

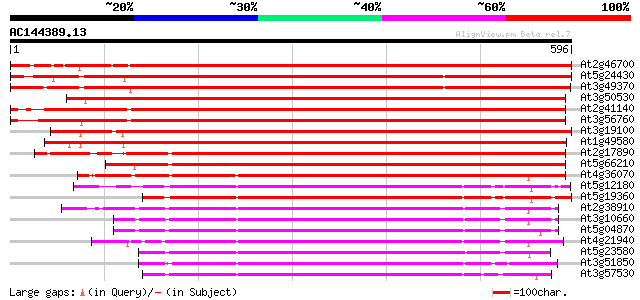
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144389.13 - phase: 0
(596 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g46700 calcium/calmodulin-dependent protein kinase CaMK4 922 0.0
At5g24430 calcium dependent protein kinase CP4 778 0.0
At3g49370 calcium dependent protein kinase - like 758 0.0
At3g50530 calcium/calmodulin-dependent protein kinase CaMK1 723 0.0
At2g41140 CDPK-related kinase 1 (CRK1) 706 0.0
At3g56760 calcium-dependent protein kinase-like 697 0.0
At3g19100 CDPK-related kinase 675 0.0
At1g49580 CDPK-related protein kinase, putative 671 0.0
At2g17890 putative calcium-dependent ser/thr protein kinase 436 e-122
At5g66210 calcium-dependent protein kinase 413 e-115
At4g36070 Calcium-dependent serine/threonine protein kinase 407 e-114
At5g12180 calcium-dependent protein kinase 340 1e-93
At5g19360 calcium-dependent protein kinase - like 331 8e-91
At2g38910 putative calcium-dependent protein kinase 330 1e-90
At3g10660 calmodulin-domain protein kinase CDPK isoform 2 323 2e-88
At5g04870 calcium-dependent protein kinase 322 3e-88
At4g21940 calcium-dependent protein kinase like protein 317 1e-86
At5g23580 calcium-dependent protein kinase (pir||S71196) 315 3e-86
At3g51850 calcium-dependent like protein kinase 313 2e-85
At3g57530 calcium-dependent protein kinase 311 9e-85
>At2g46700 calcium/calmodulin-dependent protein kinase CaMK4
Length = 595
Score = 922 bits (2382), Expect = 0.0
Identities = 463/603 (76%), Positives = 524/603 (86%), Gaps = 15/603 (2%)
Query: 1 MGQCYGKTVPGAGSHRQQNTTTIIDTSAGDGDGSRT-PLPSFANGIPSVKNTPARSSSHA 59
MGQCYGK + ++ TT T GDG++ PL G KNTPARSS+
Sbjct: 1 MGQCYGKVNQSKQNGEEEANTT---TYVVSGDGNQIQPLTPVNYG--RAKNTPARSSN-P 54
Query: 60 SPWPSPYPHG-VNP-----SPSPARASTPKRFFRRPFPPPSPAKHIRESLAKRLGKAKPK 113
SPWPSP+PHG +P SPSPAR STP+RFFRRPFPPPSPAKHI+ SL KRLG KPK
Sbjct: 55 SPWPSPFPHGSASPLPSGVSPSPARTSTPRRFFRRPFPPPSPAKHIKASLIKRLG-VKPK 113
Query: 114 EGPIPEEHGVETDSQSLDKSFGYSKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKDQPV 173
EGPIPEE G E + QSLDKSFGY KNFGAKYELGKEVGRGHFGHTCS R KKGD+KD P+
Sbjct: 114 EGPIPEERGTEPE-QSLDKSFGYGKNFGAKYELGKEVGRGHFGHTCSGRGKKGDIKDHPI 172
Query: 174 AVKIISKSKMTSAIAIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGEL 233
AVKIISK+KMT+AIAIEDVRREVKLLK+LSGHK+LIK++DACEDANNVYIVMELC+GGEL
Sbjct: 173 AVKIISKAKMTTAIAIEDVRREVKLLKSLSGHKYLIKYYDACEDANNVYIVMELCDGGEL 232
Query: 234 LDRILSRGGRYTEEDAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLID 293
LDRIL+RGG+Y E+DAKAIV+QIL+V +FCHLQGVVHRDLKPENFLFTS ED+D+KLID
Sbjct: 233 LDRILARGGKYPEDDAKAIVVQILTVVSFCHLQGVVHRDLKPENFLFTSSREDSDLKLID 292
Query: 294 FGLSDFIRPEERLNDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTE 353
FGLSDFIRP+ERLNDIVGSAYYVAPEVLHRSYS+EADIWSIGVITYILLCGSRPFWARTE
Sbjct: 293 FGLSDFIRPDERLNDIVGSAYYVAPEVLHRSYSLEADIWSIGVITYILLCGSRPFWARTE 352
Query: 354 SGIFRTVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWLRDESRPI 413
SGIFRTVLR +PN+DD+PWPS S EGKDFVKRLLNKDYRKRM+A QALTHPWLRD+SR I
Sbjct: 353 SGIFRTVLRTEPNYDDVPWPSCSSEGKDFVKRLLNKDYRKRMSAVQALTHPWLRDDSRVI 412
Query: 414 PLDILIYKLVKSYLHATPFKRAAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDNFK 473
PLDILIYKLVK+YLHATP +RAA+KAL+KALT+++LVYLRAQF LL PN+DG VSL+NFK
Sbjct: 413 PLDILIYKLVKAYLHATPLRRAALKALAKALTENELVYLRAQFMLLGPNKDGSVSLENFK 472
Query: 474 MALARHATDAMRESRVLDIIHTMEPLAYRKMDFEEFCAAATSTYQLEALDGWEDIACAAF 533
AL ++ATDAMRESRV +I+HTME LAYRKM FEEFCAAA S +QLEA+D WE+IA A F
Sbjct: 473 TALMQNATDAMRESRVPEILHTMESLAYRKMYFEEFCAAAISIHQLEAVDAWEEIATAGF 532
Query: 534 EHFESEGNRVISIEELARELNLGPSAYSVLRDWIRNTDGKLSLLGYTKFLHGVTLRSSLP 593
+HFE+EGNRVI+IEELARELN+G SAY LRDW+R++DGKLS LG+TKFLHGVTLR++
Sbjct: 533 QHFETEGNRVITIEELARELNVGASAYGHLRDWVRSSDGKLSYLGFTKFLHGVTLRAAHA 592
Query: 594 RPR 596
RPR
Sbjct: 593 RPR 595
>At5g24430 calcium dependent protein kinase CP4
Length = 594
Score = 778 bits (2009), Expect = 0.0
Identities = 393/608 (64%), Positives = 482/608 (78%), Gaps = 26/608 (4%)
Query: 1 MGQCYGKTVPGAGSHRQQNTTTIIDTSAGDGDGSRTPLPSFANGI----PSVKNTPARSS 56
MG CY + + + G+G+ S P + + P TP +S
Sbjct: 1 MGHCYSRNISAVEDD---------EIPTGNGEVSNQPSQNHRHASIPQSPVASGTPEVNS 51
Query: 57 SHASPWPSPYPHGVNPSPSPARASTPKRFFRRPFPPPSPAKHIRESLAKRLGKA-KPKEG 115
+ SP+ SP P GV PSP A TP R F+ PFPPPSPAK I +L +R G +P++
Sbjct: 52 YNISPFQSPLPAGVAPSP----ARTPGRKFKWPFPPPSPAKPIMAALRRRRGAPPQPRDE 107
Query: 116 PIPEE------HGVETDS-QSLDKSFGYSKNFGAKYELGKEVGRGHFGHTCSARAKKGDL 168
PIPE+ HG ++ + LDK+FG+ KNF KYELGKEVGRGHFGHTC A+AKKG +
Sbjct: 108 PIPEDSEDVVDHGGDSGGGERLDKNFGFGKNFEGKYELGKEVGRGHFGHTCWAKAKKGKM 167
Query: 169 KDQPVAVKIISKSKMTSAIAIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELC 228
K+Q VAVKIISK+KMTS ++IEDVRREVKLLKALSGH+H++KF+D EDA+NV++VMELC
Sbjct: 168 KNQTVAVKIISKAKMTSTLSIEDVRREVKLLKALSGHRHMVKFYDVYEDADNVFVVMELC 227
Query: 229 EGGELLDRILSRGGRYTEEDAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDAD 288
