

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144459.2 - phase: 0 /pseudo
(824 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
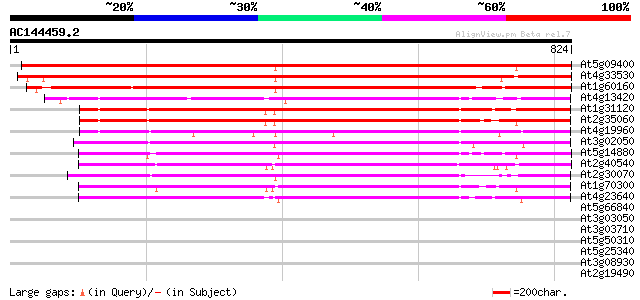
Score E
Sequences producing significant alignments: (bits) Value
At5g09400 potassium transport protein-like 1202 0.0
At4g33530 putative potassium transporter AtKT5p (AtKT5) 1117 0.0
At1g60160 unknown protein 796 0.0
At4g13420 K+ transporter HAK5 (HAK5) 576 e-164
At1g31120 putative potassium transporter 573 e-163
At2g35060 putative potassium transporter 570 e-162
At4g19960 potassium transporter-like protein 538 e-153
At3g02050 putative potassium transporter 524 e-149
At5g14880 HAK8 507 e-144
At2g40540 putative potassium transporter AtKT2p (AtKT2) 503 e-142
At2g30070 high affinity K+ transporter (AtKUP1/AtKT1p) 496 e-140
At1g70300 high affinity potassium transporter AtHAK6 (At1g70300) 490 e-138
At4g23640 potassium transport like protein 487 e-137
At5g66840 unknown protein 35 0.17
At3g03050 putative cellulose synthase catalytic subunit 33 0.65
At3g03710 putative polynucleotide phosphorylase 32 1.4
At5g50310 unknown protein 32 1.9
At5g25340 unknown protein 31 2.5
At3g08930 unknown protein 31 2.5
At2g19490 putative recA protein 31 2.5
>At5g09400 potassium transport protein-like
Length = 858
Score = 1202 bits (3110), Expect = 0.0
Identities = 611/836 (73%), Positives = 701/836 (83%), Gaps = 30/836 (3%)
Query: 18 GSALSFDSTESRWVFQEDEDPSEIEDYDASDMRHQSMFDSEDEDNAEMKLIRTGPRIDSF 77
G S DS ESRWV Q+D+D SEI D +D + +S++++ E +LIRTGPR+DSF
Sbjct: 23 GDMASMDSIESRWVIQDDDD-SEIGVDDDNDGFDGTGLESDEDEIPEHRLIRTGPRVDSF 81
Query: 78 DVEALEVPGAHTHHYEDMTTGKKIVLAFQTLGVVFGDVGTSPLYTFSVMFRKAPINDNED 137
DVEALEVPGA + YED+T G+K++LAFQTLGVVFGDVGTSPLYTFSVMF K+P+ + ED
Sbjct: 82 DVEALEVPGAPRNDYEDLTVGRKVLLAFQTLGVVFGDVGTSPLYTFSVMFSKSPVQEKED 141
Query: 138 ILGALSLVLYTLILIPFLKYVLVVLWANDDGEGGTFALYSLICRNAKVNLLPNQLPSDAR 197
++GALSLVLYTL+L+P +KYVLVVLWANDDGEGGTFALYSLI R+AK++L+PNQL SD R
Sbjct: 142 VIGALSLVLYTLLLVPLIKYVLVVLWANDDGEGGTFALYSLISRHAKISLIPNQLRSDTR 201
Query: 198 ISGFRLKVPSAELERSLKLKERLESSFTLKKILLLLVLAGTSMVIANGVVTPAMSVLSSV 257
IS FRLKVP ELERSLKLKE+LE+S LKKILL+LVLAGTSMVIA+GVVTPAMSV+S+V
Sbjct: 202 ISSFRLKVPCPELERSLKLKEKLENSLILKKILLVLVLAGTSMVIADGVVTPAMSVMSAV 261
Query: 258 NGLKVGVDAIQQDEVVMISVACLVVLFSLQKYGTSKVGLAVGPALFIWFCSLAGNGVYNL 317
GLKVGVD ++QD+VVMISVA LV+LFSLQKYGTSK+GL VGPAL IWFCSLAG G+YNL
Sbjct: 262 GGLKVGVDVVEQDQVVMISVAFLVILFSLQKYGTSKMGLVVGPALLIWFCSLAGIGIYNL 321
Query: 318 VKYDSSVFRAFNPIHIYYFFARNSTKAWYSLGGCLLCATGSEAMFADLCYFSVRSVQPII 377
+KYDSSV+RAFNP+HIYYFF RNS AWY+LGGC+LCATGSEA+FADLCYFSVRSVQ
Sbjct: 322 IKYDSSVYRAFNPVHIYYFFKRNSINAWYALGGCILCATGSEALFADLCYFSVRSVQLTF 381
Query: 378 GLFGSSCIPYG----------------------TPC*CW*SLFFFCS--KLIASRTMTTA 413
C+ G P + + F + LIASRTMTTA
Sbjct: 382 VCLVLPCLMLGYMGQAAYLMENHADASQAFFSSVPGSAFWPVLFIANIAALIASRTMTTA 441
Query: 414 TFSCIKQSTALGCFPRLKIIHTSRKFMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAY 473
TFSCIKQSTALGCFPRLKIIHTSRKFMGQIYIPV+NWFLLAV LV VC+ISSIDEIGNAY
Sbjct: 442 TFSCIKQSTALGCFPRLKIIHTSRKFMGQIYIPVLNWFLLAVCLVVVCSISSIDEIGNAY 501
Query: 474 GIAELGVMMMTTILVTLVMLLIWQMHIIIVMSFLGVFLGLELVFFSSVLWSITDGSWIIL 533
G+AELGVMM TTILVTL+MLLIWQ++I+IV++FL VFLG+ELVFFSSV+ S+ DGSWIIL
Sbjct: 502 GMAELGVMMTTTILVTLIMLLIWQINIVIVIAFLVVFLGVELVFFSSVIASVGDGSWIIL 561
Query: 534 VFAAIMFFIMFIWNYGSKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGI 593
VFA IMF IM+IWNYGSKL+YETEV+QKLS DLMRELGCNLGTIRAPGIGLLYNELVKG+
Sbjct: 562 VFAVIMFGIMYIWNYGSKLRYETEVEQKLSMDLMRELGCNLGTIRAPGIGLLYNELVKGV 621
Query: 594 PGIFGHFLTTLPAIHSMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKD 653
P IFGHFLTTLPAIHSM+IFV IKYVPV +VPQ+ERFLFRRVC +SYHLFRCIARYGYKD
Sbjct: 622 PAIFGHFLTTLPAIHSMVIFVCIKYVPVPVVPQNERFLFRRVCTKSYHLFRCIARYGYKD 681
Query: 654 ARKENHQAFEQLLMESLEKFIRREAQERSLESDGDEDTELEDEYAGSRVLIAPNGSVYSL 713
ARKE HQAFEQLL+ESLEKFIRREAQERSLESDG++D++ E+++ GSRV+I PNGS+YS+
Sbjct: 682 ARKETHQAFEQLLIESLEKFIRREAQERSLESDGNDDSDSEEDFPGSRVVIGPNGSMYSM 741
Query: 714 GVPLLADFNESFMPSFEPSTSEEAGPPSP-----KPLVLDAEQLLERELSFIRNAKESGL 768
GVPLL+++ + P E +TS + P V +AEQ LERELSFI AKESG+
Sbjct: 742 GVPLLSEYRDLNKPIMEMNTSSDHTNHHPFDTSSDSSVSEAEQSLERELSFIHKAKESGV 801
Query: 769 VYLLGHGDIRARKDSWFIKKLVINYFYAFLRKNCRRGVTNLSVPHSHLMQVGMTYM 824
VYLLGHGDIRARKDSWFIKKLVINYFY FLRKNCRRG+ NLSVP SHLMQVGMTYM
Sbjct: 802 VYLLGHGDIRARKDSWFIKKLVINYFYTFLRKNCRRGIANLSVPQSHLMQVGMTYM 857
>At4g33530 putative potassium transporter AtKT5p (AtKT5)
Length = 855
Score = 1117 bits (2889), Expect = 0.0
Identities = 581/854 (68%), Positives = 669/854 (78%), Gaps = 45/854 (5%)
Query: 12 EEEDDPGSALSFD---------STESRWVFQEDEDPSEIEDYDAS---DMRHQSMFDSED 59
EEE G D ++ SRWVF E +D EDYD + H M E+
Sbjct: 5 EEESSGGDGSEIDEEFGGDDSTTSLSRWVFDEKDDYEVNEDYDDDGYDEHNHPEMDSDEE 64
Query: 60 EDNAEMKLIRTGPRIDSFDVEALEVPGAHTHHYEDMTTGKKIVLAFQTLGVVFGDVGTSP 119
+DN E +LIRT P +DSFDV+ALE+PG + ED GKK++LA QTLGVVFGD+GTSP
Sbjct: 65 DDNVEQRLIRTSPAVDSFDVDALEIPGTQKNEIEDTGIGKKLILALQTLGVVFGDIGTSP 124
Query: 120 LYTFSVMFRKAPINDNEDILGALSLVLYTLILIPFLKYVLVVLWANDDGEGGTFALYSLI 179
LYTF+VMFR++PIND EDI+GALSLV+YTLILIP +KYV VLWANDDGEGGTFALYSLI
Sbjct: 125 LYTFTVMFRRSPINDKEDIIGALSLVIYTLILIPLVKYVHFVLWANDDGEGGTFALYSLI 184
Query: 180 CRNAKVNLLPNQLPSDARISGFRLKVPSAELERSLKLKERLESSFTLKKILLLLVLAGTS 239
CR+A V+L+PNQLPSDARISGF LKVPS ELERSL +KERLE+S LKK+LL+LVLAGT+
