (8%)
Query: 397 WYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATAEEDEYEDEEEDYQQ 444
W+TGE M+ +F E + + D + E + +D EEDE +D++ED ++
Sbjct: 294 WFTGEAMEAEDF-EIDDDEEDDIDEDEDEED---EEDEEDDDDEDEEE 337
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,525,866
Number of Sequences: 26719
Number of extensions: 466035
Number of successful extensions: 3040
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 2502
Number of HSP's gapped (non-prelim): 277
length of query: 449
length of database: 11,318,596
effective HSP length: 103
effective length of query: 346
effective length of database: 8,566,539
effective search space: 2964022494
effective search space used: 2964022494
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144474.3