EGGELLDRIL+RGGRY E DAK I++QILS AF HLQGVVHRDLKPENFLFTSR+EDA
Sbjct: 228 EGGELLDRILARGGRYPEVDAKRILVQILSATAFFHLQGVVHRDLKPENFLFTSRNEDAI 287
Query: 289 MKLIDFGLSDFIRPEERLNDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPF 348
+K+IDFGLSDFIR ++RLND+VGSAYYVAPEVLHRSYS EAD+WSIGVI+YILLCGSRPF
Sbjct: 288 LKVIDFGLSDFIRYDQRLNDVVGSAYYVAPEVLHRSYSTEADMWSIGVISYILLCGSRPF 347
Query: 349 WARTESGIFRTVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWLRD 408
+ RTES IFR VLRA+PNF+D+PWPS+SP KDFVKRLLNKD+RKRMTAAQAL HPWLRD
Sbjct: 348 YGRTESAIFRCVLRANPNFEDMPWPSISPTAKDFVKRLLNKDHRKRMTAAQALAHPWLRD 407
Query: 409 ESRPIPLDILIYKLVKSYLHATPFKRAAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVS 468
E+ + LD +YKLVKSY+ A+PF+R+A+KALSKA+ D++LV+L+AQF LL+P +DG +S
Sbjct: 408 ENPGLLLDFSVYKLVKSYIRASPFRRSALKALSKAIPDEELVFLKAQFMLLDP-KDGGLS 466
Query: 469 LDNFKMALARHATDAMRESRVLDIIHTMEPLAYRKMDFEEFCAAATSTYQLEALDGWEDI 528
L+ F MAL R+ATDAM ESR+ DI++TM+PLA +K+DFEEFCAAA S YQLEAL+ WE I
Sbjct: 467 LNCFTMALTRYATDAMMESRLPDILNTMQPLAQKKLDFEEFCAAAVSVYQLEALEEWEQI 526
Query: 529 ACAAFEHFESEGNRVISIEELARELNLGPSAYSVLRDWIRNTDGKLSLLGYTKFLHGVTL 588
A +AFEHFE EGNR+IS++ELA E+++GPSAY +L+DWIR++DGKLS LGY KFLHGVT+
Sbjct: 527 ATSAFEHFEHEGNRIISVQELAGEMSVGPSAYPLLKDWIRSSDGKLSFLGYAKFLHGVTV 586
Query: 589 RSSLPRPR 596
RSS RPR
Sbjct: 587 RSSSSRPR 594
>At3g49370 calcium dependent protein kinase - like
Length = 594
Score = 758 bits (1958), Expect = 0.0
Identities = 385/600 (64%), Positives = 470/600 (78%), Gaps = 13/600 (2%)
Query: 1 MGQCYGKTVPGAGSHRQQNTTTIIDTSAGDGDGSRTPLPSFANGIPSVKNTPARSSSHAS 60
MG CY + + + + T + +T S ++ IP T + + S
Sbjct: 1 MGHCYSRNISTVDDDDEIPSATAQLPHRSHQNHHQT---SSSSSIPQSPATSEVNPYNIS 57
Query: 61 PWPSPYPHGVNPSPSPARASTPKRFFRRPFPPPSPAKHIRESLAKRLGKAK-PKEGPIPE 119
P+ SP P GV PSP A TP R F+ PFPPPSPAK I +L +R G A P++GPIPE
Sbjct: 58 PFQSPLPAGVAPSP----ARTPGRKFKWPFPPPSPAKPIMAALRRRRGTAPHPRDGPIPE 113
Query: 120 EHGVETDS----QSLDKSFGYSKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKDQPVAV 175
+ + LDK+FG++KNF KYELG+EVGRGHFGHTC A+AKKG +K Q VAV
Sbjct: 114 DSEAGGSGGGIGERLDKNFGFAKNFEGKYELGREVGRGHFGHTCWAKAKKGKIKGQTVAV 173
Query: 176 KIISKSKMTSAIAIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELLD 235
KIISKSKMTSA++IEDVRREVKLLKALSGH H++KF+D ED++NV++VMELCEGGELLD
Sbjct: 174 KIISKSKMTSALSIEDVRREVKLLKALSGHSHMVKFYDVFEDSDNVFVVMELCEGGELLD 233
Query: 236 RILSRGGRYTEEDAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFG 295
IL+RGGRY E +AK I++QILS AF HLQGVVHRDLKPENFLFTS++EDA +K+IDFG
Sbjct: 234 SILARGGRYPEAEAKRILVQILSATAFFHLQGVVHRDLKPENFLFTSKNEDAVLKVIDFG 293
Query: 296 LSDFIRPEERLNDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTESG 355
LSD+ R ++RLND+VGSAYYVAPEVLHRSYS EADIWSIGVI+YILLCGSRPF+ RTES
Sbjct: 294 LSDYARFDQRLNDVVGSAYYVAPEVLHRSYSTEADIWSIGVISYILLCGSRPFYGRTESA 353
Query: 356 IFRTVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWLRDESRPIPL 415
IFR VLRA+PNFDDLPWPS+SP KDFVKRLLNKD+RKRMTAAQAL HPWLRDE+ + L
Sbjct: 354 IFRCVLRANPNFDDLPWPSISPIAKDFVKRLLNKDHRKRMTAAQALAHPWLRDENPGLLL 413
Query: 416 DILIYKLVKSYLHATPFKRAAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDNFKMA 475
D IYKLVKSY+ A+PF+RAA+K+LSKA+ +++LV+L+AQF LLEP DG + L NF A
Sbjct: 414 DFSIYKLVKSYIRASPFRRAALKSLSKAIPEEELVFLKAQFMLLEP-EDGGLHLHNFTTA 472
Query: 476 LARHATDAMRESRVLDIIHTMEPLAYRKMDFEEFCAAATSTYQLEALDGWEDIACAAFEH 535
L R+ATDAM ESR+ DI++ M+PLA++K+DFEEFCAA+ S YQLEAL+ WE IA AFEH
Sbjct: 473 LTRYATDAMIESRLPDILNMMQPLAHKKLDFEEFCAASVSVYQLEALEEWEQIATVAFEH 532
Query: 536 FESEGNRVISIEELARELNLGPSAYSVLRDWIRNTDGKLSLLGYTKFLHGVTLRSSLPRP 595
FESEG+R IS++ELA E++LGP+AY +L+DWIR+ DGKL+ LGY KFLHGVT+RSS RP
Sbjct: 533 FESEGSRAISVQELAEEMSLGPNAYPLLKDWIRSLDGKLNFLGYAKFLHGVTVRSSSSRP 592
>At3g50530 calcium/calmodulin-dependent protein kinase CaMK1
Length = 601
Score = 723 bits (1866), Expect = 0.0
Identities = 358/541 (66%), Positives = 436/541 (80%), Gaps = 11/541 (2%)
Query: 61 PWPSPYP-HGVNPSPSPARA-------STPKRFFRRPFPPPSPAKHIRESLAKRLGKAKP 112
P+ SP P H +PAR+ STPKRFF+RPFPPPSPAKHIR LA+R G KP
Sbjct: 57 PFYSPSPAHYFFSKKTPARSPATNSTNSTPKRFFKRPFPPPSPAKHIRAVLARRHGSVKP 116
Query: 113 KEGPIPEEHGVETDSQSLDKSFGYSKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKDQP 172
IPE E LDKSFG+SK+F +KYELG EVGRGHFG+TC+A+ KKGD K Q
Sbjct: 117 NSSAIPEGSEAEGGGVGLDKSFGFSKSFASKYELGDEVGRGHFGYTCAAKFKKGDNKGQQ 176
Query: 173 VAVKIISKSKMTSAIAIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGE 232
VAVK+I K+KMT+AIAIEDVRREVK+L+ALSGH +L F+DA ED +NVYIVMELCEGGE
Sbjct: 177 VAVKVIPKAKMTTAIAIEDVRREVKILRALSGHNNLPHFYDAYEDHDNVYIVMELCEGGE 236
Query: 233 LLDRILSRGGRYTEEDAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLI 292
LLDRILSRGG+YTEEDAK +++QIL+V AFCHLQGVVHRDLKPENFLFTS+ + + +K I
Sbjct: 237 LLDRILSRGGKYTEEDAKTVMIQILNVVAFCHLQGVVHRDLKPENFLFTSKEDTSQLKAI 296
Query: 293 DFGLSDFIRPEERLNDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWART 352
