

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144431.4 - phase: 0 /pseudo
(1510 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
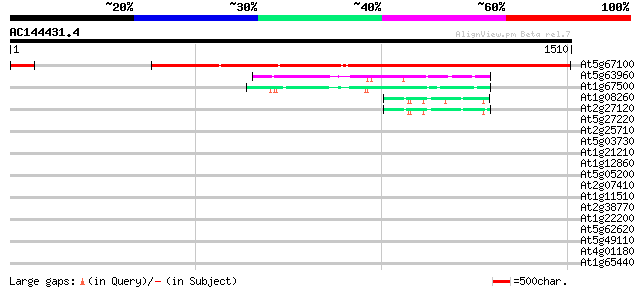
Score E
Sequences producing significant alignments: (bits) Value
At5g67100 DNA polymerase alpha 1 1545 0.0
At5g63960 DNA polymerase III catalytic subunit 199 8e-51
At1g67500 putative DNA polymerase zeta catalytic subunit 122 2e-27
At1g08260 hypothetical protein 45 2e-04
At2g27120 putative DNA polymerase epsilon catalytic subunit 44 7e-04
At5g27220 putative protein 35 0.43
At2g25710 biotin holocarboxylase synthetase 33 0.96
At5g03730 SERINE/THREONINE-PROTEIN KINASE CTR1 33 1.3
At1g21210 hypothetical protein 33 1.6
At1g12860 bHLH transcription factor (bHLH033) 33 1.6
At5g05200 unknown protein 32 2.1
At2g07410 putative retroelement pol polyprotein 32 2.8
At1g11510 hypthetical protein 32 3.6
At2g38770 unknown protein 31 4.8
At1g22200 unknown protein 31 4.8
At5g62620 unknown protein 31 6.2
At5g49110 unknown protein 31 6.2
At4g01180 hypothetical protein 30 8.1
At1g65440 30 8.1
>At5g67100 DNA polymerase alpha 1
Length = 1492
Score = 1545 bits (3999), Expect = 0.0
Identities = 771/1131 (68%), Positives = 925/1131 (81%), Gaps = 11/1131 (0%)
Query: 382 VWQGTLYQSCCVVVKNMQRCVYAIPSHPLHSTNEMIQLEKDVQESRISPADFRKKLQDAV 441
V G Y+SCCVVVKN+QRCVYAIP+ + ++E+I LE++V++SR+SP FR KL +
Sbjct: 370 VKMGDTYKSCCVVVKNIQRCVYAIPNDSIFPSHELIMLEQEVKDSRLSPESFRGKLHEMA 429
Query: 442 SDTKNEIAKHLVDLGVSSFSMAPVKRKYAFERTDIPAGENYVVKINYLFKDTALPVDLKG 501
S KNEIA+ L+ L VS+FSMAPVKR YAFER D+PAGE YV+KINY FKD LP DLKG
Sbjct: 430 SKLKNEIAQELLQLNVSNFSMAPVKRNYAFERPDVPAGEQYVLKINYSFKDRPLPEDLKG 489
Query: 502 KSFCALLGARNSALELFLIKRKIKGPSWLQVSNFSTCSASQRVSWCKFEVIVDSPKDIRA 561
+SF ALLG+ SALE F++KRKI GP WL++S+FSTCS S+ VSWCKFEV V SPKDI
Sbjct: 490 ESFSALLGSHTSALEHFILKRKIMGPCWLKISSFSTCSPSEGVSWCKFEVTVQSPKDITI 549
Query: 562 SSPSSSKITLVNPPVVVTAINLKTTINEKQNINEIVSASVVSCNMVKIDTPMLASEWKRP 621
S+ +V+PP VVTAINLKT +NEKQNI+EIVSASV+ + KID PM A E KR
Sbjct: 550 LV---SEEKVVHPPAVVTAINLKTIVNEKQNISEIVSASVLCFHNAKIDVPMPAPERKRS 606
Query: 622 GMLTHFTVIRKLDGNIFPMGFNTEVTDRNIKAGSNVLCVESSERALLNRLMLQLHKMDSD 681
G+L+HFTV+R +G +P+G+ EV+DRN K G NVL +E+SERALLNRL L+L+K+DSD
Sbjct: 607 GILSHFTVVRNPEGTGYPIGWKKEVSDRNSKNGCNVLSIENSERALLNRLFLELNKLDSD 666
Query: 682 VLVGHNISGFDLDVLLHRSQACRVPSSMWSKLGRLNRSTMPKLDRRGKTFGFGADPAIMS 741
+LVGHNISGFDLDVLL R+QAC+V SSMWSK+GRL RS MPKL + +G GA P +MS
Sbjct: 667 ILVGHNISGFDLDVLLQRAQACKVQSSMWSKIGRLKRSFMPKL-KGNSNYGSGATPGLMS 725
Query: 742 CVAGRLLCDTYLCSRDLLKEVSYSLTHLAKTQLNQSRKEVAPHEVPKMFQTAKSLMELIE 801
C+AGRLLCDT LCSRDLLKEVSYSLT L+KTQLN+ RKE+AP+++PKMFQ++K+L+ELIE
Sbjct: 726 CIAGRLLCDTDLCSRDLLKEVSYSLTDLSKTQLNRDRKEIAPNDIPKMFQSSKTLVELIE 785
Query: 802 YGETDAWLSMELMFYLSVLPLTRQLTNLSGNLWGKTLQGARAQRVEYLLLHEFHKKKYIV 861
GETDAWLSMELMF+LSVLPLT QLTN+SGNLWGKTLQGARAQR+EY LLH FH KK+I+