Sbjct: 185 CRHANVSLIPNQLPSDARISGFGLKVPSPELERSLIIKERLEASMALKKLLLILVLAGTA 244
Query: 240 MVIANGVVTPAMSVLSSVNGLKVGVDAIQQDEVVMISVACLVVLFSLQKYGTSKVGLAVG 299
MVIA+ VVTPAMSV+S++ GLKVGV I+QD+VV+ISV+ LV+LFS+QKYGTSK+GL +G
Sbjct: 245 MVIADAVVTPAMSVMSAIGGLKVGVGVIEQDQVVVISVSFLVILFSVQKYGTSKLGLVLG 304
Query: 300 PALFIWFCSLAGNGVYNLVKYDSSVFRAFNPIHIYYFFARNSTKAWYSLGGCLLCATGSE 359
PAL +WF LAG G+YNLVKYDSSVF+AFNP +IY+FF RNS AWY+LGGC+LCATGSE
Sbjct: 305 PALLLWFFCLAGIGIYNLVKYDSSVFKAFNPAYIYFFFKRNSVNAWYALGGCVLCATGSE 364
Query: 360 AMFADLCYFSVRSVQPIIGLFGSSCIPYG----------------------TPC*CW*SL 397
AMFADL YFSV S+Q L C+ G P + +
Sbjct: 365 AMFADLSYFSVHSIQLTFILLVLPCLLLGYLGQAAYLSENFSAAGDAFFSSVPSSLFWPV 424
Query: 398 FFFCS--KLIASRTMTTATFSCIKQSTALGCFPRLKIIHTSRKFMGQIYIPVINWFLLAV 455
F + LIASR MTTATF+CIKQS ALGCFPRLKIIHTS+KF+GQIYIPV+NW LL V
Sbjct: 425 FLISNVAALIASRAMTTATFTCIKQSIALGCFPRLKIIHTSKKFIGQIYIPVLNWSLLVV 484
Query: 456 SLVFVCTISSIDEIGNAYGIAELGVMMMTTILVTLVMLLIWQMHIIIVMSFLGVFLGLEL 515
L+ VC+ S+I IGNAYGIAELG+MM TTILVTL+MLLIWQ +II+V F V L +EL
Sbjct: 485 CLIVVCSTSNIFAIGNAYGIAELGIMMTTTILVTLIMLLIWQTNIIVVSMFAIVSLIVEL 544
Query: 516 VFFSSVLWSITDGSWIILVFAAIMFFIMFIWNYGSKLKYETEVKQKLSPDLMRELGCNLG 575
VFFSSV S+ DGSWIILVFA IMF IMF+WNYGSKLKYETEV++KL DL+RELG NLG
Sbjct: 545 VFFSSVCSSVADGSWIILVFATIMFLIMFVWNYGSKLKYETEVQKKLPMDLLRELGSNLG 604
Query: 576 TIRAPGIGLLYNELVKGIPGIFGHFLTTLPAIHSMIIFVSIKYVPVAMVPQSERFLFRRV 635
TIRAPGIGLLYNEL KG+P IFGHFLTTLPAIHSM+IFV IKYVPV VPQ+ERFLFRRV
Sbjct: 605 TIRAPGIGLLYNELAKGVPAIFGHFLTTLPAIHSMVIFVCIKYVPVPSVPQTERFLFRRV 664
Query: 636 CQRSYHLFRCIARYGYKDARKENHQAFEQLLMESLEKFIRREAQERSLESDGD-EDTELE 694
C RSYHLFRC+ARYGYKD RKE+HQAFEQ+L+ESLEKFIR+EAQER+LESDGD DT+ E
Sbjct: 665 CPRSYHLFRCVARYGYKDVRKESHQAFEQILIESLEKFIRKEAQERALESDGDHNDTDSE 724
Query: 695 DEYAGSRVLIAPNGSVYSLGVPLLADF----NESFMPSFEPSTSEEAGPPSPKPLVLDAE 750
D+ SRVLIAPNGSVYSLGVPLLA+ N+ M + S AGP S LD E
Sbjct: 725 DDTTLSRVLIAPNGSVYSLGVPLLAEHMNSSNKRPMERRKASIDFGAGPSS----ALDVE 780
Query: 751 QLLERELSFIRNAKESGLVYLLGHGDIRARKDSWFIKKLVINYFYAFLRKNCRRGVTNLS 810
Q LE+ELSFI AKESG+VYLLGHGDIRA KDSWF+KKLVINY YAFLRKN RRG+TNLS
Sbjct: 781 QSLEKELSFIHKAKESGVVYLLGHGDIRATKDSWFLKKLVINYLYAFLRKNSRRGITNLS 840
Query: 811 VPHSHLMQVGMTYM 824
VPH+HLMQVGMTYM
Sbjct: 841 VPHTHLMQVGMTYM 854
>At1g60160 unknown protein
Length = 826
Score = 796 bits (2057), Expect = 0.0
Identities = 428/829 (51%), Positives = 555/829 (66%), Gaps = 52/829 (6%)
Query: 25 STESRWVFQEDEDP-----SEIEDYDASDMRHQSMFDSEDEDNAEMKLIRTGPRIDSFDV 79
S++ RWV + D SEI D D S N +L++ R DS DV
Sbjct: 20 SSDRRWVDGSEVDSETPLFSEIRDRDYSF------------GNLRRRLMKKPKRADSLDV 67
Query: 80 EALEVPGAHTHHYEDMTTGKKIVLAFQTLGVVFGDVGTSPLYTFSVMFRKAPINDNEDIL 139
EA+E+ G+H H+ +D++ + +AFQTLGVV+GD+GTSPLY FS +F K PI D+L
Sbjct: 68 EAMEIAGSHGHNLKDLSLLTTLGIAFQTLGVVYGDMGTSPLYVFSDVFSKVPIRSEVDVL 127
Query: 140 GALSLVLYTLILIPFLKYVLVVLWANDDGEGGTFALYSLICRNAKVNLLPNQLPSDARIS 199
GALSLV+YT+ +IP KYV VVL AND+GEG A L+ + AKVN LPNQ P+D +IS
Sbjct: 128 GALSLVIYTIAVIPLAKYVFVVLKANDNGEGNASAPMCLV-KYAKVNKLPNQQPADEQIS 186
Query: 200 GFRLKVPSAELERSLKLKERLESSFTLKKILLLLVLAGTSMVIANGVVTPAMSVLSSVNG 259
FRLK+P+ ELER+L +KE LE+ LK +LLLLVL GTSM+I +G++TPAMSV+S+++G
Sbjct: 187 SFRLKLPTPELERALGIKEALETKGYLKTLLLLLVLMGTSMIIGDGILTPAMSVMSAMSG 246
Query: 260 LKVGVDAIQQDEVVMISVACLVVLFSLQKYGTSKVGLAVGPALFIWFCSLAGNGVYNLVK 319
L+ V + +VM S+ LV LFS+Q++GT KVG P L +WF SL G+YNL+K
Sbjct: 247 LQGEVKGFGTNALVMSSIVILVALFSIQRFGTGKVGFLFAPVLALWFFSLGAIGIYNLLK 306
Query: 320 YDSSVFRAFNPIHIYYFFARNSTKAWYSLGGCLLCATGSEAMFADLCYFSVRSVQPIIGL 379
YD +V RA NP +I FF +NS +AW +LGGC+LC TG+EAMFADL +FSVRS+Q
Sbjct: 307 YDFTVIRALNPFYIVLFFNKNSKQAWSALGGCVLCITGAEAMFADLGHFSVRSIQMAFTC 366
Query: 380 FGSSCIPYG----------------------TPC*CW*SLFFFCS--KLIASRTMTTATF 415
C+ P + +F + +IAS+ M +ATF
Sbjct: 367 VVFPCLLLAYMGQAAYLTKHPEASARIFYDSVPKSLFWPVFVIATLAAMIASQAMISATF 426
Query: 416 SCIKQSTALGCFPRLKIIHTSRKFMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAYGI 475
SC+KQ+ ALGCFPRLKIIHTS+K +GQIYIPVINWFL+ + ++ V S I NAYGI
Sbjct: 427 SCVKQAMALGCFPRLKIIHTSKKRIGQIYIPVINWFLMIMCILVVSIFRSTTHIANAYGI 486
Query: 476 AELGVMMMTTILVTLVMLLIWQMHIIIVMSFLGVFLGLELVFFSSVLWSITDGSWIILVF 535
AE+GVMM++T+LVTLVMLLIWQ +I + + F +F +E ++ +VL I +G W+ LVF
Sbjct: 487 AEVGVMMVSTVLVTLVMLLIWQTNIFLALCFPLIFGSVETIYLLAVLTKILEGGWVPLVF 546
Query: 536 AAIMFFIMFIWNYGSKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGIPG 595
A +M+IWNYGS LKY++EV++++S D MRELG LGTIR PGIGLLYNELV+GIP
Sbjct: 547 ATFFLTVMYIWNYGSVLKYQSEVRERISMDFMRELGSTLGTIRIPGIGLLYNELVQGIPS 606
Query: 596 IFGHFLTTLPAIHSMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDAR 655
IFG FL TLPAIHS IIFV IKYVPV +VPQ ERFLFRRVC + YH+FRCIARYGYKD R
Sbjct: 607 IFGQFLLTLPAIHSTIIFVCIKYVPVPVVPQEERFLFRRVCPKDYHMFRCIARYGYKDVR 666
Query: 656 KENHQAFEQLLMESLEKFIRREAQERSLESDGDEDTELEDEYAGSRVLIAPNGSVYSLGV 715
KE+ + FEQLL+ESLEKF+R EA E +LES +++ RV +A + L
Sbjct: 667 KEDSRVFEQLLIESLEKFLRCEALEDALES-------TLNDFDPDRVSVASDTYTDDLMA 719
Query: 716 PLLADFNESFMPSFEPSTSEEAGPPSPKPLVLDAEQLLERELSFIRNAKESGLVYLLGHG 775
PL+ S E E P S ++ + LE EL+ +R A +SGL YLL HG
Sbjct: 720 PLIHRAKRS---EPEQELDSEVLPSSSVGSSMEEDPALEYELAALREATDSGLTYLLAHG 776
Query: 776 DIRARKDSWFIKKLVINYFYAFLRKNCRRGVTNLSVPHSHLMQVGMTYM 824
D+RA+K+S F+KKLVINYFYAFLR+NCR G NL+VPH +++Q GMTYM
Sbjct: 777 DVRAKKNSIFVKKLVINYFYAFLRRNCRAGAANLTVPHMNILQAGMTYM 825
>At4g13420 K+ transporter HAK5 (HAK5)
Length = 785
Score = 576 bits (1484), Expect = e-164