DFGLSD++RP+ERLNDIVGSAYYVAPEVLHRSYS EADIWS+GVI YILLCGSRPFWART
Sbjct: 297 DFGLSDYVRPDERLNDIVGSAYYVAPEVLHRSYSTEADIWSVGVIVYILLCGSRPFWART 356
Query: 353 ESGIFRTVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWLRDES-R 411
ESGIFR VL+ADP+FDD PWP +S E +DFVKRLLNKD RKR+TAAQAL+HPW++D +
Sbjct: 357 ESGIFRAVLKADPSFDDPPWPLLSSEARDFVKRLLNKDPRKRLTAAQALSHPWIKDSNDA 416
Query: 412 PIPLDILIYKLVKSYLHATPFKRAAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDN 471
+P+DIL++KL+++YL ++ ++AA++ALSK LT D+L YLR QF LLEP+++G +SL+N
Sbjct: 417 KVPMDILVFKLMRAYLRSSSLRKAALRALSKTLTVDELFYLREQFALLEPSKNGTISLEN 476
Query: 472 FKMALARHATDAMRESRVLDIIHTMEPLAYRKMDFEEFCAAATSTYQLEALDGWEDIACA 531
K AL + ATDAM++SR+ + + + L YR+MDFEEFCAAA S +QLEALD WE A
Sbjct: 477 IKSALMKMATDAMKDSRIPEFLGQLSALQYRRMDFEEFCAAALSVHQLEALDRWEQHARC 536
Query: 532 AFEHFESEGNRVISIEELARELNLGPS--AYSVLRDWIRNTDGKLSLLGYTKFLHGVTLR 589
A+E FE EGNR I I+ELA EL LGPS ++VL DW+R+TDGKLS LG+ K LHGV+ R
Sbjct: 537 AYELFEKEGNRPIMIDELASELGLGPSVPVHAVLHDWLRHTDGKLSFLGFVKLLHGVSSR 596
Query: 590 S 590
+
Sbjct: 597 T 597
>At2g41140 CDPK-related kinase 1 (CRK1)
Length = 576
Score = 706 bits (1821), Expect = 0.0
Identities = 356/595 (59%), Positives = 446/595 (74%), Gaps = 28/595 (4%)
Query: 1 MGQCYGKTVPGAGSHRQQNTTTIIDTSAGDGDGSRTPLPSFANGIPSVKNTPARSSSHAS 60
MG C+GK V +Q + ++ P+ N P+ PA+SS
Sbjct: 1 MGICHGKPV-------EQQSKSL-------------PVSGETNEAPTNSQPPAKSSGFPF 40
Query: 61 PWPSPYPHGVNPSPSPARA--STPKRFFRRPFPPPSPAKHIRESLAKRLGKAKPKEGPIP 118
PSP P SPS + + STP R F+RPFPPPSPAKHIR LA+R G KP E IP
Sbjct: 41 YSPSPVPSLFKSSPSVSSSVSSTPLRIFKRPFPPPSPAKHIRAFLARRYGSVKPNEVSIP 100
Query: 119 EEHGVETDSQSLDKSFGYSKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKII 178
E E LDKSFG+SK F + YE+ EVGRGHFG+TCSA+ KKG LK Q VAVK+I
Sbjct: 101 EGKECEI---GLDKSFGFSKQFASHYEIDGEVGRGHFGYTCSAKGKKGSLKGQEVAVKVI 157
Query: 179 SKSKMTSAIAIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRIL 238
KSKMT+AIAIEDV REVK+L+AL+GHK+L++F+DA ED NVYIVMELC+GGELLD+IL
Sbjct: 158 PKSKMTTAIAIEDVSREVKMLRALTGHKNLVQFYDAFEDDENVYIVMELCKGGELLDKIL 217
Query: 239 SRGGRYTEEDAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSD 298
RGG+Y+E+DAK +++QILSV A+CHLQGVVHRDLKPENFLF+++ E + +K IDFGLSD
Sbjct: 218 QRGGKYSEDDAKKVMVQILSVVAYCHLQGVVHRDLKPENFLFSTKDETSPLKAIDFGLSD 277
Query: 299 FIRPEERLNDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTESGIFR 358
+++P+ERLNDIVGSAYYVAPEVLHR+Y EAD+WSIGVI YILLCGSRPFWARTESGIFR
Sbjct: 278 YVKPDERLNDIVGSAYYVAPEVLHRTYGTEADMWSIGVIAYILLCGSRPFWARTESGIFR 337
Query: 359 TVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWL-RDESRPIPLDI 417
VL+A+PNF++ PWPS+SPE DFVKRLLNKDYRKR+TAAQAL HPWL IP D+
Sbjct: 338 AVLKAEPNFEEAPWPSLSPEAVDFVKRLLNKDYRKRLTAAQALCHPWLVGSHELKIPSDM 397
Query: 418 LIYKLVKSYLHATPFKRAAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDNFKMALA 477
+IYKLVK Y+ +T +++A+ AL+K LT QL YLR QF LL P+++G++S+ N+K A+
Sbjct: 398 IIYKLVKVYIMSTSLRKSALAALAKTLTVPQLAYLREQFTLLGPSKNGYISMQNYKTAIL 457
Query: 478 RHATDAMRESRVLDIIHTMEPLAYRKMDFEEFCAAATSTYQLEALDGWEDIACAAFEHFE 537
+ +TDAM++SRV D +H + L Y+K+DFEEFCA+A S YQLEA++ WE A A+E FE
Sbjct: 458 KSSTDAMKDSRVFDFVHMISCLQYKKLDFEEFCASALSVYQLEAMETWEQHARRAYELFE 517
Query: 538 SEGNRVISIEELARELNLGPS--AYSVLRDWIRNTDGKLSLLGYTKFLHGVTLRS 590
+GNR I IEELA EL LGPS + VL+DWIR++DGKLS LG+ + LHGV+ R+
Sbjct: 518 KDGNRPIMIEELASELGLGPSVPVHVVLQDWIRHSDGKLSFLGFVRLLHGVSSRT 572
>At3g56760 calcium-dependent protein kinase-like
Length = 577
Score = 697 bits (1799), Expect = 0.0
Identities = 349/596 (58%), Positives = 441/596 (73%), Gaps = 29/596 (4%)
Query: 1 MGQCYGKTVPGAGSHRQQNTTTIIDTSAGDGDGSRTPLPSFANGIPSVKNTPARSSSHAS 60
MG C+GK + + P+ + P + A+SS
Sbjct: 1 MGLCHGKPI--------------------EQQSKNLPISNEIEETPKNSSQKAKSSGFPF 40
Query: 61 PWPSPYPHGVNPSP---SPARASTPKRFFRRPFPPPSPAKHIRESLAKRLGKAKPKEGPI 117
PSP P SP S + +STP R F+RPFPPPSPAKHIR LA+R G KP E I
Sbjct: 41 YSPSPLPSLFKTSPAVSSSSVSSTPLRIFKRPFPPPSPAKHIRALLARRHGSVKPNEASI 100
Query: 118 PEEHGVETDSQSLDKSFGYSKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKI 177
PE E LDK FG+SK F + YE+ EVGRGHFG+TCSA+ KKG LK Q VAVK+
Sbjct: 101 PEGSECEV---GLDKKFGFSKQFASHYEIDGEVGRGHFGYTCSAKGKKGSLKGQDVAVKV 157
Query: 178 ISKSKMTSAIAIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRI 237
I KSKMT+AIAIEDVRREVK+L+AL+GHK+L++F+DA ED NVYIVMELC+GGELLD+I
Sbjct: 158 IPKSKMTTAIAIEDVRREVKILRALTGHKNLVQFYDAFEDDENVYIVMELCQGGELLDKI 217
Query: 238 LSRGGRYTEEDAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLS 297
L RGG+Y+E DAK +++QILSV A+CHLQGVVHRDLKPENFLFT++ E + +K IDFGLS
Sbjct: 218 LQRGGKYSEVDAKKVMIQILSVVAYCHLQGVVHRDLKPENFLFTTKDESSPLKAIDFGLS 277
Query: 298 DFIRPEERLNDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTESGIF 357
D++RP+ERLNDIVGSAYYVAPEVLHR+Y EAD+WSIGVI YILLCGSRPFWAR+ESGIF
Sbjct: 278 DYVRPDERLNDIVGSAYYVAPEVLHRTYGTEADMWSIGVIAYILLCGSRPFWARSESGIF 337
Query: 358 RTVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWL-RDESRPIPLD 416
