

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
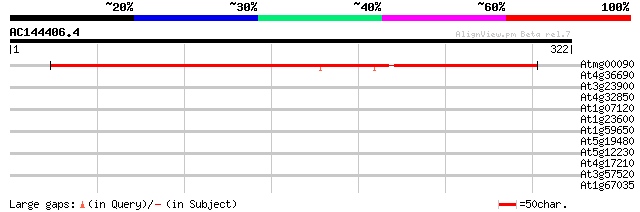
Query= AC144406.4 - phase: 1 /pseudo
(322 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
Atmg00090 -mitochondrial genome- ribosomal protein S3 (RPS3) 477 e-135
At4g36690 splicing factor like protein 35 0.072
At3g23900 unknown protein 33 0.16
At4g32850 poly(A) polymerase like protein 31 0.79
At1g07120 hypothetical protein 30 2.3
At1g23600 putative OBP32pep protein 29 3.0
At1g59650 unknown protein 29 3.9
At5g19480 putative protein 28 5.1
At5g12230 unknown protein 28 5.1
At4g17210 putative protein 28 5.1
At3g57520 imbibition protein homolog 28 5.1
At1g67035 putative protein 28 5.1
>Atmg00090 -mitochondrial genome- ribosomal protein S3 (RPS3)
Length = 556
Score = 477 bits (1228), Expect = e-135
Identities = 241/286 (84%), Positives = 254/286 (88%), Gaps = 8/286 (2%)
Query: 24 SIYYYGKLVYQDVNLRSYFGSIRPPARLTFGFRLGRCILIHFPKRTFIHFFLPRRPRRLK 83
S YYYGK VYQDVNLRSYFGSIRPP RLTFGFRLGRCI++HFPKRTFIHFFLPRRPRRLK
Sbjct: 24 SEYYYGKFVYQDVNLRSYFGSIRPPTRLTFGFRLGRCIILHFPKRTFIHFFLPRRPRRLK 83
Query: 84 RRDKSRPGKEKGRWW-AFGKVGPIGCLHSSNKKDEERNEVRGRRAGKRVESIRLDDREKQ 142
RR+K+RPGKEKGRWW FGK GPI CLHSS+ +EERNEVRGR A KRVESIRLDDR+KQ
Sbjct: 84 RREKTRPGKEKGRWWTTFGKAGPIECLHSSDDTEEERNEVRGRGARKRVESIRLDDRKKQ 143
Query: 143 NEIRIWPKKKQRYGYHDRSPSIKKNLSKSLRVSGA--HPKYAGAVNDIAFLIENYDSFRK 200
NEIR WPKKKQRYGYHDR PSIKKNLSKSLR+SGA HPKYAG VNDIAFLIEN DSF+K
Sbjct: 144 NEIRGWPKKKQRYGYHDRLPSIKKNLSKSLRISGAFKHPKYAGVVNDIAFLIENDDSFKK 203
Query: 201 TKFFKLFF---PRSDGPTGHLFKRTLPAVRPSLNYSVMQYFFCRKNQMHFDPVVVLNHFV 257
TK FKLFF RSDGPT +L RTLPAVRPSLN+ VMQYFF KNQ++FDPVVVLNHFV
Sbjct: 204 TKLFKLFFQNKSRSDGPTSYL--RTLPAVRPSLNFLVMQYFFNTKNQINFDPVVVLNHFV 261
Query: 258 APGVAEPSTMGGTNAQGRSLDKRIRSRIAFFVESSTSEKKCLAEAK 303
APG AEPSTMG NAQGRSL KRIRSRIAFFVESSTSEKKCLAEAK
Sbjct: 262 APGAAEPSTMGRANAQGRSLQKRIRSRIAFFVESSTSEKKCLAEAK 307
>At4g36690 splicing factor like protein
Length = 573
Score = 34.7 bits (78), Expect = 0.072
Identities = 29/103 (28%), Positives = 41/103 (39%), Gaps = 3/103 (2%)
Query: 77 RRPRRLKRRDKSRPGKEKGRWWAFGKVGPIGCLHSSNKKDEERNEVRGRRAGKRVESIRL 136
R R + RD SR E+G G S ++ + R + RG R +R S
Sbjct: 96 RHHRSSRHRDHSR---ERGERRERGGRDDDDYRRSRDRDHDRRRDDRGGRRSRRSRSRSK 152
Query: 137 DDREKQNEIRIWPKKKQRYGYHDRSPSIKKNLSKSLRVSGAHP 179
D E++ R K KQR D +P L+ V+G P
Sbjct: 153 DRSERRTRSRSPSKSKQRVSGFDMAPPASAMLAAGAAVTGQVP 195
>At3g23900 unknown protein
Length = 989
Score = 33.5 bits (75), Expect = 0.16
Identities = 32/99 (32%), Positives = 46/99 (46%), Gaps = 23/99 (23%)
