

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144430.5 - phase: 0 /pseudo
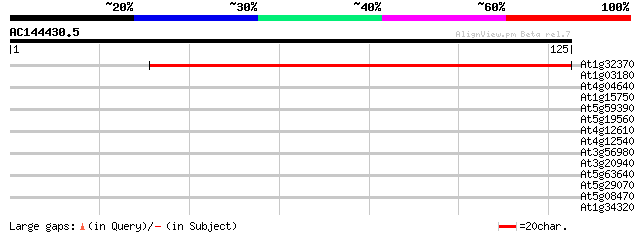
(125 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g32370 unknown protein 119 4e-28
At1g03180 hypothetical protein 29 0.53
At4g04640 ATPase gamma-subunit (AtpC1) 28 0.91
At1g15750 unknown protein (At1g15750) 28 1.2
At5g59390 transcriptional regulator - like protein 27 3.4
At5g19560 putative protein 26 4.5
At4g12610 putative protein 26 4.5
At4g12540 hypothetical protein 26 4.5
At3g56980 putative bHLH transcription factor (bHLH039) 26 4.5
At3g20940 cytochrome P450, putative 26 4.5
At5g63640 unknown protein (At5g63640) 25 7.7
At5g29070 putative protein 25 7.7
At5g08470 putative protein 25 7.7
At1g34320 hypothetical protein 25 7.7
>At1g32370 unknown protein
Length = 125
Score = 119 bits (298), Expect = 4e-28
Identities = 63/94 (67%), Positives = 74/94 (78%)
Query: 32 TGGDNSAKAVVAEQISQTVQSTSNLLHLMQHSSPAQAKLVKLPKNLLAKVSTVKNTQQVL 91
+GGD +AKAVVA+QISQ V ST+NLLHLM+ SS +QA+L KLPKNLLAK S K T Q L
Sbjct: 12 SGGDRTAKAVVADQISQAVNSTANLLHLMRQSSSSQAQLAKLPKNLLAKASLTKATGQAL 71
Query: 92 EQLPRVISSLDAHMENGLQNVPQLKTVVQLLANM 125
QLP+VISSLDAH+E+GL + L TV QLL NM
Sbjct: 72 AQLPQVISSLDAHIESGLHSGVHLNTVTQLLENM 105
>At1g03180 hypothetical protein
Length = 275
Score = 29.3 bits (64), Expect = 0.53
Identities = 15/48 (31%), Positives = 30/48 (62%)
Query: 75 KNLLAKVSTVKNTQQVLEQLPRVISSLDAHMENGLQNVPQLKTVVQLL 122
+ L+++ VKN + LE+L + ISSL + ++ ++ P ++ VV +L
Sbjct: 82 RKLMSRKREVKNEIKKLEKLMKTISSLRSALQLMIREAPGIQKVVLIL 129
>At4g04640 ATPase gamma-subunit (AtpC1)
Length = 373
Score = 28.5 bits (62), Expect = 0.91
Identities = 19/63 (30%), Positives = 35/63 (55%), Gaps = 5/63 (7%)
Query: 64 SPAQAKLVKLPKNLLAKVSTVKNTQQVLEQLPRVISSLDAHMENGLQN-VPQLKTVVQLL 122
SP QA L + L ++ +VKNTQ++ E + V ++ + + N P +T+V++L
Sbjct: 47 SPLQASL----RELRDRIDSVKNTQKITEAMKLVAAAKVRRAQEAVVNGRPFSETLVEVL 102
Query: 123 ANM 125
N+
Sbjct: 103 YNI 105
>At1g15750 unknown protein (At1g15750)
Length = 1131
Score = 28.1 bits (61), Expect = 1.2
Identities = 16/50 (32%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Query: 34 GDNSAKAVVAEQISQTVQSTSNLLHLMQHSSPAQAKLVKLPKNL-LAKVS 82
GD+ V I++ S + L + S P+Q + ++LP+NL +AK+S
Sbjct: 723 GDSRNMVDVKPVITEESNDKSKIWKLTEVSEPSQCRSLRLPENLRVAKIS 772
>At5g59390 transcriptional regulator - like protein
Length = 561
Score = 26.6 bits (57), Expect = 3.4
Identities = 16/66 (24%), Positives = 32/66 (48%), Gaps = 4/66 (6%)
Query: 43 AEQISQTVQSTSNLLHLMQHSSPAQAKLVKLPKNLLAKVSTVKNTQQVLEQ----LPRVI 98
++ + + V+ +L + Q + K+V L +N+ + K ++Q LEQ R +
Sbjct: 145 SDTVGKNVKKKRDLKSISQIVEEDERKMVHLVENMSQTIEKKKQSKQELEQKVDETSRFL 204
Query: 99 SSLDAH 104
SL+ H
Sbjct: 205 ESLELH 210
>At5g19560 putative protein
Length = 493
Score = 26.2 bits (56), Expect = 4.5
Identities = 23/104 (22%), Positives = 45/104 (43%), Gaps = 11/104 (10%)
Query: 21 LWRPMSTQNAATGGDNSAKAVVAEQISQTVQSTSNLLHLMQHSSPAQAKLVKLPKNLLAK 80
+W+ Q G V E+ +L +++ P +P++ L +
Sbjct: 299 IWKRKMCQKEKDGKSQWGSTVSLEKRELFEVRAETILVMLKQQFPG------IPQSSL-E 351
Query: 81 VSTVKNT----QQVLEQLPRVISSLDAHMENGLQNVPQLKTVVQ 120
VS +KN Q +LE RV+ SL + + + +++V + +VQ