R VL+A+PNF++ PWPS+SP+ DFVKRLLNKDYRKR+TAAQAL HPWL IP D
Sbjct: 338 RAVLKAEPNFEEAPWPSLSPDAVDFVKRLLNKDYRKRLTAAQALCHPWLVGSHELKIPSD 397
Query: 417 ILIYKLVKSYLHATPFKRAAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDNFKMAL 476
++IYKLVK Y+ ++ +++A+ AL+K LT QL YL+ QF LL P+++G++S+ N+K A+
Sbjct: 398 MIIYKLVKVYIMSSSLRKSALAALAKTLTVPQLTYLQEQFNLLGPSKNGYISMQNYKTAI 457
Query: 477 ARHATDAMRESRVLDIIHTMEPLAYRKMDFEEFCAAATSTYQLEALDGWEDIACAAFEHF 536
+ +T+A ++SRVLD +H + L Y+K+DFEEFCA+A S YQLEA++ WE A A+E +
Sbjct: 458 LKSSTEATKDSRVLDFVHMISCLQYKKLDFEEFCASALSVYQLEAMETWEQHARRAYELY 517
Query: 537 ESEGNRVISIEELARELNLGPS--AYSVLRDWIRNTDGKLSLLGYTKFLHGVTLRS 590
E +GNRVI IEELA EL LGPS + VL+DWIR++DGKLS LG+ + LHGV+ R+
Sbjct: 518 EKDGNRVIMIEELATELGLGPSVPVHVVLQDWIRHSDGKLSFLGFVRLLHGVSSRT 573
>At3g19100 CDPK-related kinase
Length = 599
Score = 675 bits (1741), Expect = 0.0
Identities = 346/568 (60%), Positives = 438/568 (76%), Gaps = 18/568 (3%)
Query: 44 GIPSVKNTPARSSSHASPW--PSPYPHGVNPS------PSPARASTPKRFFRRPFPPPSP 95
G V+++PA +S P+ PSP H N S S + STP R R F PPSP
Sbjct: 35 GKSPVRSSPAVKASPFFPFYTPSPARHRRNKSRDGGGGESKSVTSTPLRQLARAFHPPSP 94
Query: 96 AKHIRESLAKRLGKAKPKEGPIP----EEHGVETDSQSLDKSFGYSKNFGAKYELGKEVG 151
A+HIR+ L +R K KE +P ++ E + LDK FG+SK ++ ELG+E+G
Sbjct: 95 ARHIRDVLRRRKEK---KEAALPAARQQKEEEEREEVGLDKRFGFSKELQSRIELGEEIG 151
Query: 152 RGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLLKALSGHKHLIKF 211
RGHFG+TCSA+ KKG+LKDQ VAVK+I KSKMTSAI+IEDVRREVK+L+ALSGH++L++F
Sbjct: 152 RGHFGYTCSAKFKKGELKDQEVAVKVIPKSKMTSAISIEDVRREVKILRALSGHQNLVQF 211
Query: 212 HDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSVAAFCHLQGVVHR 271
+DA ED NVYIVMELC GGELLDRIL+RGG+Y+E+DAKA+++QIL+V AFCHLQGVVHR
Sbjct: 212 YDAFEDNANVYIVMELCGGGELLDRILARGGKYSEDDAKAVLIQILNVVAFCHLQGVVHR 271
Query: 272 DLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYYVAPEVLHRSYSVEADI 331
DLKPENFL+TS+ E++ +K+IDFGLSDF+RP+ERLNDIVGSAYYVAPEVLHRSY+ EAD+
Sbjct: 272 DLKPENFLYTSKEENSMLKVIDFGLSDFVRPDERLNDIVGSAYYVAPEVLHRSYTTEADV 331
Query: 332 WSIGVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDY 391
WSIGVI YILLCGSRPFWARTESGIFR VL+ADP+FD+ PWPS+S E KDFVKRLL KD
Sbjct: 332 WSIGVIAYILLCGSRPFWARTESGIFRAVLKADPSFDEPPWPSLSFEAKDFVKRLLYKDP 391
Query: 392 RKRMTAAQALTHPWLRDESR-PIPLDILIYKLVKSYLHATPFKRAAVKALSKALTDDQLV 450
RKRMTA+QAL HPW+ + IP DILI+K +K+YL ++ ++AA+ ALSK LT D+L+
Sbjct: 392 RKRMTASQALMHPWIAGYKKIDIPFDILIFKQIKAYLRSSSLRKAALMALSKTLTTDELL 451
Query: 451 YLRAQFRLLEPNRDGHVSLDNFKMALARHATDAMRESRVLDIIHTMEPLAYRKMDFEEFC 510
YL+AQF L PN++G ++LD+ ++ALA +AT+AM+ESR+ D + + L Y+ MDFEEFC
Sbjct: 452 YLKAQFAHLAPNKNGLITLDSIRLALATNATEAMKESRIPDFLALLNGLQYKGMDFEEFC 511
Query: 511 AAATSTYQLEALDGWEDIACAAFEHFESEGNRVISIEELARELNLGPS--AYSVLRDWIR 568
AA+ S +Q E+LD WE A+E FE GNRVI IEELA EL +G S +++L DWIR
Sbjct: 512 AASISVHQHESLDCWEQSIRHAYELFEMNGNRVIVIEELASELGVGSSIPVHTILNDWIR 571
Query: 569 NTDGKLSLLGYTKFLHGVTLRSSLPRPR 596
+TDGKLS LG+ K LHGV+ R SL + R
Sbjct: 572 HTDGKLSFLGFVKLLHGVSTRQSLAKTR 599
>At1g49580 CDPK-related protein kinase, putative
Length = 606
Score = 671 bits (1732), Expect = 0.0
Identities = 341/575 (59%), Positives = 431/575 (74%), Gaps = 21/575 (3%)
Query: 38 LPSFANGIPSVKNTPARSSSHASPW-----PSPYPHGVNPS-------PSPARASTPKRF 85
+ FA P P S++ ASP+ PSP H N S S + STP R
Sbjct: 27 IDDFAPDHPGKSPIPTPSAAKASPFFPFYTPSPARHRRNKSRDVGGGGESKSLTSTPLRQ 86
Query: 86 FRRPFPPPSPAKHIRESLAKRLGKAKPKEGPIPE-----EHGVETDSQSLDKSFGYSKNF 140
RR F PPSPAKHIR +L +R GK + + + E + LDK FG+SK F
Sbjct: 87 LRRAFHPPSPAKHIRAALRRRKGKKEAALSGVTQLTTEVPQREEEEEVGLDKRFGFSKEF 146
Query: 141 GAKYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLLK 200
++ ELG+E+GRGHFG+TCSA+ KKG+LK Q VAVKII KSKMT+AIAIEDVRREVK+L+
Sbjct: 147 HSRVELGEEIGRGHFGYTCSAKFKKGELKGQVVAVKIIPKSKMTTAIAIEDVRREVKILQ 206
Query: 201 ALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSVA 260
ALSGHK+L++F+DA ED NVYI MELCEGGELLDRIL+RGG+Y+E DAK +++QIL+V
Sbjct: 207 ALSGHKNLVQFYDAFEDNANVYIAMELCEGGELLDRILARGGKYSENDAKPVIIQILNVV 266
Query: 261 AFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYYVAPEV 320
AFCH QGVVHRDLKPENFL+TS+ E++ +K IDFGLSDF+RP+ERLNDIVGSAYYVAPEV
Sbjct: 267 AFCHFQGVVHRDLKPENFLYTSKEENSQLKAIDFGLSDFVRPDERLNDIVGSAYYVAPEV 326
Query: 321 LHRSYSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPWPSVSPEGK 380
LHRSY+ EAD+WSIGVI YILLCGSRPFWARTESGIFR VL+ADP+FD+ PWP +S + K
Sbjct: 327 LHRSYTTEADVWSIGVIAYILLCGSRPFWARTESGIFRAVLKADPSFDEPPWPFLSSDAK 386
Query: 381 DFVKRLLNKDYRKRMTAAQALTHPWLR--DESRPIPLDILIYKLVKSYLHATPFKRAAVK 438
DFVKRLL KD R+RM+A+QAL HPW+R + IP DILI++ +K+YL ++ ++AA++
Sbjct: 387 DFVKRLLFKDPRRRMSASQALMHPWIRAYNTDMNIPFDILIFRQMKAYLRSSSLRKAALR 446
Query: 439 ALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDNFKMALARHATDAMRESRVLDIIHTMEP 498
ALSK L D+++YL+ QF LL PN+DG +++D +MALA +AT+AM+ESR+ + + +
Sbjct: 447 ALSKTLIKDEILYLKTQFSLLAPNKDGLITMDTIRMALASNATEAMKESRIPEFLALLNG 506