Query: 80 RRLKRRDKSRPGKEKGRWWAFGKVGPIGCLHSSNKKDEERNEVRGRRAGKRVESIRLDDR 139
RR + R KS GK S NK+ R++ RR+G+R S + +
Sbjct: 787 RRRRSRSKSVEGKR-----------------SYNKETRSRDKKSKRRSGRRSRSPSSEGK 829
Query: 140 EKQNEIRIWP----KKKQRYGYHDRSPSI-KKNLSKSLR 173
+ + +IR P +KK R+ H RS SI KKN S+ R
Sbjct: 830 QGR-DIRSSPGYSDEKKSRHKRHSRSRSIEKKNSSRDKR 867
Score = 32.0 bits (71), Expect = 0.46
Identities = 23/65 (35%), Positives = 33/65 (50%), Gaps = 5/65 (7%)
Query: 113 NKKDEERN--EVRGRRAGKRVESIRLDDREKQNEIRIWPKKKQRYGYHDRSPSIKKNLSK 170
NK DE+RN R R K VE R ++E ++ + K K+R G RSPS + +
Sbjct: 776 NKLDEDRNTGSRRRRSRSKSVEGKRSYNKETRSRDK---KSKRRSGRRSRSPSSEGKQGR 832
Query: 171 SLRVS 175
+R S
Sbjct: 833 DIRSS 837
>At4g32850 poly(A) polymerase like protein
Length = 741
Score = 31.2 bits (69), Expect = 0.79
Identities = 25/101 (24%), Positives = 42/101 (40%), Gaps = 6/101 (5%)
Query: 34 QDVNLRSYFGSIRPPARLTFGFRLGRCILI-HFPKRTFIHFFLP---RRPRRLKRRDKSR 89
Q ++R R + ++ G + + H +R F P RRPR+ R ++
Sbjct: 452 QQFDIRGTVDEFRQEVNMYMFWKPGMDVFVSHVRRRQLPPFVFPNGYRRPRQ--SRHQNL 509
Query: 90 PGKEKGRWWAFGKVGPIGCLHSSNKKDEERNEVRGRRAGKR 130
PG + G + G + H+ K D E +VR + KR
Sbjct: 510 PGGKSGEDGSVSHSGSVVERHAKRKNDSEMMDVRPEKPEKR 550
>At1g07120 hypothetical protein
Length = 392
Score = 29.6 bits (65), Expect = 2.3
Identities = 18/75 (24%), Positives = 35/75 (46%), Gaps = 6/75 (8%)
Query: 112 SNKKDEERNEVRGRRAGKRVESIRLDDREKQNEIRIWPKKKQRY------GYHDRSPSIK 165
++K ++E +E+R A R + L E + + +W K + Y G + ++P
Sbjct: 25 NDKLEKENHELRQEVARLRAQVSNLKSHENERKSMLWKKLQSSYDGSNTDGSNLKAPESV 84
Query: 166 KNLSKSLRVSGAHPK 180
K+ +K V +PK
Sbjct: 85 KSNTKGQEVRNPNPK 99
>At1g23600 putative OBP32pep protein
Length = 270
Score = 29.3 bits (64), Expect = 3.0
Identities = 11/59 (18%), Positives = 31/59 (51%)
Query: 114 KKDEERNEVRGRRAGKRVESIRLDDREKQNEIRIWPKKKQRYGYHDRSPSIKKNLSKSL 172
K + +R+E + +++ +D E+ ++ +W ++++ ++D P +K + K L
Sbjct: 20 KGESKRSENVKSNSNTDMDTNNTEDDEEIKQLNLWSDAEKKHPWYDPPPKVKVTMKKGL 78
>At1g59650 unknown protein
Length = 492
Score = 28.9 bits (63), Expect = 3.9
Identities = 33/123 (26%), Positives = 47/123 (37%), Gaps = 8/123 (6%)
Query: 197 SFRKTKFFKLFFPRSDG-PTGHLFKRTLPAVRPSLNYSVMQYFFCRKNQMHFDPVVVLNH 255
S RK KL F +G PTG LF T+ RP V FC + FD +
Sbjct: 185 STRKKAAVKLSFKWREGHPTGPLFSTTMQLQRPMAGSQVP---FCPLEKKMFDSWSI--- 238
Query: 256 FVAPGVAEPSTMGGTNAQGRSLDKRIRSRIAFFVESSTSEKKCLAEAKKVAIPSVVCKPP 315
+ PG + + + L + F V+ S++K A+ V +P V P
Sbjct: 239 -IEPGSFRVRSKTYFRDKKKELAPNYAAYNPFGVDVFLSQRKVNHIAQYVELPVVTTTPT 297
Query: 316 LHP 318
P
Sbjct: 298 KLP 300