Sbjct: 786 CGETDAWLSMELMFHLSVLPLTLQLTNISGNLWGKTLQGARAQRIEYYLLHTFHSKKFIL 845
Query: 862 PDKFSNYAKETKLTKRRVTHGVDDGNFDDADINDANYHNDASESDHKKNKKAASYAGGLV 921
PDK S KE K +KRR+ + +D N D+ D D ND S+ K KK +YAGGLV
Sbjct: 846 PDKISQRMKEIKSSKRRMDYAPEDRNVDELDA-DLTLENDPSKGS--KTKKGPAYAGGLV 902
Query: 922 LEPKKGLYDKYILLLDFNSLYPSIIQEYNICFTTVERSSDDSFPRLPSSKTTGVLPELLK 981
LEPK+GLYDKY+LLLDFNSLYPSIIQEYNICFTT+ RS +D PRLPSS+T G+LP+L++
Sbjct: 903 LEPKRGLYDKYVLLLDFNSLYPSIIQEYNICFTTIPRS-EDGVPRLPSSQTPGILPKLME 961
Query: 982 KLVKLRREKKTWMKTASGLKRQQLDIEQQALKLTANSMYGCLGFSNSRFYAKPLAELITL 1041
LV +R+ K MK +GLK +LDI QQALKLTANSMYGCLGFSNSRFYAKPLAELITL
Sbjct: 962 HLVSIRKSVKLKMKKETGLKYWELDIRQQALKLTANSMYGCLGFSNSRFYAKPLAELITL 1021
Query: 1042 QGREILQSTVDLVQNNLNLEVIYGDTDSIMIYSGLDDIAKATSISKKVIQEVNKKYRCLE 1101
QGR+ILQ TVDLVQN+LNLEVIYGDTDSIMI+SGLDDI + +I KVIQEVNKKYRCL+
Sbjct: 1022 QGRDILQRTVDLVQNHLNLEVIYGDTDSIMIHSGLDDIEEVKAIKSKVIQEVNKKYRCLK 1081
Query: 1102 IDLDGLYKRMLLLKKKKYAAVKVQFKDGTPYEVIERKGLDIVRRDWSLLAKDLGDFCLTQ 1161
ID DG+YKRMLLL+KKKYAAVK+QFKDG P E IERKG+D+VRRDWSLL+K++GD CL++
Sbjct: 1082 IDCDGIYKRMLLLRKKKYAAVKLQFKDGKPCEDIERKGVDMVRRDWSLLSKEIGDLCLSK 1141
Query: 1162 ILSGGSCEDVVESIHNSLMKVQEEMRNGQVALEKYVITKTLTKPPEAYPDAKNQPHVLVA 1221
IL GGSCEDVVE+IHN LMK++EEMRNGQVALEKYVITKTLTKPP AYPD+K+QPHV VA
Sbjct: 1142 ILYGGSCEDVVEAIHNELMKIKEEMRNGQVALEKYVITKTLTKPPAAYPDSKSQPHVQVA 1201
Query: 1222 QRLKQQGYTSGCSVGDTIPYVICCEQG-GSSGSATGIALRARHPDELKQEQGTWLIDIDY 1280
R++Q+GY G + DT+PY+IC EQG SS S+ GIA RARHPDE+K E WL+DIDY
Sbjct: 1202 LRMRQRGYKEGFNAKDTVPYIICYEQGNASSASSAGIAERARHPDEVKSEGSRWLVDIDY 1261
Query: 1281 YLSQQIHPVISRLCASIQGTSPERLADCLGLDTSKFQHKSSEA-SDDPTSSLLFAGDDEE 1339
YL+QQIHPV+SRLCA IQGTSPERLA+CLGLD SK++ KS++A S DP++SLLFA DEE
Sbjct: 1262 YLAQQIHPVVSRLCAEIQGTSPERLAECLGLDPSKYRSKSNDATSSDPSTSLLFATSDEE 1321
Query: 1340 SEKPTSSGTDESDYNFWRKLCCPKC-FENGAGRISAAMIANQVKRQAEKFVLMYYRGLLM 1398
S+KP + T+ESD FW KL CPKC E+ G IS AMIANQVKRQ + FV MYY+G+++
Sbjct: 1322 SKKPATPETEESDSTFWLKLHCPKCQQEDSTGIISPAMIANQVKRQIDGFVSMYYKGIMV 1381
Query: 1399 CDDETCKHTTRSVSFRLVGDSERGTVCPNYPRCNGHLNRKYTEADLYKQLSYFCHVFDTV 1458
C+DE+CKHTTRS +FRL+G+ ERGTVCPNYP CNG L RKYTEADLYKQLSYFCH+ DT
Sbjct: 1382 CEDESCKHTTRSPNFRLLGERERGTVCPNYPNCNGTLLRKYTEADLYKQLSYFCHILDTQ 1441
Query: 1459 CYIEKMEAKSRIPIEKELIKIRPIVDLAASTIQKIRDRCAFGWVKLQDLVV 1509
C +EKM+ RI +EK + KIRP V AA+ + RDRCA+GW++L D+V+
Sbjct: 1442 CSLEKMDVGVRIQVEKAMTKIRPAVKSAAAITRSSRDRCAYGWMQLTDIVI 1492
Score = 53.9 bits (128), Expect = 7e-07
Identities = 28/69 (40%), Positives = 44/69 (63%), Gaps = 2/69 (2%)
Query: 1 MADDDGSSRKRRPTRGPDATARSQALELLRSRRSNGPRSTTTP--QIRLENPIYDTIPED 58
M+ D+ + RR +RG +A++R LE L++ R G RS + IRL+ PI+DT+ ++
Sbjct: 1 MSGDNSTETGRRRSRGAEASSRKDTLERLKAIRQGGIRSASGGGYDIRLQKPIFDTVDDE 60
Query: 59 EYTALVASR 67
EY ALV+ R
Sbjct: 61 EYDALVSRR 69
>At5g63960 DNA polymerase III catalytic subunit
Length = 1081
Score = 199 bits (507), Expect = 8e-51