Query: 499 LAYRKMDFEEFCAAATSTYQLEALDGWEDIACAAFEHFESEGNRVISIEELARELNLGPS 558
L YR MDFEEFCAAA + +Q E+LD WE A+E F+ GNR I IEELA EL +GPS
Sbjct: 507 LQYRGMDFEEFCAAAINVHQHESLDCWEQSIRHAYELFDKNGNRAIVIEELASELGVGPS 566
Query: 559 --AYSVLRDWIRNTDGKLSLLGYTKFLHGVTLRSS 591
+SVL DWIR+TDGKLS G+ K LHGV++R+S
Sbjct: 567 IPVHSVLHDWIRHTDGKLSFFGFVKLLHGVSVRAS 601
>At2g17890 putative calcium-dependent ser/thr protein kinase
Length = 571
Score = 436 bits (1122), Expect = e-122
Identities = 250/570 (43%), Positives = 353/570 (61%), Gaps = 30/570 (5%)
Query: 27 SAGDGDGSRTPLPSFANGIPSVKNTPARSSSHASPWPSPYPHGVNPSPSPARASTPKRFF 86
S+G SR P P + VK+ P RS H P + TP R
Sbjct: 11 SSGHNRSSRNPHPH--PPLTVVKSRPPRSPCSFMAVTIQKDHRTQPRRNATAKKTPTRH- 67
Query: 87 RRPFPPPSPAKHIRESLAKRLGKAKPKEGPIPEEHGVETDSQSLDKSFGYSKNFGAKYEL 146
P +RE + G+ HG ET FGY+K+F +Y +
Sbjct: 68 ------TPPHGKVREKVISNNGR----------RHG-ETIPYGKRVDFGYAKDFDHRYTI 110
Query: 147 GKEVGRGHFGHTCSARAKK-GDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLLKALSGH 205
GK +G G FG+T A KK GD VAVK I K+KMT IA+EDV+REVK+L+AL+GH
Sbjct: 111 GKLLGHGQFGYTYVATDKKTGDR----VAVKKIDKAKMTIPIAVEDVKREVKILQALTGH 166
Query: 206 KHLIKFHDACEDANNVYIVMELCEGGELLDRILSR-GGRYTEEDAKAIVLQILSVAAFCH 264
+++++F++A ED N+VYIVMELCEGGELLDRIL+R RY+E DA +V Q+L VAA CH
Sbjct: 167 ENVVRFYNAFEDKNSVYIVMELCEGGELLDRILARKDSRYSERDAAVVVRQMLKVAAECH 226
Query: 265 LQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYYVAPEVLHRS 324
L+G+VHRD+KPENFLF S ED+ +K DFGLSDFI+P ++ +DIVGSAYYVAPEVL R
Sbjct: 227 LRGLVHRDMKPENFLFKSTEEDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRR 286
Query: 325 YSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPWPSVSPEGKDFVK 384
E+D+WSIGVI+YILLCG RPFW +TE GIF+ VL+ P+F PWP++S KDFVK
Sbjct: 287 SGPESDVWSIGVISYILLCGRRPFWDKTEDGIFKEVLKNKPDFRRKPWPTISNSAKDFVK 346
Query: 385 RLLNKDYRKRMTAAQALTHPWLRD--ESRPIPLDILIYKLVKSYLHATPFKRAAVKALSK 442
+LL KD R R+TAAQAL+HPW+R+ ++ IP+DI + ++ ++ + K+ A++AL+
Sbjct: 347 KLLVKDPRARLTAAQALSHPWVREGGDASEIPIDISVLNNMRQFVKFSRLKQFALRALAT 406
Query: 443 ALTDDQLVYLRAQFRLLEPNRDGHVSLDNFKMALARHATDAMRESRVLDIIHTMEPLAYR 502
L +++L LR QF ++ +++G +SL+ + ALA+ ++++RV +I+ ++
Sbjct: 407 TLDEEELADLRDQFDAIDVDKNGVISLEEMRQALAKDHPWKLKDARVAEILQAIDSNTDG 466
Query: 503 KMDFEEFCAAATSTYQLEALDG--WEDIACAAFEHFESEGNRVISIEELARELNLGPSAY 560
+DF EF AAA QLE D W+ + AAFE F+ +G+ I+ EEL L S
Sbjct: 467 FVDFGEFVAAALHVNQLEEHDSEKWQQRSRAAFEKFDIDGDGFITAEELRMHTGLKGSIE 526
Query: 561 SVLRDWIRNTDGKLSLLGYTKFLHGVTLRS 590
+L + + DGK+SL + + L +++S
Sbjct: 527 PLLEEADIDNDGKISLQEFRRLLRTASIKS 556
>At5g66210 calcium-dependent protein kinase
Length = 523
Score = 413 bits (1062), Expect = e-115
Identities = 231/501 (46%), Positives = 323/501 (64%), Gaps = 16/501 (3%)
Query: 102 SLAKRLGKAKPKEGPIPEEHGVETDSQSLD------KSFGYSKNFGAKYELGKEVGRGHF 155
S ++R + K K P P + T ++ FGYSK+F Y +GK +G G F
Sbjct: 14 SSSRRSSQTKSKAAPTPIDTKASTKRRTGSIPCGKRTDFGYSKDFHDHYTIGKLLGHGQF 73
Query: 156 GHTCSA-RAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLLKALSGHKHLIKFHDA 214
G+T A GD VAVK + KSKM IA+EDV+REV++L ALSGH+++++FH+A
Sbjct: 74 GYTYVAIHRPNGDR----VAVKRLDKSKMVLPIAVEDVKREVQILIALSGHENVVQFHNA 129
Query: 215 CEDANNVYIVMELCEGGELLDRILSR-GGRYTEEDAKAIVLQILSVAAFCHLQGVVHRDL 273
ED + VYIVMELCEGGELLDRILS+ G RY+E+DA +V Q+L VA CHL G+VHRD+
Sbjct: 130 FEDDDYVYIVMELCEGGELLDRILSKKGNRYSEKDAAVVVRQMLKVAGECHLHGLVHRDM 189
Query: 274 KPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYYVAPEVLHRSYSVEADIWS 333
KPENFLF S D+ +K DFGLSDFI+P +R +DIVGSAYYVAPEVL R E+D+WS
Sbjct: 190 KPENFLFKSAQLDSPLKATDFGLSDFIKPGKRFHDIVGSAYYVAPEVLKRRSGPESDVWS 249
Query: 334 IGVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDYRK 393
IGVITYILLCG RPFW RTE GIF+ VLR P+F PW ++S KDFVK+LL KD R
Sbjct: 250 IGVITYILLCGRRPFWDRTEDGIFKEVLRNKPDFSRKPWATISDSAKDFVKKLLVKDPRA 309
Query: 394 RMTAAQALTHPWLRD--ESRPIPLDILIYKLVKSYLHATPFKRAAVKALSKALTDDQLVY 451
R+TAAQAL+H W+R+ + IP+DI + ++ ++ + K+ A++AL+ L + ++
Sbjct: 310 RLTAAQALSHAWVREGGNATDIPVDISVLNNLRQFVRYSRLKQFALRALASTLDEAEISD 369
Query: 452 LRAQFRLLEPNRDGHVSLDNFKMALARHATDAMRESRVLDIIHTMEPLAYRKMDFEEFCA 511
LR QF ++ +++G +SL+ + ALA+ +++SRV +I+ ++ +DF EF A
Sbjct: 370 LRDQFDAIDVDKNGVISLEEMRQALAKDLPWKLKDSRVAEILEAIDSNTDGLVDFTEFVA 429
Query: 512 AATSTYQLEALDG--WEDIACAAFEHFESEGNRVISIEELARELNLGPSAYSVLRDWIRN 569
AA +QLE D W+ + AAFE F+ + + I+ EEL L S +L + +
Sbjct: 430 AALHVHQLEEHDSEKWQLRSRAAFEKFDLDKDGYITPEELRMHTGLRGSIDPLLDEADID 489
Query: 570 TDGKLSLLGYTKFLHGVTLRS 590
DGK+SL + + L ++ S
Sbjct: 490 RDGKISLHEFRRLLRTASISS 510
>At4g36070 Calcium-dependent serine/threonine protein kinase
Length = 536
Score = 407 bits (1047), Expect = e-114
Identities = 225/527 (42%), Positives = 329/527 (61%), Gaps = 21/527 (3%)
Query: 73 SPSPARASTPKRFFRRPFPPPSPAKHIRESLAKRLGKAKPKEGPIPEEHGVETDSQSLDK 132
SP R T R P P SP + K K+ + + G+ +
Sbjct: 7 SPKATRRGTGSR---NP-NPDSPTQGKASEKVSNKNKKNTKKIQLRHQGGIPYGKRI--- 59
Query: 133 SFGYSKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDV 192