Identities = 319/809 (39%), Positives = 485/809 (59%), Gaps = 67/809 (8%)
Query: 52 QSMFDSEDEDNAEMKLIRTGP------RIDSFDVEALEVPGAHTHHYEDMTTGKKIVLAF 105
+ D ++ +N E KL R DSF +EA + P +T M+ + LAF
Sbjct: 5 EHQIDGDEVNNHENKLNEKKKSWGKLYRPDSFIIEAGQTP-TNTGRRSLMSWRTTMSLAF 63
Query: 106 QTLGVVFGDVGTSPLYTFSVMFRKAPINDNEDILGALSLVLYTLILIPFLKYVLVVLWAN 165
Q+LGVV+GD+GTSPLY ++ F IND +D++G LSL++YT+ L+ LKYV +VL AN
Sbjct: 64 QSLGVVYGDIGTSPLYVYASTFTDG-INDKDDVVGVLSLIIYTITLVALLKYVFIVLQAN 122
Query: 166 DDGEGGTFALYSLICRNAKVNLLPNQLPSDARISGFRLKVPSAELERSLKLKERLESSFT 225
D+GEGGTFALYSLICR AK+ L+PNQ P D +S + L++P+ +L R+ +KE+LE+S
Sbjct: 123 DNGEGGTFALYSLICRYAKMGLIPNQEPEDVELSNYTLELPTTQLRRAHMIKEKLENSKF 182
Query: 226 LKKILLLLVLAGTSMVIANGVVTPAMSVLSSVNGLKVGVDAIQQDEVVMISVACLVVLFS 285
K IL L+ + GTSMVI +G++TP++SVLS+V+G+K ++ Q+ VV +SVA L+VLF+
Sbjct: 183 AKIILFLVTIMGTSMVIGDGILTPSISVLSAVSGIK----SLGQNTVVGVSVAILIVLFA 238
Query: 286 LQKYGTSKVGLAVGPALFIWFCSLAGNGVYNLVKYDSSVFRAFNPIHIYYFFARNSTKAW 345
Q++GT KVG + P + +WF L G G++NL K+D +V +A NP++I Y+F R + W
Sbjct: 239 FQRFGTDKVGFSFAPIILVWFTFLIGIGLFNLFKHDITVLKALNPLYIIYYFRRTGRQGW 298
Query: 346 YSLGGCLLCATGSEAMFADLCYFSVRSVQPIIGLFGSSCIPYGTPC*CW*SLFFFCSK-- 403
SLGG LC TG+EAMFADL +FSVR+VQ SC+ Y + + +K
Sbjct: 299 ISLGGVFLCITGTEAMFADLGHFSVRAVQ-----ISFSCVAYPALVTIYCGQAAYLTKHT 353
Query: 404 ---------------------------LIASRTMTTATFSCIKQSTALGCFPRLKIIHTS 436
+IAS+ M + FS I QS +GCFPR+K++HTS
Sbjct: 354 YNVSNTFYDSIPDPLYWPTFVVAVAASIIASQAMISGAFSVISQSLRMGCFPRVKVVHTS 413
Query: 437 RKFMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAYGIAELGVMMMTTILVTLVMLLIW 496
K+ GQ+YIP IN+ L+ + + ++IG+AYGIA + VM++TT++VTL+ML+IW
Sbjct: 414 AKYEGQVYIPEINYLLMLACIAVTLAFRTTEKIGHAYGIAVVTVMVITTLMVTLIMLVIW 473
Query: 497 QMHIIIVMSFLGVFLGLELVFFSSVLWSITDGSWIILVFAAIMFFIMFIWNYGSKLKYET 556
+ +I+ + FL VF +E+++ SSV++ T G ++ L ++ +M IW Y LKY
Sbjct: 474 KTNIVWIAIFLVVFGSIEMLYLSSVMYKFTSGGYLPLTITVVLMAMMAIWQYVHVLKYRY 533
Query: 557 EVKQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPAIHSMIIFVSI 616
E+++K+S + ++ + R PGIGL Y ELV GI +F H+++ L ++HS+ + +SI
Sbjct: 534 ELREKISRENAIQMATSPDVNRVPGIGLFYTELVNGITPLFSHYISNLSSVHSVFVLISI 593
Query: 617 KYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKENHQAFEQLLMESLEKFIRR 676
K +PV V SERF FR V + +FRC+ RYGYK+ +E + FE+ + L++FI
Sbjct: 594 KTLPVNRVTSSERFFFRYVGPKDSGMFRCVVRYGYKEDIEEPDE-FERHFVYYLKEFIH- 651
Query: 677 EAQERSLESDGDE--DTELEDEYAGSRVLIAPNGSVYSLGVPLLADFNESFMPSFEPSTS 734
E + G E +T+ E+E ++ + V S G + S S+S
Sbjct: 652 --HEHFMSGGGGEVDETDKEEEPNAETTVVPSSNYVPSSG----------RIGSAHSSSS 699
Query: 735 EEAGPPSPKPLVLDAEQLLERELSFIRNAKESGLVYLLGHGDIRARKDSWFIKKLVINYF 794
++ + Q +E + + A+E G+VYL+G +I A K+S KK ++N+
Sbjct: 700 DKIRSGRVVQV-----QSVEDQTELVEKAREKGMVYLMGETEITAEKESSLFKKFIVNHA 754
Query: 795 YAFLRKNCRRGVTNLSVPHSHLMQVGMTY 823
Y FL+KNCR G L++P S L++VGMTY
Sbjct: 755 YNFLKKNCREGDKALAIPRSKLLKVGMTY 783
>At1g31120 putative potassium transporter
Length = 796
Score = 573 bits (1477), Expect = e-163
Identities = 312/747 (41%), Positives = 466/747 (61%), Gaps = 37/747 (4%)
Query: 103 LAFQTLGVVFGDVGTSPLYTFSVMFRKAPINDNEDILGALSLVLYTLILIPFLKYVLVVL 162
L+FQ+LGVV+GD+GTSPLY F F + I D EDI+GALSL++Y+L LIP LKYV VV
Sbjct: 59 LSFQSLGVVYGDLGTSPLYVFYNTFPRG-IKDPEDIIGALSLIIYSLTLIPLLKYVFVVC 117
Query: 163 WANDDGEGGTFALYSLICRNAKVNLLPNQLPSDARISGFRLKVPSAELERSLKLKERLES 222
AND+G+GGTFALYSL+CR+AKV+ +PNQ +D ++ + + E + K K LE+
Sbjct: 118 KANDNGQGGTFALYSLLCRHAKVSTIPNQHRTDEELTTYS-RTTFHERSFAAKTKRWLEN 176
Query: 223 SFTLKKILLLLVLAGTSMVIANGVVTPAMSVLSSVNGLKVGVDAIQQDEVVMISVACLVV 282
+ K LL+LVL GT MVI +G++TPA+SVLS+ GL+V + I VV+++V LV
Sbjct: 177 GTSRKNALLILVLVGTCMVIGDGILTPAISVLSAAGGLRVNLPHINNGIVVVVAVVILVS 236
Query: 283 LFSLQKYGTSKVGLAVGPALFIWFCSLAGNGVYNLVKYDSSVFRAFNPIHIYYFFARNST 342
LFS+Q YGT +VG P +F+WF +A G++N+ K+D SV +AF+P++I+ +F R
Sbjct: 237 LFSVQHYGTDRVGWLFAPIVFLWFLFIASIGMFNIWKHDPSVLKAFSPVYIFRYFKRGGQ 296
Query: 343 KAWYSLGGCLLCATGSEAMFADLCYFSVRSVQ--------PIIGLFGSSCIPY------- 387
W SLGG +L TG EA+FADL +F V +VQ P + L S Y
Sbjct: 297 DRWTSLGGIMLSITGIEALFADLSHFPVSAVQFAFTVIVFPCLLLAYSGQAAYLRKYPHH 356
Query: 388 -------GTPC*CW*SLFFFCSK--LIASRTMTTATFSCIKQSTALGCFPRLKIIHTSRK 438
P + +F + ++AS+ +ATFS IKQ+ A GCFPR+K++HTSRK
Sbjct: 357 VEDAFYQSIPSLVYWPMFIIATAAAIVASQATISATFSLIKQALAHGCFPRVKVVHTSRK 416
Query: 439 FMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAYGIAELGVMMMTTILVTLVMLLIWQM 498
F+GQIY+P INW L+ + + + ++IGNAYG A + VM++TT+L+ L+M+L+W+
Sbjct: 417 FLGQIYVPDINWILMILCIAVTAGFKNQNQIGNAYGTAVVIVMLVTTLLMMLIMILVWRC 476
Query: 499 HIIIVMSFLGVFLGLELVFFSSVLWSITDGSWIILVFAAIMFFIMFIWNYGSKLKYETEV 558
H ++V+ F + L +E +FS+VL+ + G W+ LV AA IM++W+YG+ +YE E+
Sbjct: 477 HWVLVLLFTLLSLVVECTYFSAVLFKVNQGGWVPLVIAAAFLVIMYVWHYGTLKRYEFEM 536
Query: 559 KQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPAIHSMIIFVSIKY 618
K+S + LG +LG +R PGIGL+Y EL G+P IF HF+T LPA HS++IFV +K
Sbjct: 537 HSKVSMAWILGLGPSLGLVRVPGIGLVYTELASGVPHIFSHFITNLPATHSVVIFVCVKN 596
Query: 619 VPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKENHQAFEQLLMESLEKFIRREA 678
+PV VPQ ERFL +R+ +++H+FRC+ARYGY+D K++ FE+ L ESL F+R E+
Sbjct: 597 LPVYTVPQEERFLVKRIGPKNFHMFRCVARYGYRDLHKKDDD-FEKRLFESLFLFLRLES 655
Query: 679 QERSLESDGDEDTELEDEYAGSRVLIAPNGSVYSLGVPLLADFNESFMPSFEPSTSEEAG 738
SD ++ + + SR + NG+ + D +S P+T++
Sbjct: 656 MMEGC-SDSEDYSVCGSQQRQSRDGVNGNGN--EIRNVSTFDTFDSIESVIAPTTTKRT- 711