Identities = 180/671 (26%), Positives = 314/671 (45%), Gaps = 96/671 (14%)
Query: 654 GSNVLCVESSERALLNRLMLQLHKMDSDVLVGHNISGFDLDVLLHRSQACRVPSSMWSKL 713
G +V+ E+ LL L + +D D+++G+NI FDL L+ R+ + + L
Sbjct: 359 GVDVMSFETEREVLLAWRDL-IRDVDPDIIIGYNICKFDLPYLIERAATLGIEE--FPLL 415
Query: 714 GRLNRSTMPKLDRRGKTFGFGADPAIMSCVAGRLLCDTYLCSRDLLKEVSYSLTHLAKTQ 773
GR+ S + D + G + + + GR D K SYSL ++
Sbjct: 416 GRVKNSRVRVRDSTFSSRQQGIRESKETTIEGRFQFDLIQAIHRDHKLSSYSLNSVSAHF 475
Query: 774 LNQSRKEVAPHEVPKMFQT--AKSLMELIEYGETDAWLSMELMFYLSVLPLTRQLTNLSG 831
L++ +++V H + Q A++ L Y DA+L L+ L + ++ ++G
Sbjct: 476 LSEQKEDVH-HSIITDLQNGNAETRRRLAVYCLKDAYLPQRLLDKLMFIYNYVEMARVTG 534
Query: 832 NLWGKTLQGARAQRVEYLLLHEFHKKKYIVPDKFSNYAKETKLTKRRVTHGVDDGNFDDA 891
L ++ +V LL + +K ++P+ AK++ G + G
Sbjct: 535 VPISFLLARGQSIKVLSQLLRKGKQKNLVLPN-----AKQS---------GSEQG----- 575
Query: 892 DINDANYHNDASESDHKKNKKAASYAGGLVLEPKKGLYDKYILLLDFNSLYPSIIQEYNI 951
+Y G VLE + G Y+K I LDF SLYPSI+ YN+
Sbjct: 576 -----------------------TYEGATVLEARTGFYEKPIATLDFASLYPSIMMAYNL 612
Query: 952 CFTTVERSSD--------DSFPRLPSSKT-------TGVLPELLKKLVKLRREKKTWMKT 996
C+ T+ D + + PS +T G+LPE+L++L+ R+ K +K
Sbjct: 613 CYCTLVTPEDVRKLNLPPEHVTKTPSGETFVKQTLQKGILPEILEELLTARKRAKADLKE 672
Query: 997 ASG-LKRQQLDIEQQALKLTANSMYGCLGFSNSRFYAKPLAELITLQGREILQSTVDLVQ 1055
A L++ LD Q ALK++ANS+YG G + + ++ +T GR++++ T LV+
Sbjct: 673 AKDPLEKAVLDGRQLALKISANSVYGFTGATVGQLPCLEISSSVTSYGRQMIEQTKKLVE 732
Query: 1056 NNL--------NLEVIYGDTDSIMIYSGLDDIAKATSISKKVIQEVNKKY-RCLEIDLDG 1106
+ N EVIYGDTDS+M+ G+ D+ A ++ ++ + ++ + + ++++ +
Sbjct: 733 DKFTTLGGYQYNAEVIYGDTDSVMVQFGVSDVEAAMTLGREAAEHISGTFIKPIKLEFEK 792
Query: 1107 LYKRMLLLKKKKYAAVKVQFKDGTPYEVIERKGLDIVRRDWSLLAKDLGDFCLTQILSG- 1165
+Y LL+ KK+YA + + + ++ ++ KG++ VRRD LL K+L L +IL
Sbjct: 793 VYFPYLLINKKRYAG--LLWTNPQQFDKMDTKGIETVRRDNCLLVKNLVTESLNKILIDR 850
Query: 1166 ---GSCEDVVESIHNSLMKVQEEMRNGQVALEKYVITKTLTKPPEAYPDAKNQPHVLVAQ 1222
G+ E+V ++I + LM ++ L VITK LTK + Y H +A+
Sbjct: 851 DVPGAAENVKKTISDLLM--------NRIDLSLLVITKGLTKTGDDY--EVKSAHGELAE 900
Query: 1223 RLKQQGYTSGCSVGDTIPYVICCEQGGSSGSATGIALRARHPDELKQEQGTWLIDIDYYL 1282
R++++ + +VGD +PYVI +A G R D + Q ID +YYL
Sbjct: 901 RMRKRDAATAPNVGDRVPYVII-------KAAKGAKAYERSEDPIYVLQNNIPIDPNYYL 953
Query: 1283 SQQIHPVISRL 1293
QI + R+
Sbjct: 954 ENQISKPLLRI 964
>At1g67500 putative DNA polymerase zeta catalytic subunit
Length = 1871
Score = 122 bits (305), Expect = 2e-27
Identities = 167/727 (22%), Positives = 291/727 (39%), Gaps = 128/727 (17%)
Query: 637 IFPMGFNTEVTD-RNIKAGSNV-LCVESSERALLNRLMLQLHKMDSDVLVGHNISGFDLD 694
+F + F+ + D RN+ S L V ER L + L K D DVL+G +I G +
Sbjct: 1078 VFVLLFSPDSIDQRNVDGLSGCKLSVFLEERQLFRYFIETLCKWDPDVLLGWDIQGGSIG 1137
Query: 695 VLLHRS---------QACRVPSSMWS-------KLGR--------LNRSTMPKL---DRR 727
L R+ R PS + KLG N + + ++ D
Sbjct: 1138 FLAERAAQLGIRFLNNISRTPSPTTTNNSDNKRKLGNNLLPDPLVANPAQVEEVVIEDEW 1197