FGY+K+F +Y +GK +G G FG T A + VAVK I K+KMT I +EDV
Sbjct: 60 DFGYAKDFDNRYTIGKLLGHGQFGFTYVATDNNNGNR---VAVKRIDKAKMTQPIEVEDV 116
Query: 193 RREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAI 252
+REVK+L+AL GH++++ FH+A ED +YIVMELC+GGELLDRIL+ RYTE+DA +
Sbjct: 117 KREVKILQALGGHENVVGFHNAFEDKTYIYIVMELCDGGELLDRILAN--RYTEKDAAVV 174
Query: 253 VLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGS 312
V Q+L VAA CHL+G+VHRD+KPENFLF S E + +K DFGLSDFI+P + DIVGS
Sbjct: 175 VRQMLKVAAECHLRGLVHRDMKPENFLFKSTEEGSSLKATDFGLSDFIKPGVKFQDIVGS 234
Query: 313 AYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPW 372
AYYVAPEVL R E+D+WSIGVITYILLCG RPFW +T+ GIF V+R P+F ++PW
Sbjct: 235 AYYVAPEVLKRRSGPESDVWSIGVITYILLCGRRPFWDKTQDGIFNEVMRKKPDFREVPW 294
Query: 373 PSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWLRD--ESRPIPLDILIYKLVKSYLHAT 430
P++S KDFVK+LL K+ R R+TAAQAL+H W+++ E+ +P+DI + ++ ++ +
Sbjct: 295 PTISNGAKDFVKKLLVKEPRARLTAAQALSHSWVKEGGEASEVPIDISVLNNMRQFVKFS 354
Query: 431 PFKRAAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDNFKMALARHATDAMRESRVL 490
K+ A++AL+K + +D+L LR QF ++ +++G +SL+ + ALA+ ++++RV
Sbjct: 355 RLKQIALRALAKTINEDELDDLRDQFDAIDIDKNGSISLEEMRQALAKDVPWKLKDARVA 414
Query: 491 DIIHTMEPLAYRKMDFEEFCAAATSTYQLEALDG--WEDIACAAFEHFESEGNRVISIEE 548
+I+ + +DF EF AA QLE D W+ + AAF+ F+ +G+ I+ EE
Sbjct: 415 EILQANDSNTDGLVDFTEFVVAALHVNQLEEHDSEKWQQRSRAAFDKFDIDGDGFITPEE 474
Query: 549 L-----ARELNLGPSAYSVLRDWIRNTDGKLSLLGYTKFLHGVTLRS 590
L ++ L S +L + + DG++S+ + + L +L+S
Sbjct: 475 LRLNQCLQQTGLKGSIEPLLEEADVDEDGRISINEFRRLLRSASLKS 521
>At5g12180 calcium-dependent protein kinase
Length = 528
Score = 340 bits (872), Expect = 1e-93
Identities = 196/537 (36%), Positives = 304/537 (56%), Gaps = 54/537 (10%)
Query: 68 HGVNPSPSPARASTPKRFFRRPFPPPSPAKHIRESLAKRLGKAKPKEGPIPEEHGVETDS 127
+ N + A AS P+ P PPP+ K+GPI G +
Sbjct: 26 NAANSTGPTAEASVPQSKHAPPSPPPAT-----------------KQGPIGPVLGRPMED 68
Query: 128 QSLDKSFGYSKNFGAKYELGKEVGRGHFG--HTCSARAKKGDLKDQPVAVKIISKSKMTS 185
A Y LGKE+GRG FG H C+ +A A K I+K K+ +
Sbjct: 69 VK------------ASYSLGKELGRGQFGVTHLCTQKAT-----GHQFACKTIAKRKLVN 111
Query: 186 AIAIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYT 245
IEDVRREV+++ L+G ++++ A ED ++V++VMELC GGEL DRI+++G Y+
Sbjct: 112 KEDIEDVRREVQIMHHLTGQPNIVELKGAYEDKHSVHLVMELCAGGELFDRIIAKG-HYS 170
Query: 246 EEDAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEER 305
E A +++ I+ + CH GV+HRDLKPENFL ++ E++ +K DFGLS F +P E
Sbjct: 171 ERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLNKDENSPLKATDFGLSVFYKPGEV 230
Query: 306 LNDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADP 365
DIVGSAYY+APEVL R Y EADIWSIGV+ YILLCG PFWA +E+GIF +LR
Sbjct: 231 FKDIVGSAYYIAPEVLKRKYGPEADIWSIGVMLYILLCGVPPFWAESENGIFNAILRGHV 290
Query: 366 NFDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWLRD--ESRPIPLDILIYKLV 423
+F PWPS+SP+ KD VK++LN D ++R+TAAQ L HPW+++ E+ +PLD + +
Sbjct: 291 DFSSDPWPSISPQAKDLVKKMLNSDPKQRLTAAQVLNHPWIKEDGEAPDVPLDNAVMSRL 350
Query: 424 KSYLHATPFKRAAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDNFKMALARHATDA 483
K + FK+ A++ ++ L++++++ L+ F+ ++ + G ++L+ + LA+ T
Sbjct: 351 KQFKAMNNFKKVALRVIAGCLSEEEIMGLKEMFKGMDTDSSGTITLEELRQGLAKQGT-R 409
Query: 484 MRESRVLDIIHTMEPLAYRKMDFEEFCAAATSTYQLEALDGWEDIACAAFEHFESEGNRV 543
+ E V ++ + +D+ EF AA T + LD E+ +AF+HF+ + +
Sbjct: 410 LSEYEVQQLMEAADADGNGTIDYGEFIAA---TMHINRLDR-EEHLYSAFQHFDKDNSGY 465
Query: 544 ISIEELAREL-----NLGPSAYSVLRDWIRNTDGKLSLLGYTKFLHGVTLRSSLPRP 595
I++EEL + L N G ++ + + DG+++ Y +F+ +R P P
Sbjct: 466 ITMEELEQALREFGMNDGRDIKEIISEVDGDNDGRIN---YDEFV--AMMRKGNPDP 517
>At5g19360 calcium-dependent protein kinase - like
Length = 523
Score = 331 bits (848), Expect = 8e-91
Identities = 178/464 (38%), Positives = 282/464 (60%), Gaps = 23/464 (4%)
Query: 142 AKYELGKEVGRGHFG--HTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLL 199
+ Y LGKE+GRG FG H C+ +A A K I+K K+ + IEDVRREV+++
Sbjct: 66 SSYTLGKELGRGQFGVTHLCTQKAT-----GLQFACKTIAKRKLVNKEDIEDVRREVQIM 120
Query: 200 KALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSV 259
L+G ++++ A ED ++V++VMELC GGEL DRI+++G Y+E A +++ I+ +
Sbjct: 121 HHLTGQPNIVELKGAYEDKHSVHLVMELCAGGELFDRIIAKG-HYSERAAASLLRTIVQI 179
Query: 260 AAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYYVAPE 319
CH GV+HRDLKPENFL S+ E++ +K DFGLS F +P E DIVGSAYY+APE
Sbjct: 180 IHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKPGEVFKDIVGSAYYIAPE 239
Query: 320 VLHRSYSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPWPSVSPEG 379
VL R Y EADIWSIGV+ YILLCG PFWA +E+GIF +L +F PWP +SP+
Sbjct: 240 VLRRKYGPEADIWSIGVMLYILLCGVPPFWAESENGIFNAILSGQVDFSSDPWPVISPQA 299
Query: 380 KDFVKRLLNKDYRKRMTAAQALTHPWLRD--ESRPIPLDILIYKLVKSYLHATPFKRAAV 437
KD V+++LN D ++R+TAAQ L HPW+++ E+ +PLD + +K + FK+ A+
Sbjct: 300 KDLVRKMLNSDPKQRLTAAQVLNHPWIKEDGEAPDVPLDNAVMSRLKQFKAMNNFKKVAL 359