Query: 739 PPSPKPLVLDAEQLL--ERELSFIRNAKESGLVYLLGHGDIRARKDSWFIKKLVINYFYA 796
V + Q+ E+ FI +++G+V+++G+ +RAR+++ F K++ I+Y YA
Sbjct: 712 ----SHTVTGSSQMSGGGDEVEFINGCRDAGVVHIMGNTVVRARREARFYKRIAIDYVYA 767
Query: 797 FLRKNCRRGVTNLSVPHSHLMQVGMTY 823
FLRK CR +VP L+ VG +
Sbjct: 768 FLRKICRENSAIFNVPQESLLNVGQIF 794
>At2g35060 putative potassium transporter
Length = 792
Score = 570 bits (1468), Expect = e-162
Identities = 312/749 (41%), Positives = 465/749 (61%), Gaps = 46/749 (6%)
Query: 103 LAFQTLGVVFGDVGTSPLYTFSVMFRKAPINDNEDILGALSLVLYTLILIPFLKYVLVVL 162
L+FQ+LGVV+GD+GTSPLY F F I D EDI+GALSL++Y+L LIP LKYV VV
Sbjct: 60 LSFQSLGVVYGDLGTSPLYVFYNTFPHG-IKDPEDIIGALSLIIYSLTLIPLLKYVFVVC 118
Query: 163 WANDDGEGGTFALYSLICRNAKVNLLPNQLPSDARISGFRLKVPSAELERSLKLKERLES 222
AND+G+GGTFALYSL+CR+AKV + NQ +D ++ + + E + K K LE
Sbjct: 119 KANDNGQGGTFALYSLLCRHAKVKTIQNQHRTDEELTTYS-RTTFHEHSFAAKTKRWLEK 177
Query: 223 SFTLKKILLLLVLAGTSMVIANGVVTPAMSVLSSVNGLKVGVDAIQQDEVVMISVACLVV 282
+ K LL+LVL GT MVI +G++TPA+SVLS+ GL+V + I VV ++V LV
Sbjct: 178 RTSRKTALLILVLVGTCMVIGDGILTPAISVLSAAGGLRVNLPHISNGVVVFVAVVILVS 237
Query: 283 LFSLQKYGTSKVGLAVGPALFIWFCSLAGNGVYNLVKYDSSVFRAFNPIHIYYFFARNST 342
LFS+Q YGT +VG P +F+WF S+A G+YN+ K+D+SV +AF+P++IY +F R
Sbjct: 238 LFSVQHYGTDRVGWLFAPIVFLWFLSIASIGMYNIWKHDTSVLKAFSPVYIYRYFKRGGR 297
Query: 343 KAWYSLGGCLLCATGSEAMFADLCYFSVRSVQ--------PIIGLFGSSCIPY------- 387
W SLGG +L TG EA+FADL +F V +VQ P + L S Y
Sbjct: 298 DRWTSLGGIMLSITGIEALFADLSHFPVSAVQIAFTVIVFPCLLLAYSGQAAYIRRYPDH 357
Query: 388 -------GTPC*CW*SLFFFCSK--LIASRTMTTATFSCIKQSTALGCFPRLKIIHTSRK 438
P + +F + ++AS+ +ATFS +KQ+ A GCFPR+K++HTSRK
Sbjct: 358 VADAFYRSIPGSVYWPMFIIATAAAIVASQATISATFSLVKQALAHGCFPRVKVVHTSRK 417
Query: 439 FMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAYGIAELGVMMMTTILVTLVMLLIWQM 498
F+GQIY+P INW L+ + + + +IGNAYG A + VM++TT+L+TL+M+L+W+
Sbjct: 418 FLGQIYVPDINWILMILCIAVTAGFKNQSQIGNAYGTAVVIVMLVTTLLMTLIMILVWRC 477
Query: 499 HIIIVMSFLGVFLGLELVFFSSVLWSITDGSWIILVFAAIMFFIMFIWNYGSKLKYETEV 558
H ++V+ F + L +E +FS++L+ I G W+ LV AA IM++W+YG+ +YE E+
Sbjct: 478 HWVLVLIFTVLSLVVECTYFSAMLFKIDQGGWVPLVIAAAFLLIMWVWHYGTLKRYEFEM 537
Query: 559 KQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPAIHSMIIFVSIKY 618
++S + LG +LG +R PG+GL+Y EL G+P IF HF+T LPAIHS+++FV +K
Sbjct: 538 HCRVSMAWILGLGPSLGLVRVPGVGLVYTELASGVPHIFSHFITNLPAIHSVVVFVCVKN 597
Query: 619 VPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKENHQAFEQLLMESLEKFIRREA 678
+PV VP+ ERFL +R+ +++H+FRC+ARYGY+D K++ FE+ L ESL ++R E+
Sbjct: 598 LPVYTVPEEERFLVKRIGPKNFHMFRCVARYGYRDLHKKDDD-FEKRLFESLFLYVRLES 656
Query: 679 QERSLESDGDEDTELEDEYAGSRVLIAPNGSVYSLGVPLLADFNESF-MPSFEPSTSEEA 737
SD D+ + GS+ + L + NE+ + +F+ S E+
Sbjct: 657 MMEGGCSDSDDYS-----ICGSQQQLKDT----------LGNGNENENLATFDTFDSIES 701
Query: 738 GPPSPK---PLVLDAEQLLERELSFIRNAKESGLVYLLGHGDIRARKDSWFIKKLVINYF 794
P + + ++ EL FI +++G+V+++G+ +RAR+++ F KK+ I+Y
Sbjct: 702 ITPVKRVSNTVTASSQMSGVDELEFINGCRDAGVVHIMGNTVVRARREARFYKKIAIDYV 761
Query: 795 YAFLRKNCRRGVTNLSVPHSHLMQVGMTY 823
YAFLRK CR +VP L+ VG +
Sbjct: 762 YAFLRKICREHSVIYNVPQESLLNVGQIF 790
>At4g19960 potassium transporter-like protein
Length = 842
Score = 538 bits (1387), Expect = e-153
Identities = 303/786 (38%), Positives = 454/786 (57%), Gaps = 70/786 (8%)
Query: 103 LAFQTLGVVFGDVGTSPLYTFSVMFRKAPINDNEDILGALSLVLYTLILIPFLKYVLVVL 162
L+FQ+LG+V+GD+GTSPLY F F I+D+ED++GALSL++Y+L+LIP +KYV +V
Sbjct: 60 LSFQSLGIVYGDLGTSPLYVFYNTFPDG-IDDSEDVIGALSLIIYSLLLIPLIKYVFIVC 118
Query: 163 WANDDGEGGTFALYSLICRNAKVNLLPNQLPSDARISGFRLKVPSAELERSLKLKERLES 222
AND+G+GGT A+YSL+CR+AKV L+PNQ SD ++ + V SAE + K K+ LE
Sbjct: 119 KANDNGQGGTLAIYSLLCRHAKVKLIPNQHRSDEDLTTYSRTV-SAEGSFAAKTKKWLEG 177
Query: 223 SFTLKKILLLLVLAGTSMVIANGVVTPAMSVLSS--VNGLKVGVDAIQ------QDEVVM 274
K+ LL++VL GT M+I +G++TPA+S VN K+ I D VV+
Sbjct: 178 KEWRKRALLVVVLLGTCMMIGDGILTPAISATGGIKVNNPKMSGGNIHFTLFSTSDIVVL 237
Query: 275 ISVACLVVLFSLQKYGTSKVGLAVGPALFIWFCSLAGNGVYNLVKYDSSVFRAFNPIHIY 334
+++ L+ LFS+Q YGT KVG P + IWF + G+YN+ KYD+SV +AF+P +IY
Sbjct: 238 VAIVILIGLFSMQHYGTDKVGWLFAPIVLIWFLFIGATGMYNICKYDTSVLKAFSPTYIY 297
Query: 335 YFFARNSTKAWYSLGGCLLCAT-----------GSEAMFADLCYFSVRSVQPIIGLFGSS 383
+F R W SLGG LL T G+EA++AD+ YF + ++Q F
Sbjct: 298 LYFKRRGRDGWISLGGILLSITAISKCYFLNIAGTEALYADIAYFPLLAIQLAFTFFVFP 357
Query: 384 CIPYG----------------------TPC*CW*SLFFFCS--KLIASRTMTTATFSCIK 419
C+ P + +F + ++ S+ + T+S +K
Sbjct: 358 CLLLAYCGQAAYLVIHKEHYQDAFYASIPDSVYWPMFIVATGAAIVGSQATISGTYSIVK 417
Query: 420 QSTALGCFPRLKIIHTSRKFMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAYG----- 474
Q+ A GCFPR+KI+HTS+KF+GQIY P INW L+ + + +IGNAYG
Sbjct: 418 QAVAHGCFPRVKIVHTSKKFLGQIYCPDINWILMLGCIAVTASFKKQSQIGNAYGKMTTT 477
Query: 475 -----------IAELGVMMMTTILVTLVMLLIWQMHIIIVMSFLGVFLGLELVFFSSVLW 523
A + VM++TT+L+ L+MLL+W H I+V+ F + +EL +FS+V++
Sbjct: 478 SKYKKNYFSQWTAVVLVMLVTTLLMVLIMLLVWHCHWILVLIFTFLSFFVELSYFSAVIF 537
Query: 524 SITDGSWIILVFAAIMFFIMFIWNYGSKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIG 583
I +G W+ L+ AAI +M +W+Y + KYE E+ K+S + LG +LG +R PGIG
Sbjct: 538 KIDEGGWVPLIIAAISLLVMSVWHYATVKKYEFEMHSKVSMSWILGLGPSLGLVRVPGIG 597
Query: 584 LLYNELVKGIPGIFGHFLTTLPAIHSMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLF 643
L+Y EL G+P IF HF+T LPAIHS+++FV +KY+PV VP+ ERFL +R+ +++ +F
Sbjct: 598 LVYTELASGVPHIFSHFITNLPAIHSVVVFVCVKYLPVYTVPEEERFLVKRIGPKTFRMF 657