Query: 728 GKTFGFGADPAIMSCVAGRLLCDTYLCSRDLLKEVSYSLTHLAKTQLNQSRKEVAPHEVP 787
G+T G V GR++ + + R +K Y++ +++ L Q + +
Sbjct: 1198 GRTHASGVH------VGGRIVLNAWRLIRGEVKLNMYTIEAVSEAVLRQKVPSIPYKVLT 1251
Query: 788 KMFQT--AKSLMELIEYGETDAWLSMELMFYLSVLPLTRQLTNLSGNLWGKTLQGARAQR 845
+ F + A + IEY A L++E+M L ++ T +L + G + L R
Sbjct: 1252 EWFSSGPAGARYRCIEYVIRRANLNLEIMSQLDMINRTSELARVFGIDFFSVLSRGSQYR 1311
Query: 846 VEYLLLHEFHKKKYIVPDKFSNYAKETKLTKRRVTHGVDDGNFDDADINDANYHNDASES 905
VE +LL H + Y+ + GN A
Sbjct: 1312 VESMLLRLAHTQNYLA---------------------ISPGNQQVAS------------- 1337
Query: 906 DHKKNKKAASYAGGLVLEPKKGLYDKYILLLDFNSLYPSIIQEYNICFTT---------- 955
+ A LV+EP+ YD +++LDF SLYPS+I YN+CF+T
Sbjct: 1338 ------QPAMECVPLVMEPESAFYDDPVIVLDFQSLYPSMIIAYNLCFSTCLGKLAHLKM 1391
Query: 956 ----VERSSDD---------------SFPRLPSSKTTGVLPELLKKLVKLRREKKTWMKT 996
V S D S +P G+LP LL++++ R K MK
Sbjct: 1392 NTLGVSSYSLDLDVLQDLNQILQTPNSVMYVPPEVRRGILPRLLEEILSTRIMVKKAMKK 1451
Query: 997 ---ASGLKRQQLDIEQQALKLTANSMYG--CLGFSNSRFYAKPLAELITLQGREILQSTV 1051
+ + + + Q ALKL AN YG GFS R LA+ I GR L+ +
Sbjct: 1452 LTPSEAVLHRIFNARQLALKLIANVTYGYTAAGFSG-RMPCAELADSIVQCGRSTLEKAI 1510
Query: 1052 DLVQ--NNLNLEVIYGDTDSIMIYSGLDDIAKATSISKKVIQEVNK-KYRCLEIDLDGLY 1108
V +N N V+YGDTDS+ + + +A + +++ + + + + ++ +Y
Sbjct: 1511 SFVNANDNWNARVVYGDTDSMFVLLKGRTVKEAFVVGQEIASAITEMNPHPVTLKMEKVY 1570
Query: 1109 KRMLLLKKKKYAAVKVQFKDGTPYEVIERKGLDIVRRD-WSLLAKDLGDFCLTQILSGGS 1167
LL KK+Y + + + KG++ VRRD +AK + Q L
Sbjct: 1571 HPCFLLTKKRYVGYSYE-SPNQREPIFDAKGIETVRRDTCEAVAK-----TMEQSLRLFF 1624
Query: 1168 CEDVVESIHNSLMKVQEEMRNGQVALEKYVITKTLTKPPEAYPDAK-NQPHVLVAQRLKQ 1226
+ + + + L + + + +G+V+L+ ++ K + + D+ P +VA + +
Sbjct: 1625 EQKNISKVKSYLYRQWKRILSGRVSLQDFIFAKEVRLGTYSTRDSSLLPPAAIVATKSMK 1684
Query: 1227 QGYTSGCSVGDTIPYVICCEQGGSSGSATGIALRARHPDELKQEQGTWLIDIDYYLSQQI 1286
+ + +PYV+ + G+ + P L + ++ YY+++QI
Sbjct: 1685 ADPRTEPRYAERVPYVVIHGEPGAR-----LVDMVVDPLVLLDVDTPYRLNDLYYINKQI 1739
Query: 1287 HPVISRL 1293
P + R+
Sbjct: 1740 IPALQRV 1746
>At1g08260 hypothetical protein
Length = 2271
Score = 45.4 bits (106), Expect = 2e-04
Identities = 86/374 (22%), Positives = 146/374 (38%), Gaps = 99/374 (26%)
Query: 1006 DIEQQALKLTANSMYGCLGFSNSRFYAKPLAELITLQGREILQSTVDLVQN-NLNLEVIY 1064
D Q A K NS YG + +R+Y+ +A ++T G +I+Q+ L++ LE+
Sbjct: 774 DSLQLAHKCILNSFYGYVMRKGARWYSMEMAGVVTYTGAKIIQNARLLIERIGKPLEL-- 831
Query: 1065 GDTDSI-----------MIYSGLD---------------DIAKATSISK--KVIQEVNKK 1096
DTD I + +D D+AK + + ++ V K
Sbjct: 832 -DTDGIWCCLPGSFPENFTFKTIDMKKLTISYPCVMLNVDVAKNNTNDQYQTLVDPVRKT 890
Query: 1097 YRC-----LEIDLDGLYKRML--------LLKKKKYAAVKVQFKDGTPYEVIERKGLDIV 1143
Y+ +E ++DG YK M+ +L KK+YA DGT + E KG +I
Sbjct: 891 YKSHSECSIEFEVDGPYKAMIIPASKEEGILIKKRYAVFN---HDGT---LAELKGFEIK 944
Query: 1144 RRDWSLLAKDLGDFCLTQILSGGSCED-----------------------------VVES 1174
RR L K + L G + E+ ++