Query: 438 KALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDNFKMALARHATDAMRESRVLDIIHTME 497
+ ++ L++++++ L+ F+ ++ + G ++L+ + LA+ T + E V ++ +
Sbjct: 360 RVIAGCLSEEEIMGLKEMFKGMDTDNSGTITLEELRQGLAKQGT-RLSEYEVQQLMEAAD 418
Query: 498 PLAYRKMDFEEFCAAATSTYQLEALDGWEDIACAAFEHFESEGNRVISIEELAREL---- 553
+D+ EF AA T + LD E+ +AF+HF+ + + I+ EEL + L
Sbjct: 419 ADGNGTIDYGEFIAA---TMHINRLDR-EEHLYSAFQHFDKDNSGYITTEELEQALREFG 474
Query: 554 -NLGPSAYSVLRDWIRNTDGKLSLLGYTKFLHGVTLRSSLPRPR 596
N G ++ + + DG+++ Y +F+ + + P P+
Sbjct: 475 MNDGRDIKEIISEVDGDNDGRIN---YEEFVAMMRKGNPDPNPK 515
>At2g38910 putative calcium-dependent protein kinase
Length = 583
Score = 330 bits (846), Expect = 1e-90
Identities = 200/536 (37%), Positives = 300/536 (55%), Gaps = 35/536 (6%)
Query: 56 SSHASPWPSPYPHGVNPSPSPARASTPKRFFRRPFPPPSPAKHIRESLAKRLGKAKPKEG 115
SS P PS P P P A+ PPP P I E+ K + KE
Sbjct: 59 SSTVDPAPSTLPTPSTPPPPVKMANEE--------PPPKP---ITENKEDPNSKPQKKEA 107
Query: 116 PIPE--EHGVETDSQSLDKSFGYSKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKDQPV 173
+ G++ DS K+ +N Y +G+++G+G FG T KK +
Sbjct: 108 HMKRMASAGLQIDSVLGRKT----ENLKDIYSVGRKLGQGQFGTTFLCVDKK---TGKEF 160
Query: 174 AVKIISKSKMTSAIAIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGEL 233
A K I+K K+T+ +EDVRRE++++ LSGH ++I+ A EDA V++VME+C GGEL
Sbjct: 161 ACKTIAKRKLTTPEDVEDVRREIQIMHHLSGHPNVIQIVGAYEDAVAVHVVMEICAGGEL 220
Query: 234 LDRILSRGGRYTEEDAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLID 293
DRI+ RG YTE+ A + I+ V CH GV+HRDLKPENFLF S E+A +K ID
Sbjct: 221 FDRIIQRG-HYTEKKAAELARIIVGVIEACHSLGVMHRDLKPENFLFVSGDEEAALKTID 279
Query: 294 FGLSDFIRPEERLNDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTE 353
FGLS F +P E D+VGS YYVAPEVL + YS E D+WS GVI YILL G PFW TE
Sbjct: 280 FGLSVFFKPGETFTDVVGSPYYVAPEVLRKHYSHECDVWSAGVIIYILLSGVPPFWDETE 339
Query: 354 SGIFRTVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWLRDESRPI 413
GIF VL+ D +F PWPSVS KD V+R+L +D +KRMT + L HPW R + +
Sbjct: 340 QGIFEQVLKGDLDFISEPWPSVSESAKDLVRRMLIRDPKKRMTTHEVLCHPWARVDGVAL 399
Query: 414 --PLDILIYKLVKSYLHATPFKRAAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDN 471
PLD + ++ + K+ A+K ++++L+++++ L+ F++++ + GH++L+
Sbjct: 400 DKPLDSAVLSRLQQFSAMNKLKKIAIKVIAESLSEEEIAGLKEMFKMIDTDNSGHITLEE 459
Query: 472 FKMALARHATDAMRESRVLDIIHTMEPLAYRKMDFEEFCAAATSTYQLEALDGWEDIACA 531
K L R D +++S +L ++ + +D+ EF AA ++E ED
Sbjct: 460 LKKGLDRVGAD-LKDSEILGLMQAADIDNSGTIDYGEFIAAMVHLNKIEK----EDHLFT 514
Query: 532 AFEHFESEGNRVISIEEL---ARELNLGP-SAYSVLRDWIRNTDGKLSLLGYTKFL 583
AF +F+ +G+ I+ +EL ++ L +LR+ ++ DG++ Y++F+
Sbjct: 515 AFSYFDQDGSGYITRDELQQACKQFGLADVHLDDILREVDKDNDGRID---YSEFV 567
>At3g10660 calmodulin-domain protein kinase CDPK isoform 2
Length = 646
Score = 323 bits (827), Expect = 2e-88
Identities = 183/479 (38%), Positives = 280/479 (58%), Gaps = 22/479 (4%)
Query: 111 KPKEGPIPEEHGVETDSQSLDKSFGYSKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKD 170
KPK G+ T+S K+ +NF Y LG+++G+G FG T K
Sbjct: 157 KPKHMRRVSSAGLRTESVLQRKT----ENFKEFYSLGRKLGQGQFGTTFLCLEKG---TG 209
Query: 171 QPVAVKIISKSKMTSAIAIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEG 230
A K ISK K+ + +EDVRRE++++ L+GH ++I A ED V++VMELC G
Sbjct: 210 NEYACKSISKRKLLTDEDVEDVRREIQIMHHLAGHPNVISIKGAYEDVVAVHLVMELCSG 269
Query: 231 GELLDRILSRGGRYTEEDAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMK 290
GEL DRI+ RG YTE A + I+ V CH GV+HRDLKPENFLF SR ED+ +K
Sbjct: 270 GELFDRIIQRG-HYTERKAAELARTIVGVLEACHSLGVMHRDLKPENFLFVSREEDSLLK 328
Query: 291 LIDFGLSDFIRPEERLNDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWA 350
IDFGLS F +P+E D+VGS YYVAPEVL + Y E+D+WS GVI YILL G PFWA
Sbjct: 329 TIDFGLSMFFKPDEVFTDVVGSPYYVAPEVLRKRYGPESDVWSAGVIVYILLSGVPPFWA 388
Query: 351 RTESGIFRTVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWLRDE- 409
TE GIF VL D +F PWPS+S KD V+++L +D ++R+TA Q L HPW++ +
Sbjct: 389 ETEQGIFEQVLHGDLDFSSDPWPSISESAKDLVRKMLVRDPKRRLTAHQVLCHPWVQIDG 448
Query: 410 -SRPIPLDILIYKLVKSYLHATPFKRAAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVS 468
+ PLD + +K + FK+ A++ ++++L+++++ L+ F++++ + G ++
Sbjct: 449 VAPDKPLDSAVLSRMKQFSAMNKFKKMALRVIAESLSEEEIAGLKQMFKMIDADNSGQIT 508
Query: 469 LDNFKMALARHATDAMRESRVLDIIHTMEPLAYRKMDFEEFCAAATSTYQLEALDGWEDI 528
+ K L R + ++ES +LD++ + +D++EF AA ++E ED
Sbjct: 509 FEELKAGLKRVGAN-LKESEILDLMQAADVDNSGTIDYKEFIAATLHLNKIER----EDH 563
Query: 529 ACAAFEHFESEGNRVISIEEL---ARELNLGPSAY-SVLRDWIRNTDGKLSLLGYTKFL 583
AAF +F+ + + I+ +EL E + + ++RD ++ DG++ Y +F+
Sbjct: 564 LFAAFSYFDKDESGFITPDELQQACEEFGVEDARIEEMMRDVDQDKDGRID---YNEFV 619
>At5g04870 calcium-dependent protein kinase
Length = 610
Score = 322 bits (826), Expect = 3e-88
Identities = 182/479 (37%), Positives = 280/479 (57%), Gaps = 22/479 (4%)
Query: 111 KPKEGPIPEEHGVETDSQSLDKSFGYSKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKD 170
KPK G+ T+S K+ +NF Y LG+++G+G FG T K