Query: 644 RCIARYGYKDARKENHQAFEQLLMESLEKFIRREA-QERSLESDGDEDTELEDEYAGSRV 702
RC+ARYGYKD K++ FE L+ L FIR E E + S T + S V
Sbjct: 658 RCVARYGYKDLHKKDDD-FENKLLTKLSSFIRIETMMEPTSNSSTYSSTYSVNHTQDSTV 716
Query: 703 LIAPNGSVYSLGVPL-----LADFNESFMPSFEPSTSEEAGPPSPKPLVLDAEQLLEREL 757
+ N + ++ + + D+ S + + + S + ++ E+ EL
Sbjct: 717 DLIHNNNNHNHNNNMDMFSSMVDYTVSTLDTIVSAESLHNTVSFSQDNTVEEEE--TDEL 774
Query: 758 SFIRNAKESGLVYLLGHGDIRARKDSWFIKKLVINYFYAFLRKNCRRGVTNLSVPHSHLM 817
F++ KESG+V+++G+ ++AR SW KK+ I+Y YAFL K CR L VPH L+
Sbjct: 775 EFLKTCKESGVVHIMGNTVVKARTGSWLPKKIAIDYVYAFLAKICRANSVILHVPHETLL 834
Query: 818 QVGMTY 823
VG +
Sbjct: 835 NVGQVF 840
>At3g02050 putative potassium transporter
Length = 789
Score = 524 bits (1350), Expect = e-149
Identities = 293/771 (38%), Positives = 438/771 (56%), Gaps = 45/771 (5%)
Query: 95 MTTGKKIVLAFQTLGVVFGDVGTSPLYTF-SVMFRKAPINDNED-ILGALSLVLYTLILI 152
M ++LA+Q+ GVV+GD+ TSPLY F S K + NED + GA SL+ +TL LI
Sbjct: 20 MNLSSNLILAYQSFGVVYGDLSTSPLYVFPSTFIGKLHKHHNEDAVFGAFSLIFWTLTLI 79
Query: 153 PFLKYVLVVLWANDDGEGGTFALYSLICRNAKVNLLPNQLPSDARISGFRLKVPSAELER 212
P LKY+LV+L A+D+GEGGTFALYSL+CR+AK++LLPNQ +D +S ++ PS +
Sbjct: 80 PLLKYLLVLLSADDNGEGGTFALYSLLCRHAKLSLLPNQQAADEELSAYKFG-PSTDTVT 138
Query: 213 SLKLKERLESSFTLKKILLLLVLAGTSMVIANGVVTPAMSVLSSVNGLKVGVDAIQQDEV 272
S + LE L+ LLL+VL G +MVI +GV+TPA+SVLSS++GL+ + E+
Sbjct: 139 SSPFRTFLEKHKRLRTALLLVVLFGAAMVIGDGVLTPALSVLSSLSGLQATEKNVTDGEL 198
Query: 273 VMISVACLVVLFSLQKYGTSKVGLAVGPALFIWFCSLAGNGVYNLVKYDSSVFRAFNPIH 332
++++ LV LF+LQ GT +V P + IW S+ G+YN+++++ + A +P++
Sbjct: 199 LVLACVILVGLFALQHCGTHRVAFMFAPIVIIWLISIFFIGLYNIIRWNPKIIHAVSPLY 258
Query: 333 IYYFFARNSTKAWYSLGGCLLCATGSEAMFADLCYFSVRSVQPIIGLFGSSCIPY----- 387
I FF W SLGG LL TG+EAMFA+L +F+ S++ + C+
Sbjct: 259 IIKFFRVTGQDGWISLGGVLLSVTGTEAMFANLGHFTSVSIRVAFAVVVYPCLVVQYMGQ 318
Query: 388 -----------------GTPC*CW*SLFFFCS--KLIASRTMTTATFSCIKQSTALGCFP 428
P + +F + ++ S+ + T TFS IKQ ALGCFP
Sbjct: 319 AAFLSKNLGSIPNSFYDSVPDPVFWPVFVIATLAAIVGSQAVITTTFSIIKQCHALGCFP 378
Query: 429 RLKIIHTSRKFMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAYGIAELGVMMMTTILV 488
R+K++HTS+ GQIYIP INW L+ ++L IGNAYGIA + VM +TT +
Sbjct: 379 RIKVVHTSKHIYGQIYIPEINWILMILTLAMAIGFRDTTLIGNAYGIACMVVMFITTFFM 438
Query: 489 TLVMLLIWQMHIIIVMSFLGVFLGLELVFFSSVLWSITDGSWIILVFAAIMFFIMFIWNY 548
LV++++WQ + FLG +E V+ S+ L +T+G W+ V I M++W+Y
Sbjct: 439 ALVIVVVWQKSCFLAALFLGTLWIIEGVYLSAALMKVTEGGWVPFVLTFIFMIAMYVWHY 498
Query: 549 GSKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPAIH 608
G++ KY ++ K+S + LG +LG +R PGIGL+Y+EL G+P IF HF+T LPA H
Sbjct: 499 GTRRKYSFDLHNKVSLKWLLGLGPSLGIVRVPGIGLVYSELATGVPAIFSHFVTNLPAFH 558
Query: 609 SMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKENHQAFEQLLME 668
+++FV +K VPV V ERFL RVC + Y ++RCI RYGYKD ++E+ FE L++
Sbjct: 559 KVVVFVCVKSVPVPHVSPEERFLIGRVCPKPYRMYRCIVRYGYKDIQREDGD-FENQLVQ 617
Query: 669 SLEKFIRREAQE------RSLESDGDEDTELEDEYAGSRVLIAPNGSVYSLGVPLLADFN 722
S+ +FI+ EA + S +DG + + +L P +
Sbjct: 618 SIAEFIQMEASDLQSSASESQSNDGRMAVLSSQKSLSNSILTVSEVEEIDYADPTIQSSK 677
Query: 723 ESFMPSFEPSTSEEAGPPSPKPLVLDAEQL----------LERELSFIRNAKESGLVYLL 772
+ S S E+ P QL + EL + AKE+G+ Y++
Sbjct: 678 SMTLQSLR-SVYEDEYPQGQVRRRHVRFQLTASSGGMGSSVREELMDLIRAKEAGVAYIM 736
Query: 773 GHGDIRARKDSWFIKKLVINYFYAFLRKNCRRGVTNLSVPHSHLMQVGMTY 823
GH +++RK S ++KK+ I+ Y+FLRKNCR L++PH L++VGM Y
Sbjct: 737 GHSYVKSRKSSSWLKKMAIDIGYSFLRKNCRGPAVALNIPHISLIEVGMIY 787
>At5g14880 HAK8
Length = 781
Score = 507 bits (1306), Expect = e-144
Identities = 298/777 (38%), Positives = 447/777 (57%), Gaps = 70/777 (9%)
Query: 101 IVLAFQTLGVVFGDVGTSPLYTFSVMFRKAPIND--NEDILGALSLVLYTLILIPFLKYV 158
+ LA+Q+LGVV+GD+ TSPLY + F + + NE+I G LSL+ +TL LIP +KYV
Sbjct: 21 LTLAYQSLGVVYGDLATSPLYVYKSTFAEDITHSETNEEIFGVLSLIFWTLTLIPLVKYV 80
Query: 159 LVVLWANDDGEGGTFALYSLICRNAKVNLLPNQLPSDARISGF---------RLKVPSAE 209
+VL A+D+GEGGTFALYSL+CR+A+++ LPN +D +S + RLKVP
Sbjct: 81 FIVLRADDNGEGGTFALYSLLCRHARISSLPNFQLADEDLSEYKKNSGENPMRLKVPG-- 138
Query: 210 LERSLKLKERLESSFTLKKILLLLVLAGTSMVIANGVVTPAMSVLSSVNGLKVGVDAIQQ 269
LK LE L+ +LL+L L GT MVI +GV+TPA+SV S+V+GL++ + QQ
Sbjct: 139 ----WSLKNTLEKHKFLQNMLLVLALIGTCMVIGDGVLTPAISVFSAVSGLELSMSK-QQ 193
Query: 270 DEVVMISVAC--LVVLFSLQKYGTSKVGLAVGPALFIWFCSLAGNGVYNLVKYDSSVFRA 327
+ V + V C L++LFSLQ YGT ++G P + W ++ GVYN+ ++ V++A
Sbjct: 194 HQYVEVPVVCAILILLFSLQHYGTHRLGFVFAPIVLAWLLCISTIGVYNIFHWNPHVYKA 253
Query: 328 FNPIHIYYFFARNSTKAWYSLGGCLLCATGSEAMFADLCYFSVRSVQPII--GLFGSSCI 385
+P +IY F + + W SLGG LLC TGSEAMFADL +F+ S+Q ++ S +
Sbjct: 254 LSPYYIYKFLKKTRKRGWMSLGGILLCITGSEAMFADLGHFTQLSIQIAFTFAVYPSLIL 313
Query: 386 PYGTPC*C------------------------W*SLFF-FCSKLIASRTMTTATFSCIKQ 420
Y W L + ++ S+ + T TFS IKQ
Sbjct: 314 AYMGQAAYLSKHHVLQSDYRIGFYVSVPEQIRWPVLAIAILAAVVGSQAIITGTFSIIKQ 373
Query: 421 STALGCFPRLKIIHTSRKFMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAYGIAELGV 480
T+LGCFP++KI+HTS + GQIYIP INW L+ + L I NA G+A + V
Sbjct: 374 CTSLGCFPKVKIVHTSSRMHGQIYIPEINWTLMLLCLAVTVGFRDTKHISNASGLAVITV 433
Query: 481 MMMTTILVTLVMLLIWQMHIIIVMSFLGVFLGLELVFFSSVLWSITDGSWIILVFAAIMF 540
M++TT L++LV++L W+ + ++F+ F +E+++FS+ L +G+W+ + + I
Sbjct: 434 MLVTTCLMSLVIVLCWRKSSLYALAFIFFFGTIEVLYFSASLIKFLEGAWVPVALSFIFL 493
Query: 541 FIMFIWNYGSKLKYETEVKQKLSPDLMREL--GCNLGTIRAPGIGLLYNELVKGIPGIFG 598
IM++W+YG+ +YE +V+ K+S + + L NLG +R GIG++ ELV GIP IF
Sbjct: 494 LIMYVWHYGTLKRYEFDVQNKVSINWLLTLFGSSNLGIVRVHGIGVINTELVSGIPAIFS 553