Sbjct: 945 RRGELKLIKVFQAELFDKFLHGSTLEECYSAVAAVADRWLDLLDVCEISCFFWVIYLIGF 1004
Query: 1175 IHNSLMKVQEEMRNGQVALEKYVITKTLTKPPEAYPDAKNQPHVLVAQRLKQ-QGYTSGC 1233
+ + L +++ + ++ L+ + T++K Y + K+ V A+RL + G T
Sbjct: 1005 LSHFLQNQGKDIADSEL-LDYISESSTMSKSLADYGEQKSCA-VTTAKRLAEFLGVTMVK 1062
Query: 1234 SVGDTIPYVICCEQGGS--SGSATGIALRARHPDELKQEQGTW-----------LIDIDY 1280
G Y++ CE G+ S A +A+ +P+ +K W +ID Y
Sbjct: 1063 DKGLRCQYIVACEPKGTPVSERAVPVAIFTTNPEVMKFHLRKWCKTSSDVGIRLIIDWSY 1122
Query: 1281 Y---LSQQIHPVIS 1291
Y LS I VI+
Sbjct: 1123 YKQRLSSAIQKVIT 1136
>At2g27120 putative DNA polymerase epsilon catalytic subunit
Length = 2154
Score = 43.9 bits (102), Expect = 7e-04
Identities = 86/350 (24%), Positives = 137/350 (38%), Gaps = 73/350 (20%)
Query: 1006 DIEQQALKLTANSMYGCLGFSNSRFYAKPLAELITLQGREILQSTVDLVQN-NLNLEVIY 1064
D Q A K NS YG + +R+Y+ +A ++T G +I+Q+ L++ LE+
Sbjct: 767 DSLQLAHKCILNSFYGYVMRKGARWYSMEMAGVVTYTGAKIIQNARLLIERIGKPLEL-- 824
Query: 1065 GDTDSI-----------MIYSGLD---------------DIAKATSISK--KVIQEVNKK 1096
DTD I + +D D+AK S + ++ V K
Sbjct: 825 -DTDGIWCALPGSFPENFTFKTIDMKKFTISYPCVILNVDVAKNNSNDQYQTLVDPVRKT 883
Query: 1097 YRC-----LEIDLDGLYKRML--------LLKKKKYAAVKVQFKDGTPYEVIERKGLDIV 1143
Y +E ++DG YK M+ +L KK+YA DGT + E KG ++
Sbjct: 884 YNSRSECSIEFEVDGPYKAMIIPASKEEGILIKKRYAVFN---HDGT---IAELKGFEMK 937
Query: 1144 RRDWSLLAKDLGDFCLTQILSGGS---CEDVVESIHN---SLMKVQEEMRNGQVALEKYV 1197
RR L K + L G + C V ++ N L++ Q + L+
Sbjct: 938 RRGELKLIKVFQAELFDKFLHGSTLEECYSAVAAVANRWLDLLEGQGKDIADSELLDYIS 997
Query: 1198 ITKTLTKPPEAYPDAKNQPHVLVAQRLKQ-QGYTSGCSVGDTIPYVICCEQGGS--SGSA 1254
+ T++K Y K+ V A+RL G T G Y++ E G+ S A
Sbjct: 998 ESSTMSKSLADYGQQKSCA-VTTAKRLADFLGDTMVKDKGLRCQYIVAREPEGTPVSERA 1056
Query: 1255 TGIALRARHPDELKQEQGTW-----------LIDIDYYLSQQIHPVISRL 1293
+A+ E K W +ID YY Q++H I ++
Sbjct: 1057 VPVAIFQTDDPEKKFYLQKWCKISSYTGIRSIIDWMYY-KQRLHSAIQKV 1105
>At5g27220 putative protein
Length = 1181
Score = 34.7 bits (78), Expect = 0.43
Identities = 51/230 (22%), Positives = 105/230 (45%), Gaps = 23/230 (10%)
Query: 978 ELLKKLVKLRREKKTWMKTASGLKRQQLDIEQQA--LKLTANSMYGCLGFSNSRFYAKPL 1035
+L K V + EKK +T + ++ + +IE++ L L + + C + + +
Sbjct: 236 DLEKYCVDVNAEKKNLGRTQTHRRKLEEEIERKTKDLTLVMDKIAEC-----EKLFERRS 290
Query: 1036 AELITLQGREILQSTVDLVQNNLNLEVIYGDTDSIMIYSGLDDIAKATSISKKVIQEVNK 1095
ELI QG E+ L Q +++LE G+ + +M + + K+ + S+++ +E+ +
Sbjct: 291 LELIKTQG-EVELKGKQLEQMDIDLERHRGEVNVVM-----EHLEKSQTRSRELAEEIER 344
Query: 1096 KYRCLEIDLD--GLYKRMLLLKKKKYAAVKVQFKDGTPYEVIERKGLDIVRRDWSL---L 1150
K + L LD Y + + L +++ A + + V ++K LD + D L L
Sbjct: 345 KRKELTAVLDKTAEYGKTIELVEEELALQQKLLDIRSSELVSKKKELDGLSLDLELVNSL 404
Query: 1151 AKDLGDFCLTQILSGGSCEDVVESI-----HNSLMKVQEEMRNGQVALEK 1195
+L + G ED+ I HN +K+ E + ++A+++
Sbjct: 405 NNELKETVQRIESKGKELEDMERLIQERSGHNESIKLLLEEHSEELAIKE 454
Score = 34.3 bits (77), Expect = 0.56
Identities = 33/163 (20%), Positives = 78/163 (47%), Gaps = 13/163 (7%)