Sbjct: 352 VSKIKNNKDVGQAILESYSRVLESLASKIMSRIEDVLEADRLVQ 395
>At4g12610 putative protein
Length = 649
Score = 26.2 bits (56), Expect = 4.5
Identities = 15/46 (32%), Positives = 25/46 (53%), Gaps = 2/46 (4%)
Query: 8 RQDPPTAANRRRPLWRPMSTQNAATG--GDNSAKAVVAEQISQTVQ 51
+ + PT A + P P S+ +AATG ++ +AV+ E+ T Q
Sbjct: 454 KSNTPTKAVKAEPASAPASSSSAATGPVTEDEIRAVLMEKKQVTTQ 499
>At4g12540 hypothetical protein
Length = 461
Score = 26.2 bits (56), Expect = 4.5
Identities = 17/63 (26%), Positives = 28/63 (43%), Gaps = 2/63 (3%)
Query: 3 KMEQRRQDPPTAANRRRPLWRPMSTQNAATGGDNSAKAVVAEQISQTVQSTSNLLHLMQH 62
K+ +R P T AN PL T + GG++ A + + + + + NL+
Sbjct: 188 KLTAKRIFPDTVANE--PLEASKETSDDVLGGESLKTAGLGKSLVHAMDFSENLVEYKPC 245
Query: 63 SSP 65
SSP
Sbjct: 246 SSP 248
>At3g56980 putative bHLH transcription factor (bHLH039)
Length = 258
Score = 26.2 bits (56), Expect = 4.5
Identities = 10/32 (31%), Positives = 22/32 (68%)
Query: 69 KLVKLPKNLLAKVSTVKNTQQVLEQLPRVISS 100
KL+K + LL ++S +NT+ ++Q P+ +++
Sbjct: 134 KLIKKKEELLVQISGQRNTECYVKQPPKAVAN 165
>At3g20940 cytochrome P450, putative
Length = 563
Score = 26.2 bits (56), Expect = 4.5
Identities = 12/40 (30%), Positives = 23/40 (57%)
Query: 81 VSTVKNTQQVLEQLPRVISSLDAHMENGLQNVPQLKTVVQ 120
++ K Q++ E++ V+ + EN L N+P L+ VV+
Sbjct: 329 INNPKILQRLREEIESVVGNTRLIQENDLPNLPYLQAVVK 368
>At5g63640 unknown protein (At5g63640)
Length = 447
Score = 25.4 bits (54), Expect = 7.7
Identities = 9/22 (40%), Positives = 17/22 (76%)
Query: 19 RPLWRPMSTQNAATGGDNSAKA 40
RPL RP+ ++ A+ GGD+ +++
Sbjct: 370 RPLIRPLPSEEASRGGDSHSQS 391
>At5g29070 putative protein
Length = 307
Score = 25.4 bits (54), Expect = 7.7
Identities = 18/55 (32%), Positives = 27/55 (48%), Gaps = 7/55 (12%)
Query: 23 RPMSTQNAATGGDNS-------AKAVVAEQISQTVQSTSNLLHLMQHSSPAQAKL 70
RP +TQ A TG ++S + +V A Q +ST ++ M +P AKL
Sbjct: 112 RPWATQGATTGPESSPTCRPTGSCSVAAPQPQNFGRSTWSMTWSMVQHTPISAKL 166
>At5g08470 putative protein
Length = 1125
Score = 25.4 bits (54), Expect = 7.7
Identities = 15/50 (30%), Positives = 26/50 (52%)
Query: 66 AQAKLVKLPKNLLAKVSTVKNTQQVLEQLPRVISSLDAHMENGLQNVPQL 115
A AK + K+LLA V V + LE++ + L + + GL++ P +
Sbjct: 612 AAAKYFEEQKDLLAHVILVSCSTLALEKVQHIHHVLSSVIAEGLEHAPSV 661
>At1g34320 hypothetical protein
Length = 657
Score = 25.4 bits (54), Expect = 7.7
Identities = 19/60 (31%), Positives = 29/60 (47%), Gaps = 11/60 (18%)
Query: 32 TGGDNSAKAVVAEQIS-------QTVQSTSNLLHLMQHSSPAQAKLVKLP----KNLLAK 80
+GG +SA V +IS T+ +NL+H + S K V LP +NL++K
Sbjct: 141 SGGFSSATTVKGNKISILSFEVANTIVKGANLMHSLSKDSITHLKEVVLPSEGVQNLISK 200
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.122 0.327
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,313,334
Number of Sequences: 26719
Number of extensions: 70696
Number of successful extensions: 272
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 264
Number of HSP's gapped (non-prelim): 15
length of query: 125
length of database: 11,318,596
effective HSP length: 87
effective length of query: 38
effective length of database: 8,994,043
effective search space: 341773634
effective search space used: 341773634
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC144430.5