Query: 599 HFLTTLPAIHSMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKEN 658
HF+T LPA H +++F+ +K VPV V ERFL RV + Y L+RCIARYGY+D K++
Sbjct: 554 HFITNLPAFHQVVVFLCVKSVPVPHVKPEERFLVGRVGPKEYRLYRCIARYGYRDVHKDD 613
Query: 659 HQAFEQLLMESLEKFIRREAQERSLESDGDEDTELEDEYAGSRVLIAPNGSVYSLGVPLL 718
+ FE L+ S+ +FIR ++ L D + E R+ + S GV +
Sbjct: 614 VE-FENDLICSIAEFIR---SDKPLNYSPDPENE---SGINERLTVVAASSSNLEGVQIY 666
Query: 719 ADFNESFMPSFEPSTSEEA--GPPSPK---------PLVLDAEQLLERELSFIRNAKESG 767
D EPS+S E PSP+ P ++ E EL+ + A+E+G
Sbjct: 667 EDDGSD---KQEPSSSSEVIMVAPSPRFKKRVRFVLPESARIDRSAEEELTELTEAREAG 723
Query: 768 LVYLLGHGDIRARKDSWFIKKLVINYFYAFLRKNCRRGVTNLSVPHSHLMQVGMTYM 824
+ +++GH +RA+ S +KK+ IN+ Y FLR+N R LS PH+ ++VGM Y+
Sbjct: 724 MAFIMGHSYVRAKSGSSVMKKIAINFGYDFLRRNSRGPCYGLSTPHASTLEVGMVYI 780
>At2g40540 putative potassium transporter AtKT2p (AtKT2)
Length = 794
Score = 503 bits (1296), Expect = e-142
Identities = 291/782 (37%), Positives = 450/782 (57%), Gaps = 69/782 (8%)
Query: 101 IVLAFQTLGVVFGDVGTSPLYTFSVMFRK--APINDNEDILGALSLVLYTLILIPFLKYV 158
++LA+Q+LGVV+GD+ SPLY F F + NE+I G +S V +TL L+P LKYV
Sbjct: 23 LLLAYQSLGVVYGDLSISPLYVFKSTFAEDIQHSETNEEIYGVMSFVFWTLTLVPLLKYV 82
Query: 159 LVVLWANDDGEGGTFALYSLICRNAKVNLLPNQLPSDARISGFRLKVPSAELERSLKLKE 218
+VL A+D+GEGGTFALYSLICR+ KV+LLPN+ SD +S ++L+ P + S +K
Sbjct: 83 FIVLRADDNGEGGTFALYSLICRHVKVSLLPNRQVSDEALSTYKLEHPPEKNHDSC-VKR 141
Query: 219 RLESSFTLKKILLLLVLAGTSMVIANGVVTPAMSVLSSVNGLKVGVDAIQQDEVVMISVA 278
LE L LLLLVL GT MVI +G++TPA+SV S+V+GL++ + + + +I +
Sbjct: 142 YLEKHKWLHTALLLLVLLGTCMVIGDGLLTPAISVFSAVSGLELNMSK-EHHQYAVIPIT 200
Query: 279 C--LVVLFSLQKYGTSKVGLAVGPALFIWFCSLAGNGVYNLVKYDSSVFRAFNPIHIYYF 336
C LV LFSLQ +GT +VG P + W ++G G+YN+++++ +++A +P +++ F
Sbjct: 201 CFILVCLFSLQHFGTHRVGFVFAPIVLTWLLCISGIGLYNIIQWNPHIYKALSPTYMFMF 260
Query: 337 FARNSTKAWYSLGGCLLCATGSEAMFADLCYFSVRSVQP----------IIGLFGSSC-- 384
+ W SLGG LLC TG+EAMFADL +F+ ++Q I+ G +
Sbjct: 261 LRKTRVSGWMSLGGILLCITGAEAMFADLGHFNYAAIQIAFTFLVYPALILAYMGQAAYL 320
Query: 385 ---------------IPYGTPC*CW*SLFF-FCSKLIASRTMTTATFSCIKQSTALGCFP 428
+P C W L + ++ S+ + + TFS I QS +LGCFP
Sbjct: 321 SRHHHSAHAIGFYVSVP---KCLHWPVLAVAILASVVGSQAIISGTFSIINQSQSLGCFP 377
Query: 429 RLKIIHTSRKFMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAYGIAELGVMMMTTILV 488
R+K+IHTS K GQIYIP INW L+ + + + +GNA G+A + VM++TT L
Sbjct: 378 RVKVIHTSDKMHGQIYIPEINWMLMILCIAVTIGFRDVKHLGNASGLAVMAVMLVTTCLT 437
Query: 489 TLVMLLIWQMHIIIVMSFLGVFLGLELVFFSSVLWSITDGSWIILVFAAIMFFIMFIWNY 548
+LV++L W I+ ++FL F +EL++FS+ L +G+W+ ++ + I IMF+W+Y
Sbjct: 438 SLVIVLCWHKPPILALAFLLFFGSIELLYFSASLTKFREGAWLPILLSLIFMIIMFVWHY 497
Query: 549 GSKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPAIH 608
+ KYE +++ K+S + + LG +LG R PGIGL++ +L GIP F F+T LPA H
Sbjct: 498 TTIKKYEFDLQNKVSLEWLLALGPSLGISRVPGIGLVFTDLTSGIPANFSRFVTNLPAFH 557
Query: 609 SMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKENHQAFEQLLME 668
+++FV +K VPV VP +ER+L RV + +RCI RYGY+D ++ +FE L+
Sbjct: 558 RVLVFVCVKSVPVPFVPPAERYLVGRVGPVDHRSYRCIVRYGYRDVHQDV-DSFETELVS 616
Query: 669 SLEKFIRREAQERSLESDGDEDTELEDEYAGSRVLIAPNGSV------------YSLG-- 714
L FIR + +R+ + D D ++ + S +A G+V S+G
Sbjct: 617 KLADFIRYDWHKRTQQED-DNARSVQSNESSSESRLAVIGTVAYEIEDNLQPESVSIGFS 675
Query: 715 -VPLLADFNESFMPS-----------FEPSTSEEAGPPSPKPLVLDAEQLLERELSFIRN 762
V + D + P+ E ++ E+ G S +A+ L EL +
Sbjct: 676 TVESMEDVIQMAEPAPTATIRRVRFAVEENSYEDEGSTSSA----EADAELRSELRDLLA 731
Query: 763 AKESGLVYLLGHGDIRARKDSWFIKKLVINYFYAFLRKNCRRGVTNLSVPHSHLMQVGMT 822
A+E+G ++LGH ++A++ S +K+L +N+ Y FLR+NCR L VP L++VGM
Sbjct: 732 AQEAGTAFILGHSHVKAKQGSSVMKRLAVNFGYNFLRRNCRGPDVALKVPPVSLLEVGMV 791
Query: 823 YM 824
Y+
Sbjct: 792 YV 793
>At2g30070 high affinity K+ transporter (AtKUP1/AtKT1p)
Length = 712
Score = 496 bits (1276), Expect = e-140
Identities = 273/764 (35%), Positives = 422/764 (54%), Gaps = 90/764 (11%)
Query: 86 GAHTHHYEDMTTGKKIVLAFQTLGVVFGDVGTSPLYTFSVMF--RKAPINDNEDILGALS 143
G H + ++ + LA+Q+LGV++GD+ TSPLY + F + + D+E+I G S
Sbjct: 11 GISQQHLKTLSCANVLTLAYQSLGVIYGDLSTSPLYVYKTTFSGKLSLHEDDEEIFGVFS 70
Query: 144 LVLYTLILIPFLKYVLVVLWANDDGEGGTFALYSLICRNAKVNLLPNQLPSDARISGFRL 203
+ +T LI KYV +VL A+D+GEGGTFALYSL+CR AK+++LPN D ++S +
Sbjct: 71 FIFWTFTLIALFKYVFIVLSADDNGEGGTFALYSLLCRYAKLSILPNHQEMDEKLSTYAT 130
Query: 204 KVPSAELERSLKLKERLESSFTLKKILLLLVLAGTSMVIANGVVTPAMSVLSSVNGLKVG 263
P E +S +K E +K LLL VL GT M I + V+TP +SVLS+V+G+K+
Sbjct: 131 GSPG-ETRQSAAVKSFFEKHPKSQKCLLLFVLLGTCMAIGDSVLTPTISVLSAVSGVKLK 189
Query: 264 VDAIQQDEVVMISVACLVVLFSLQKYGTSKVGLAVGPALFIWFCSLAGNGVYNLVKYDSS 323
+ + ++ VV+I+ LV +FS+Q+YGT +V P W S++ GVYN +K++
Sbjct: 190 IPNLHENYVVIIACIILVAIFSVQRYGTHRVAFIFAPISTAWLLSISSIGVYNTIKWNPR 249
Query: 324 VFRAFNPIHIYYFFARNSTKAWYSLGGCLLCATGSEAMFADLCYFSVRSVQPIIGLFGSS 383
+ A +P+++Y F + W SLGG +L TG E MFADL +FS S++ F
Sbjct: 250 IVSALSPVYMYKFLRSTGVEGWVSLGGVVLSITGVETMFADLGHFSSLSIKVAFSFFVYP 309
Query: 384 CIPYG----------------------TPC*CW*SLFFFCS--KLIASRTMTTATFSCIK 419
C+ P + +F + ++ S+ + +ATFS I
Sbjct: 310 CLILAYMGEAAFLSKHHEDIQQSFYKAIPEPVFWPVFIVATFAAVVGSQAVISATFSIIS 369
Query: 420 QSTALGCFPRLKIIHTSRKFMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAYGIAELG 479
Q AL CFPR+KIIHTS K GQIYIP +NW L+ + L + + +G+AYG+A
Sbjct: 370 QCCALDCFPRVKIIHTSSKIHGQIYIPEVNWMLMCLCLAVTIGLRDTNMMGHAYGLAVTS 429
Query: 480 VMMMTTILVTLVMLLIWQMHIIIVMSFLGVFLGLELVFFSSVLWSITDGSWIILVFAAIM 539