Query: 984 VKLRREKKTWMKTASGLKRQQLDIEQQA--LKLTANSMYGCLGFSNSRFYAKPLAELITL 1041
V+++ EK+ +T +G + + +IE++ L L N + C + R + L ELI
Sbjct: 165 VEVKEEKEHLRRTDNGRRELEEEIERKTKDLTLVMNKIVDC----DKRIETRSL-ELIKT 219
Query: 1042 QGREILQSTVDLVQNNLNLEVIYGDTDSIMIYSGLDDIAKATSISKKVIQEVNKKYRCLE 1101
QG V+L + L+ I + + + + ++ + + +K+ +E+ +K + L
Sbjct: 220 QGE------VELKEKQLDQMKIDLEKYCVDVNAEKKNLGRTQTHRRKLEEEIERKTKDLT 273
Query: 1102 IDLDGLYKRMLLLKKKKYAAVKVQFKDGTPYEVIERKGLDIVR 1144
+ +D + + L +++ +K Q + + +E+ +D+ R
Sbjct: 274 LVMDKIAECEKLFERRSLELIKTQGEVELKGKQLEQMDIDLER 316
>At2g25710 biotin holocarboxylase synthetase
Length = 367
Score = 33.5 bits (75), Expect = 0.96
Identities = 30/100 (30%), Positives = 49/100 (49%), Gaps = 6/100 (6%)
Query: 1141 DIVRRDWSLLAKDLGDFCLT--QILSGGSCEDVVESIHNSLM-KVQEEMRNGQVA-LEKY 1196
D+V ++S L +G C+T Q G ++V ES LM EM +G+V L +Y
Sbjct: 125 DVVSHNFSELP--VGSVCVTDIQFKGRGRTKNVWESPKGCLMYSFTLEMEDGRVVPLIQY 182
Query: 1197 VITKTLTKPPEAYPDAKNQPHVLVAQRLKQQGYTSGCSVG 1236
V++ +T+ + D K P++ V + Y +G VG
Sbjct: 183 VVSLAVTEAVKDVCDKKGLPYIDVKIKWPNDLYVNGLKVG 222
>At5g03730 SERINE/THREONINE-PROTEIN KINASE CTR1
Length = 821
Score = 33.1 bits (74), Expect = 1.3
Identities = 37/127 (29%), Positives = 53/127 (41%), Gaps = 21/127 (16%)
Query: 526 GPSWLQVSNF-STCSASQRVSWCKFEVIVDSPKDIRASSPSSSKI-----TL------VN 573
G S L+ S F S+ SA+ W EV+ D P + ++ S I TL +N
Sbjct: 696 GLSRLKASTFLSSKSAAGTPEWMAPEVLRDEPSNEKSDVYSFGVILWELATLQQPWGNLN 755
Query: 574 PPVVVTAINLKTTINE-KQNINEIVSASVVSCNMVKIDTPMLASEWKRPGMLTHFTVIRK 632
P VV A+ K E +N+N V+A + C WKRP T ++R
Sbjct: 756 PAQVVAAVGFKCKRLEIPRNLNPQVAAIIEGC--------WTNEPWKRPSFATIMDLLRP 807
Query: 633 LDGNIFP 639
L + P
Sbjct: 808 LIKSAVP 814
>At1g21210 hypothetical protein
Length = 738
Score = 32.7 bits (73), Expect = 1.6
Identities = 18/54 (33%), Positives = 28/54 (51%)
Query: 1186 MRNGQVALEKYVITKTLTKPPEAYPDAKNQPHVLVAQRLKQQGYTSGCSVGDTI 1239
M+ LE +TKT K + YP+ ++ H++ Q+L QG TS D+I
Sbjct: 673 MKEVAAELEALRVTKTKHKWSDEYPEQEDTEHLVGVQKLSAQGETSSSIGYDSI 726
>At1g12860 bHLH transcription factor (bHLH033)
Length = 450
Score = 32.7 bits (73), Expect = 1.6
Identities = 30/130 (23%), Positives = 48/130 (36%), Gaps = 21/130 (16%)
Query: 864 KFSNYAKETKLTKRRVTHGVDDGNFDDADINDANYHNDASESDHKKNKKAASYAGGLVLE 923
K N +++ K +DD + DI+ NY +D +++ K KK A L+ E
Sbjct: 213 KMCNSESSSEMRKSSYEREIDDTSTGIIDISGLNYESDDHNTNNNKGKKKGMPAKNLMAE 272
Query: 924 ------------------PKKGLYDKYILLLDFNSLYPSIIQEYNICFTTVER---SSDD 962
PK D+ +L D ++Q N T +E SS
Sbjct: 273 RRRRKKLNDRLYMLRSVVPKISKMDRASILGDAIDYLKELLQRINDLHTELESTPPSSSS 332
Query: 963 SFPRLPSSKT 972
P P+ +T
Sbjct: 333 LHPLTPTPQT 342
>At5g05200 unknown protein
Length = 540
Score = 32.3 bits (72), Expect = 2.1
Identities = 21/81 (25%), Positives = 37/81 (44%), Gaps = 5/81 (6%)
Query: 409 PLHSTNEMIQLEKDVQESRISPADFRKKLQDAVSDTKNEIAKHLVDLGVSSFSMAPVKRK 468
P S ++ + KD++ES + DF K+ Q+ S ++L +G++ + AP K