>At5g19480 putative protein
Length = 207
Score = 28.5 bits (62), Expect = 5.1
Identities = 13/51 (25%), Positives = 27/51 (52%), Gaps = 5/51 (9%)
Query: 110 HSSNKKDEERNEVRGRRAGKRVESIRLDDREKQNEIRIWPKKKQRYGYHDR 160
H +K +E++ + K + I+ D++K + KKK++ G+HD+
Sbjct: 130 HRKHKDKKEKDREHKKHKHKHKDRIKDKDKDKDRD-----KKKEKSGHHDK 175
>At5g12230 unknown protein
Length = 221
Score = 28.5 bits (62), Expect = 5.1
Identities = 16/62 (25%), Positives = 29/62 (45%), Gaps = 3/62 (4%)
Query: 115 KDEERNEVRGRRAGKRVESIRLDDREKQNEIRIWPKKKQRYGYHDRSPSIKKNLSKSLRV 174
KD ++++ R + K R D++K + +KK + G+HD KK+ K +
Sbjct: 133 KDRDKDKDREHKKHKHKHKDRSKDKDKDKDR---DRKKDKNGHHDSGDHSKKHHDKKRKH 189
Query: 175 SG 176
G
Sbjct: 190 DG 191
>At4g17210 putative protein
Length = 527
Score = 28.5 bits (62), Expect = 5.1
Identities = 27/124 (21%), Positives = 50/124 (39%), Gaps = 22/124 (17%)
Query: 77 RRPRRLKRRDKSRPGKEKGRWW-AFGKVGPIGCLHSSNKKDEER-----NEVRGRRA--- 127
R R LK ++K R E+G W A KV I +K+ E +E+R A
Sbjct: 329 RENRELKGKEKERQEAEEGEWVEASRKVDEIMREAEKTRKEAEEMRMNVDELRREAAAKH 388
Query: 128 -------------GKRVESIRLDDREKQNEIRIWPKKKQRYGYHDRSPSIKKNLSKSLRV 174
G+ VE + ++ ++++ +KK+ + + I+ +L + +
Sbjct: 389 MVMGEAVKQLEIVGRAVEKAKTAEKRAVEDMKVLTEKKESLTHDEPDKKIRISLKEYEEL 448
Query: 175 SGAH 178
G H
Sbjct: 449 RGKH 452
>At3g57520 imbibition protein homolog
Length = 773
Score = 28.5 bits (62), Expect = 5.1
Identities = 22/85 (25%), Positives = 36/85 (41%), Gaps = 5/85 (5%)
Query: 66 PKRTFIHFFLPRRPRR-LKRRDKSRPGKEKGRWWAFGK----VGPIGCLHSSNKKDEERN 120
P + + LP RP R D +R G + W K VG C + K+ ++N
Sbjct: 524 PDGSVLRAKLPGRPTRDCLFADPARDGISLLKIWNMNKFTGIVGVFNCQGAGWCKETKKN 583
Query: 121 EVRGRRAGKRVESIRLDDREKQNEI 145
++ G SIR DD + +++
Sbjct: 584 QIHDTSPGTLTGSIRADDADLISQV 608
>At1g67035 putative protein
Length = 229
Score = 28.5 bits (62), Expect = 5.1
Identities = 12/35 (34%), Positives = 20/35 (56%)
Query: 141 KQNEIRIWPKKKQRYGYHDRSPSIKKNLSKSLRVS 175
++ EIR+ P+KK R+G++ + S SL S
Sbjct: 76 REEEIRVLPEKKTRFGFNPNQRKTSSSSSSSLAQS 110
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,788,350
Number of Sequences: 26719
Number of extensions: 339592
Number of successful extensions: 887
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 878
Number of HSP's gapped (non-prelim): 13
length of query: 322
length of database: 11,318,596
effective HSP length: 99
effective length of query: 223
effective length of database: 8,673,415
effective search space: 1934171545
effective search space used: 1934171545
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144406.4