VM++TT L+TLVM ++W+ II V++F+ F +EL++FSS ++ + +G WI ++ +
Sbjct: 430 VMLVTTCLMTLVMTIVWKQRIITVLAFVVFFGSIELLYFSSCVYKVPEGGWIPILLSLTF 489
Query: 540 FFIMFIWNYGSKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGH 599
+M+IWNYG+ K+E +V+ K+S D + LG ++G +R PGIGL+Y+ LV G+P +FGH
Sbjct: 490 MAVMYIWNYGTTKKHEFDVENKVSMDRIVSLGPSIGMVRVPGIGLVYSNLVTGVPAVFGH 549
Query: 600 FLTTLPAIHSMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKENH 659
F+T LPA H +++FV +K V V V + ERF+ RV + Y +FR + RYGY+D +E +
Sbjct: 550 FVTNLPAFHKILVFVCVKSVQVPYVGEEERFVISRVGPKEYGMFRSVVRYGYRDVPREMY 609
Query: 660 QAFEQLLMESLEKFIRREAQERSLESDGDEDTELEDEYAGSRVLIAPNGSVYSLGVPLLA 719
FE L+ + +
Sbjct: 610 D-FESRLVSA------------------------------------------------IV 620
Query: 720 DFNESFMPSFEPSTSEEAGPPSPKPLVLDAEQLLERELSFIRNAKESGLVYLLGHGDIRA 779
+F E+ EP EE + + + + E I AKE+G+ Y+LGH +A
Sbjct: 621 EFVET-----EPGLEEEE---------MSSVRRKKEECMEIMEAKEAGVAYILGHSYAKA 666
Query: 780 RKDSWFIKKLVINYFYAFLRKNCRRGVTNLSVPHSHLMQVGMTY 823
++ S +KKL +N +AF+ NCR L+VPH+ L++VGM Y
Sbjct: 667 KQSSSVLKKLAVNVVFAFMSTNCRGTDVVLNVPHTSLLEVGMVY 710
>At1g70300 high affinity potassium transporter AtHAK6 (At1g70300)
Length = 782
Score = 490 bits (1262), Expect = e-138
Identities = 281/778 (36%), Positives = 442/778 (56%), Gaps = 72/778 (9%)
Query: 101 IVLAFQTLGVVFGDVGTSPLYTFSVMFRKA--PINDNEDILGALSLVLYTLILIPFLKYV 158
+ LA+Q+LGVV+GD+ SPLY + F + NE+I G LS + +T+ L+P LKYV
Sbjct: 20 LTLAYQSLGVVYGDLSISPLYVYKSTFAEDIHHSESNEEIFGVLSFIFWTITLVPLLKYV 79
Query: 159 LVVLWANDDGEGGTFALYSLICRNAKVNLLPNQLPSDARISGFRL-KVPSAELERS---L 214
+VL A+D+GEGGTFALYSL+CR+A+VN LP+ +D ++ ++ + S+ + +S
Sbjct: 80 FIVLRADDNGEGGTFALYSLLCRHARVNSLPSCQLADEQLIEYKTDSIGSSSMPQSGFAA 139
Query: 215 KLKERLESSFTLKKILLLLVLAGTSMVIANGVVTPAMSVLSSVNGLKVGVDAIQQDEVVM 274
LK LE L+KILL+L L GT MVI +GV+TPA+SV S+V+G+++ + + + +
Sbjct: 140 SLKSTLEKHGVLQKILLVLALIGTCMVIGDGVLTPAISVFSAVSGVELSMSK-EHHKYIE 198
Query: 275 ISVACLVV--LFSLQKYGTSKVGLAVGPALFIWFCSLAGNGVYNLVKYDSSVFRAFNPIH 332
+ AC+++ LF+LQ YGT +VG P + +W ++ GVYN+ ++ V++A +P +
Sbjct: 199 LPAACVILIGLFALQHYGTHRVGFLFAPVILLWLMCISAIGVYNIFHWNPHVYQALSPYY 258
Query: 333 IYYFFARNSTKAWYSLGGCLLCATGSEAMFADLCYFSVRSVQP----------IIGLFGS 382
+Y F + ++ W SLGG LLC TGSEAMFADL +FS S++ I+ G
Sbjct: 259 MYKFLKKTQSRGWMSLGGILLCITGSEAMFADLGHFSQLSIKIAFTSLVYPSLILAYMGQ 318
Query: 383 SC-------------------IPYGTPC*CW*SLFF-FCSKLIASRTMTTATFSCIKQST 422
+ +P W L + ++ S+ + T TFS IKQ +
Sbjct: 319 AAYLSQHHIIESEYNIGFYVSVPERLR---WPVLVIAILAAVVGSQAIITGTFSIIKQCS 375
Query: 423 ALGCFPRLKIIHTSRKFMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAYGIAELGVMM 482
ALGCFP++KI+HTS K GQIYIP INW L+ + L +GNA G+A + VM+
Sbjct: 376 ALGCFPKVKIVHTSSKIHGQIYIPEINWILMVLCLAVTIGFRDTKRLGNASGLAVITVML 435
Query: 483 MTTILVTLVMLLIWQMHIIIVMSFLGVFLGLELVFFSSVLWSITDGSWIILVFAAIMFFI 542
+TT L++LV++L W +I + F+ F +E ++FS+ L +G+W+ + A
Sbjct: 436 VTTCLMSLVIVLCWHKSVIFAIVFVVFFGTIESLYFSASLIKFLEGAWVPIALAFCFLLA 495
Query: 543 MFIWNYGSKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLT 602
M W+YG+ +YE +V+ K+S + + L LG R G+GL++ ELV G+P IF HF+T
Sbjct: 496 MCTWHYGTLKRYEYDVQNKVSVNWLLSLSQTLGIARVRGLGLIHTELVSGVPAIFSHFVT 555
Query: 603 TLPAIHSMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKENHQAF 662
LPA H +++F+ +K VPV V ERFL R+ + + ++RCI R+GY+D K++ + F
Sbjct: 556 NLPAFHQVLVFLCVKSVPVPHVRPQERFLVGRIGPKEFRIYRCIVRFGYRDVHKDDFE-F 614
Query: 663 EQLLMESLEKFIRREAQ--ERSLESDGDEDTELEDEYAGSRVLIAPNGSVYSLGVPLLAD 720
E L+ S+ +FIR EA+ + E++G++D R+ + S Y G+ D
Sbjct: 615 EGDLVCSIAEFIRTEAETAATAAETNGEDD---------DRMSVVGTCSTYMQGI---ED 662
Query: 721 FNESFMPSFEPSTSEEAGPPSPK---------------PLVLDAEQLLERELSFIRNAKE 765
ES + + + E P PK P E+ +EL + A+E
Sbjct: 663 HYESDIDDPDKPGTSEIRSPKPKKKSKSKVKKRVRFVVPETPKIEKETRQELMELTEARE 722
Query: 766 SGLVYLLGHGDIRARKDSWFIKKLVINYFYAFLRKNCRRGVTNLSVPHSHLMQVGMTY 823
G+ Y++G+ ++A+ S +K+L IN Y FLR+N R L+ PH+ ++VGM Y
Sbjct: 723 GGVAYIMGNAYMKAKPGSGLLKRLAINIGYEFLRRNTRGPRNMLTSPHASTLEVGMIY 780
>At4g23640 potassium transport like protein
Length = 775
Score = 487 bits (1253), Expect = e-137
Identities = 287/781 (36%), Positives = 432/781 (54%), Gaps = 77/781 (9%)
Query: 101 IVLAFQTLGVVFGDVGTSPLYTFSVMFRKA-PINDNED-ILGALSLVLYTLILIPFLKYV 158
++LA+Q+ G+VFGD+ SPLY + F + ED I GA SL+ +T+ L+ +KY+
Sbjct: 12 LLLAYQSFGLVFGDLSISPLYVYKCTFYGGLRHHQTEDTIFGAFSLIFWTITLLSLIKYM 71
Query: 159 LVVLWANDDGEGGTFALYSLICRNAKVNLLPNQLPSDARISGFRLKVPSAELERSLKLKE 218
+ VL A+D+GEGG FALY+L+CR+A+ +LLPNQ +D IS + ++ S K
Sbjct: 72 VFVLSADDNGEGGIFALYALLCRHARFSLLPNQQAADEEISTYYGPGDASRNLPSSAFKS 131
Query: 219 RLESSFTLKKILLLLVLAGTSMVIANGVVTPAMSVLSSVNGLKVGVDAIQQDEVVMISVA 278
+E + K LL+LVL GTSMVI GV+TPA+SV SS++GL V +++ VVMI+ A
Sbjct: 132 LIERNKRSKTALLVLVLVGTSMVITIGVLTPAISVSSSIDGL-VAKTSLKHSTVVMIACA 190
Query: 279 CLVVLFSLQKYGTSKVGLAVGPALFIWFCSLAGNGVYNLVKYDSSVFRAFNPIHIYYFFA 338
LV LF LQ GT+KV P + +W +A GVYN+V ++ SV++A +P +IY FF
Sbjct: 191 LLVGLFVLQHRGTNKVAFLFAPIMILWLLIIATAGVYNIVTWNPSVYKALSPYYIYVFFR 250
Query: 339 RNSTKAWYSLGGCLLCATGSEAMFADLCYFSVRSVQPIIGLFGSSCIPYGTPC*C----- 393
W SLGG LLC TG+EA+FA+L F+ S++ F C+ Y PC
Sbjct: 251 DTGIDGWLSLGGILLCITGTEAIFAELGQFTATSIR-----FAFCCVVY--PCLVLQYMG 303
Query: 394 -------------------------W*SLFF-FCSKLIASRTMTTATFSCIKQSTALGCF 427
W L + ++AS+ + ATFS +KQ ALGCF
Sbjct: 304 QAAFLSKNFSALPSSFYSSIPDPFFWPVLMMAMLAAMVASQAVIFATFSIVKQCYALGCF 363
Query: 428 PRLKIIHTSRKFMGQIYIPVINWFLLAVSLVFVCTISSIDEIGNAYGIAELGVMMMTTIL 487