Sbjct: 253 PEFSRTSLVGIVKDIRESMLEEVDFNKEAQNIES-----FKRYLETMGLTGQATAPRVYK 307
Query: 469 YAFERTDIPAGENYVVKINYL 489
Y R + Y V + L
Sbjct: 308 YCSSRRVLTMERLYGVPLTDL 328
>At2g07410 putative retroelement pol polyprotein
Length = 411
Score = 32.0 bits (71), Expect = 2.8
Identities = 16/54 (29%), Positives = 29/54 (53%), Gaps = 5/54 (9%)
Query: 859 YIVPDKFSNYAKETKLTKRRVTHGVDDGNFDDADINDANYHNDASESDHKKNKK 912
+IV D SN + T+L+ R + N++D D +YH D +ES ++++
Sbjct: 362 HIVKDILSNKSYHTRLSTRTIM-----SNYNDGSSMDLDYHMDEAESSSSRSER 410
>At1g11510 hypthetical protein
Length = 352
Score = 31.6 bits (70), Expect = 3.6
Identities = 34/151 (22%), Positives = 68/151 (44%), Gaps = 12/151 (7%)
Query: 1063 IYGDTDSIMIYSGLDDIAKATSISK-KVIQEVNKKYRCLEIDLDGLYKRMLLLKKKKYAA 1121
++ + D I++ GL D K T +S + E+ KK ++ + L +++ L KKKY
Sbjct: 141 LWTEDDEIVVLQGLIDDKKDTGVSNTNKVYELVKKSISFDVSKNQLMEKLRAL-KKKYEN 199
Query: 1122 VKVQFKDGTPYEVI---ERKGLDIVRRDWSLLAKDLGDFCLTQILSGGSCE------DVV 1172
+ KDG + +RK ++ + W + L + S S + ++
Sbjct: 200 NLGKAKDGVEPTFVKPHDRKAFELSKLVWGGIRMALASGMKSNEKSKKSSKFESVKHELD 259
Query: 1173 ESIHNSLMKVQEE-MRNGQVALEKYVITKTL 1202
S+ NS ++E M G+V+ K + +++
Sbjct: 260 SSLPNSKNNCEDEVMDEGEVSFTKSSLVRSI 290
>At2g38770 unknown protein
Length = 1444
Score = 31.2 bits (69), Expect = 4.8
Identities = 41/175 (23%), Positives = 71/175 (40%), Gaps = 18/175 (10%)
Query: 1105 DGLYKRMLLLKKKKYAAVKVQFKDGTPYEV--IERKGLDIVRRDWSLLAKDLGDFCLTQI 1162
D L + L + + ++ ++ P+ + I +G D R WS +A + DF + Q+
Sbjct: 509 DYLLRNFNLFRLESTYEIREDIQEAVPHLLAHINNEG-DTAFRGWSRMAVPINDFKIAQV 567
Query: 1163 LSGGSCED----VVESIHNSLMKVQEEMRNGQVALEKYVITKTLTKPPEAYP-DAKNQPH 1217
E+ V + S+ + ++R+ +L+++ + L P P +
Sbjct: 568 KQPNIGEEKPSSVTAEVTFSIKSYRTQIRSEWNSLKEHDVLFLLCIRPSFEPLGPEEADK 627
Query: 1218 VLVAQRLKQQGYTSGCSVGDTIPYVICCEQGGSSGSATGIALRARHPDELKQEQG 1272
V QRL Q Y GC + D I E+G TG R DE K +G
Sbjct: 628 ATVPQRLGLQ-YVRGCEIID-----IRDEEGNLMNDFTGRVKR----DEWKPPKG 672
>At1g22200 unknown protein
Length = 386
Score = 31.2 bits (69), Expect = 4.8
Identities = 21/73 (28%), Positives = 39/73 (52%), Gaps = 4/73 (5%)
Query: 567 SKITLVNPPVVVTAINLKTTINEKQNINEIVSASVVSCNMVKIDTPMLASEWKRPGMLTH 626
S++ L PV T + + T+ EK IN V+ + C+++ +D+ ++ E R + H
Sbjct: 45 SELQLYIHPVTETQLRVDTSRGEKLRINFDVTFPALQCSIISLDSMDISGE--RHLDVRH 102
Query: 627 FTVIRKLD--GNI 637
+ R+LD GN+
Sbjct: 103 DIIKRRLDSSGNV 115
>At5g62620 unknown protein
Length = 681
Score = 30.8 bits (68), Expect = 6.2
Identities = 21/74 (28%), Positives = 32/74 (42%), Gaps = 1/74 (1%)
Query: 1321 SEASDDPTSSLLFAGDDEESEKPTSSGTDESDYNFWRKLCCPKCFENGAGRISAAMIANQ 1380
SEA PT L+ G+ KP G Y W + P + NG G I + I+
Sbjct: 542 SEAKKTPTDRSLYIGNINYYHKPLRQGKWSVTYEEWPEEDYPP-YANGPGYILSNDISRF 600
Query: 1381 VKRQAEKFVLMYYR 1394
+ ++ EK L ++
Sbjct: 601 IVKEFEKHKLRMFK 614
>At5g49110 unknown protein
Length = 1487
Score = 30.8 bits (68), Expect = 6.2
Identities = 23/83 (27%), Positives = 42/83 (49%), Gaps = 4/83 (4%)
Query: 593 INEIVSASVVSCNMVKIDTPMLASEWKRPGMLTHFTVIRKLDGNIFPMGFNTEVTDRNIK 652
IN+ SASV SC + ID+ ++ +W + + V +K NI + +TE T ++
Sbjct: 1144 INQSTSASVTSCILQSIDSAIVDMDWATKKLKNFYVVSQK---NIH-LSDDTESTVGSVL 1199
Query: 653 AGSNVLCVESSERALLNRLMLQL 675
+ ES+ R L + +++ L
Sbjct: 1200 EEALYSMAESTVRILSSFVLMNL 1222
>At4g01180 hypothetical protein
Length = 554
Score = 30.4 bits (67), Expect = 8.1
Identities = 55/279 (19%), Positives = 111/279 (39%), Gaps = 28/279 (10%)
Query: 835 GKTLQGARAQRVEYLLLHEFHKKKYIVPDKFSNYAKETKLTKRRVTHGVDDGNFDDADIN 894
GK ++ R + ++ E +K Y + + ++ K K+++ VD+ + + +
Sbjct: 147 GKNVKKKRDLKSISQIVEEDQRKLYHLFENMCQTIEKNKQRKQQLEQKVDE-TLESLEFH 205
Query: 895 DANYHNDASESDHKKNKKAASY----AGG-----LVLEPKKGLYDKYILLLDFNSLYPSI 945
+ +N E K K + GG LE K+ D+ L++ + I
Sbjct: 206 NLMLNNSYQEEIQKMEKNMQEFYQQVLGGHEKSFAELEAKREKLDERARLIEQRA----I 261
Query: 946 IQEYNICFTTVERSSDDSFPRLPSSKTTGVLPELLKKLVKLRREKKTWMKTASGLKR--- 1002
E + T +ER + E +K K ++EK+ K ++
Sbjct: 262 KNEEEMEKTRLEREMIQK----AMCEQNEANEEAMKLAEKHQKEKEKLHKRIMEMEAKLN 317
Query: 1003 --QQLDIEQQALKLTANSMYGCLGFSNSRFYAKPLAEL-ITLQGRE--ILQSTVDLVQNN 1057
Q+L++E + LK T N M +G + + +A+ I L RE + + + L +
Sbjct: 318 ETQELELEIEKLKGTTNVMKHMVGCDGDKDIVEKIAKTQIELDARETALHEKMMTLARKE 377
Query: 1058 LNLEVIYGDT--DSIMIYSGLDDIAKATSISKKVIQEVN 1094
Y D + I ++ +++ K I K++ E+N
Sbjct: 378 RATNDEYQDARKEMIKVWKANEELMKQEKIRVKIMGELN 416
>At1g65440
Length = 1703
Score = 30.4 bits (67), Expect = 8.1
Identities = 49/215 (22%), Positives = 91/215 (41%), Gaps = 44/215 (20%)
Query: 1029 RFYAKPLAELITL--QGREILQSTVDLVQNNLNLEVIYGDTDSIMI-------------- 1072
R+ PLA + TL GREIL + ++N L L+ YG + +M+
Sbjct: 914 RYLQNPLAMVATLCGPGREILSWKLHPLENFLQLDEKYGMVEQVMVDITNQVGIDINLAA 973
Query: 1073 -----YSGLDDIA-----KATSISKKVIQEVNKKYRCLEIDLDGLYKRM-------LLLK 1115
+S L I+ KA S+ + +++ + R ++ + GL K++ L ++
Sbjct: 974 SHDWFFSPLQFISGLGPRKAASLQRSLVRAGSIFVR-KDLIMHGLGKKVFVNAAGFLRIR 1032
Query: 1116 KKKYAAVKVQFKDGTPYEVIERKGLDIVRRDWSLLAKDLGDFCLTQILSGGSCEDVVESI 1175
+ AA QF D I + + + LAKD+ D + G D ++I
Sbjct: 1033 RSGLAASSSQFIDLLDDTRIHPESYSLAQE----LAKDIYDEDVR-----GDSNDDEDAI 1083
Query: 1176 HNSLMKVQEEMRN-GQVALEKYVITKTLTKPPEAY 1209
++ V++ + +V L++Y+ +K E Y
Sbjct: 1084 EMAIEHVRDRPASLRKVVLDEYLASKKRENKKETY 1118
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.337 0.147 0.483
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,427,125
Number of Sequences: 26719
Number of extensions: 1378962
Number of successful extensions: 6033
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 5989
Number of HSP's gapped (non-prelim): 41
length of query: 1510
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1398
effective length of database: 8,326,068
effective search space: 11639843064
effective search space used: 11639843064
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC144431.4