PR+KI+H R +GQIYIP INW ++ ++L I A+G+A + + +TT L
Sbjct: 364 PRVKIVHKPRWVLGQIYIPEINWVVMILTLAVTICFRDTRHIAFAFGLACMTLAFVTTWL 423
Query: 488 VTLVMLLIWQMHIIIVMSFLGVFLGLELVFFSSVLWSITDGSWIILVFAAIMFFIMFIWN 547
+ L++ +W +I+ + F+ F +EL+F +S L I G WI L+ + FI ++W+
Sbjct: 424 MPLIINFVWNRNIVFSVLFILFFGTIELIFVASALVKIPKGGWITLLLSLFFTFITYVWH 483
Query: 548 YGSKLKYETEVKQKLSPDLMRELGCNLGTIRAPGIGLLYNELVKGIPGIFGHFLTTLPAI 607
YGS+ KY + K+ + LG +LG I+ PG+GL+Y EL G+P F HFLT LPA
Sbjct: 484 YGSRKKYLCDQHNKVPMKSILSLGPSLGIIKVPGMGLIYTELASGVPATFKHFLTNLPAF 543
Query: 608 HSMIIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKENHQAFEQLLM 667
+ +++FV K VP+ VPQ ER+L R+ ++Y ++RCI R GYKD K+ FE L+
Sbjct: 544 YQVVVFVCCKTVPIPYVPQKERYLIGRIGPKTYRMYRCIIRAGYKDVNKDGDD-FEDELV 602
Query: 668 ESLEKFIRREAQERSLESDGDEDTELEDEYAGSRVLIAPNGSVYSLGVPLLADFNESFMP 727
S+ +FI+ LES+G + + G ++ + G L +E+ +
Sbjct: 603 MSIAEFIQ-------LESEGYGGSNTDRSIDGRLAVVKASN---KFGTRLSRSISEANIA 652
Query: 728 SFEPSTSEEAGPPSPKPLVLDAE-------------------------QLLERELSFIRN 762
S + SP L L AE ++ EL + N
Sbjct: 653 GSSRSQTTVTNSKSPALLKLRAEYEQELPRLSMRRMFQFRPMDTKFRQPQVKEELFDLVN 712
Query: 763 AKESGLVYLLGHGDIRARKDSWFIKKLVINYFYAFLRKNCRRGVTNLSVPHSHLMQVGMT 822
AK++ + Y++GHG ++A+++S F+K+LV+N Y+FLRKNCR L++PH L++VGM
Sbjct: 713 AKDAEVAYIVGHGHVKAKRNSVFVKQLVVNVAYSFLRKNCRSPGVMLNIPHICLIKVGMN 772
Query: 823 Y 823
Y
Sbjct: 773 Y 773
>At5g66840 unknown protein
Length = 551
Score = 35.0 bits (79), Expect = 0.17
Identities = 21/75 (28%), Positives = 38/75 (50%), Gaps = 4/75 (5%)
Query: 6 NNNSMTEEEDDPGSALSFDSTESRWVFQEDEDPSEIEDYDASDMRHQSMFDSEDEDNAEM 65
NN TEEE+D + S D ++ + D+D + D +A ++ + D DED+
Sbjct: 26 NNYDETEEEEDEDTNSSEDDSD----WSHDDDDATESDVEADEIGVKGDNDDGDEDDKVT 81
Query: 66 KLIRTGPRIDSFDVE 80
+L+ G + S +V+
Sbjct: 82 RLLTAGSDLKSVNVK 96
>At3g03050 putative cellulose synthase catalytic subunit
Length = 1145
Score = 33.1 bits (74), Expect = 0.65
Identities = 21/54 (38%), Positives = 26/54 (47%), Gaps = 1/54 (1%)
Query: 642 LFRCIARYGYKDAR-KENHQAFEQLLMESLEKFIRREAQERSLESDGDEDTELE 694
LFR IA YG+ R KE+H F +K R + RSL GD D + E
Sbjct: 707 LFRRIALYGFDPPRAKEHHPGFCSCCFSRKKKKSRVPEENRSLRMGGDSDDDEE 760
>At3g03710 putative polynucleotide phosphorylase
Length = 948
Score = 32.0 bits (71), Expect = 1.4
Identities = 42/163 (25%), Positives = 68/163 (40%), Gaps = 25/163 (15%)
Query: 171 GTFALYSLICRNAKVNLLPNQLPSDARISGFRLKVPSAELERSLKLKERLESSFTLKKIL 230
G A +L +A + + +++P+ I+G R+ + E + +KE ES
Sbjct: 231 GLHAPDALAVTSAGIAVALSEVPNAKAIAGVRVGLIGGEFIVNPTVKEMEESQ------- 283
Query: 231 LLLVLAGTSMVIAN----GVVTPAMSVLSSVNGLKVGVDAIQQDEVVMISVACLVVLFSL 286
L L LAGT I P +L +V KVG DA+Q C+ +
Sbjct: 284 LDLFLAGTDTAILTIEGYSNFLPEEMLLQAV---KVGQDAVQ--------ATCIAIEVLA 332
Query: 287 QKYGTSKVGLAV---GPALFIWFCSLAGNGVYNLVKYDSSVFR 326
+KYG K+ A+ P L+ LAG + ++ S + R
Sbjct: 333 KKYGKPKMLDAIRLPPPELYKHVKELAGEELTKALQIKSKISR 375
>At5g50310 unknown protein
Length = 666
Score = 31.6 bits (70), Expect = 1.9
Identities = 22/64 (34%), Positives = 33/64 (51%), Gaps = 15/64 (23%)
Query: 25 STESRWV-FQEDEDPSEIEDYDASDMRHQSMFDSEDEDNAEMKLIRTGPRIDSFDVEALE 83
+TE+ WV +DE+ E +D D DSEDE N+E + D +VEA++
Sbjct: 490 TTETEWVEVSDDEEGDEDDDED----------DSEDEGNSE----ESDDEDDDEEVEAMD 535
Query: 84 VPGA 87
V G+
Sbjct: 536 VDGS 539
>At5g25340 unknown protein
Length = 208
Score = 31.2 bits (69), Expect = 2.5
Identities = 17/48 (35%), Positives = 25/48 (51%), Gaps = 3/48 (6%)
Query: 645 CIARYGYKDA---RKENHQAFEQLLMESLEKFIRREAQERSLESDGDE 689
CI YG KD R +NH + +L + + +++ ER L DGDE
Sbjct: 87 CIGNYGMKDGDEVRFKNHVSGNAVLSKGYSRKSKQKNLERVLPKDGDE 134
>At3g08930 unknown protein
Length = 526
Score = 31.2 bits (69), Expect = 2.5
Identities = 38/158 (24%), Positives = 60/158 (37%), Gaps = 22/158 (13%)
Query: 467 DEIGNAYGIAELGVMMMTTI-LVTLVMLLIWQMHIIIVMSFLGVFLGLELVFFSSVLWSI 525
++ A+ LG + + +V L++ + W HIII + + L F + ++
Sbjct: 332 EQAETAWAFTVLGYLAKFILGIVGLIVSIAWVAHIIIYL-----LVDPPLSPFLNEVFIK 386
Query: 526 TDGSWIILVFAAIMFFIMFI--------WNYGSKLKYETEVKQKLSPDLMRELGCNLGTI 577
D W +L AA FF ++ G KL + T K LM N+G I
Sbjct: 387 LDDVWGLLGTAAFAFFCFYLLLAVIAGAMMLGLKLVFITIHPMKWGATLMNSFLFNVGLI 446
Query: 578 RAPGIGLL--------YNELVKGIPGIFGHFLTTLPAI 607
I ++ Y IFGH L +L I
Sbjct: 447 LLCSISVIQFCATAFGYYAQATAAQEIFGHTLQSLRGI 484
>At2g19490 putative recA protein
Length = 376
Score = 31.2 bits (69), Expect = 2.5
Identities = 25/99 (25%), Positives = 49/99 (49%), Gaps = 17/99 (17%)
Query: 611 IIFVSIKYVPVAMVPQSERFLFRRVCQRSYHLFRCIARYGYKDARKENHQAFEQLLMESL 670
II +SIK+ +A + F + ++YH + R+ K+N E+L+ +
Sbjct: 276 IIDLSIKHKFIA-----KNGTFYNLNGKNYHGKEALKRF-----LKQNESDQEELMKKLQ 325
Query: 671 EKFIRREAQERSLESDGDEDTELEDEYAGSRVLIAPNGS 709
+K I EA ++ ES+ +E+ L RV+++P+ +
Sbjct: 326 DKLIADEAADKETESESEEEDSL-------RVVVSPDNT 357
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.141 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,135,379
Number of Sequences: 26719
Number of extensions: 790838
Number of successful extensions: 3445
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 3330
Number of HSP's gapped (non-prelim): 87
length of query: 824
length of database: 11,318,596
effective HSP length: 108
effective length of query: 716
effective length of database: 8,432,944
effective search space: 6037987904
effective search space used: 6037987904
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144459.2