Sbjct: 121 KPKHMKRVSSAGLRTESVLQRKT----ENFKEFYSLGRKLGQGQFGTTFLCVEKT---TG 173
Query: 171 QPVAVKIISKSKMTSAIAIEDVRREVKLLKALSGHKHLIKFHDACEDANNVYIVMELCEG 230
+ A K I+K K+ + +EDVRRE++++ L+GH ++I A ED V++VME C G
Sbjct: 174 KEFACKSIAKRKLLTDEDVEDVRREIQIMHHLAGHPNVISIKGAYEDVVAVHLVMECCAG 233
Query: 231 GELLDRILSRGGRYTEEDAKAIVLQILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMK 290
GEL DRI+ RG YTE A + I+ V CH GV+HRDLKPENFLF S+ ED+ +K
Sbjct: 234 GELFDRIIQRG-HYTERKAAELTRTIVGVVEACHSLGVMHRDLKPENFLFVSKHEDSLLK 292
Query: 291 LIDFGLSDFIRPEERLNDIVGSAYYVAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWA 350
IDFGLS F +P++ D+VGS YYVAPEVL + Y EAD+WS GVI YILL G PFWA
Sbjct: 293 TIDFGLSMFFKPDDVFTDVVGSPYYVAPEVLRKRYGPEADVWSAGVIVYILLSGVPPFWA 352
Query: 351 RTESGIFRTVLRADPNFDDLPWPSVSPEGKDFVKRLLNKDYRKRMTAAQALTHPWLRDE- 409
TE GIF VL D +F PWPS+S KD V+++L +D +KR+TA Q L HPW++ +
Sbjct: 353 ETEQGIFEQVLHGDLDFSSDPWPSISESAKDLVRKMLVRDPKKRLTAHQVLCHPWVQVDG 412
Query: 410 -SRPIPLDILIYKLVKSYLHATPFKRAAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVS 468
+ PLD + +K + FK+ A++ ++++L+++++ L+ F +++ ++ G ++
Sbjct: 413 VAPDKPLDSAVLSRMKQFSAMNKFKKMALRVIAESLSEEEIAGLKEMFNMIDADKSGQIT 472
Query: 469 LDNFKMALARHATDAMRESRVLDIIHTMEPLAYRKMDFEEFCAAATSTYQLEALDGWEDI 528
+ K L R + ++ES +LD++ + +D++EF AA ++E ED
Sbjct: 473 FEELKAGLKRVGAN-LKESEILDLMQAADVDNSGTIDYKEFIAATLHLNKIER----EDH 527
Query: 529 ACAAFEHFESEGNRVISIEELAREL-NLGPSAYSV---LRDWIRNTDGKLSLLGYTKFL 583
AAF +F+ +G+ I+ +EL + G + +RD ++ DG++ Y +F+
Sbjct: 528 LFAAFTYFDKDGSGYITPDELQQACEEFGVEDVRIEELMRDVDQDNDGRID---YNEFV 583
>At4g21940 calcium-dependent protein kinase like protein
Length = 554
Score = 317 bits (812), Expect = 1e-86
Identities = 184/519 (35%), Positives = 298/519 (56%), Gaps = 31/519 (5%)
Query: 88 RPFPPPSPA-KHIRESLAKRLGKAKPKEGPIPEEHGV----------ETDSQSLDKSFGY 136
+P PPSP +S ++KP I ++H + ET++ L K F
Sbjct: 39 KPQKPPSPQIPTTTQSNHHHQQESKPVNQQIEKKHVLTQPLKPIVFRETET-ILGKPFEE 97
Query: 137 SKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREV 196
+ Y LGKE+GRG FG T + K + A K I K K+T I+DV+RE+
Sbjct: 98 IRKL---YTLGKELGRGQFGITYTC---KENSTGNTYACKSILKRKLTRKQDIDDVKREI 151
Query: 197 KLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQI 256
++++ LSG +++++ A ED ++++VMELC G EL DRI+++G Y+E+ A ++ +
Sbjct: 152 QIMQYLSGQENIVEIKGAYEDRQSIHLVMELCGGSELFDRIIAQG-HYSEKAAAGVIRSV 210
Query: 257 LSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYYV 316
L+V CH GV+HRDLKPENFL S E+A +K DFGLS FI + DIVGSAYYV
Sbjct: 211 LNVVQICHFMGVIHRDLKPENFLLASTDENAMLKATDFGLSVFIEEGKVYRDIVGSAYYV 270
Query: 317 APEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPWPSVS 376
APEVL RSY E DIWS G+I YILLCG PFW+ TE GIF +++ + +FD PWPS+S
Sbjct: 271 APEVLRRSYGKEIDIWSAGIILYILLCGVPPFWSETEKGIFNEIIKGEIDFDSQPWPSIS 330
Query: 377 PEGKDFVKRLLNKDYRKRMTAAQALTHPWLRDESRP-IPLDILIYKLVKSYLHATPFKRA 435
KD V++LL KD ++R++AAQAL HPW+R P P+D + +K + K+
Sbjct: 331 ESAKDLVRKLLTKDPKQRISAAQALEHPWIRGGEAPDKPIDSAVLSRMKQFRAMNKLKKL 390
Query: 436 AVKALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDNFKMALARHATDAMRESRVLDIIHT 495
A+K ++++L+++++ L+ F ++ ++ G ++ + K LA+ + + E+ V ++
Sbjct: 391 ALKVIAESLSEEEIKGLKTMFANMDTDKSGTITYEELKNGLAKLGS-KLTEAEVKQLMEA 449
Query: 496 MEPLAYRKMDFEEFCAAATSTYQLEALDGWEDIACAAFEHFESEGNRVISIEEL---ARE 552
+ +D+ EF +A Y+ + ++ AF++F+ + + I+++EL +E
Sbjct: 450 ADVDGNGTIDYIEFISATMHRYRFDR----DEHVFKAFQYFDKDNSGFITMDELESAMKE 505
Query: 553 LNLGPSA--YSVLRDWIRNTDGKLSLLGYTKFLH-GVTL 588
+G A V+ + + DG+++ + + G+TL
Sbjct: 506 YGMGDEASIKEVIAEVDTDNDGRINYEEFCAMMRSGITL 544
>At5g23580 calcium-dependent protein kinase (pir||S71196)
Length = 490
Score = 315 bits (808), Expect = 3e-86
Identities = 171/444 (38%), Positives = 267/444 (59%), Gaps = 15/444 (3%)
Query: 137 SKNFGAKYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREV 196
+KN Y LG+ +G+G FG T K+ Q +A K I K K+ +DV RE+
Sbjct: 15 TKNVEDNYFLGQVLGQGQFGTTFLCTHKQ---TGQKLACKSIPKRKLLCQEDYDDVLREI 71
Query: 197 KLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQI 256
+++ LS + ++++ A ED NV++VMELCEGGEL DRI+ RG Y+E +A ++ I
Sbjct: 72 QIMHHLSEYPNVVRIESAYEDTKNVHLVMELCEGGELFDRIVKRG-HYSEREAAKLIKTI 130
Query: 257 LSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYYV 316
+ V CH GVVHRDLKPENFLF+S EDA +K DFGLS F P E +++VGSAYYV
Sbjct: 131 VGVVEACHSLGVVHRDLKPENFLFSSSDEDASLKSTDFGLSVFCTPGEAFSELVGSAYYV 190
Query: 317 APEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPWPSVS 376
APEVLH+ Y E D+WS GVI YILLCG PFWA +E GIFR +L+ F+ PWPS+S
Sbjct: 191 APEVLHKHYGPECDVWSAGVILYILLCGFPPFWAESEIGIFRKILQGKLEFEINPWPSIS 250
Query: 377 PEGKDFVKRLLNKDYRKRMTAAQALTHPWLRDE--SRPIPLDILIYKLVKSYLHATPFKR 434
KD +K++L + +KR+TA Q L HPW+ D+ + PLD + +K + K+
Sbjct: 251 ESAKDLIKKMLESNPKKRLTAHQVLCHPWIVDDKVAPDKPLDCAVVSRLKKFSAMNKLKK 310
Query: 435 AAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDNFKMALARHATDAMRESRVLDIIH 494
A++ +++ L+++++ L+ F++++ ++ G ++ + K ++ R ++ M ES + +++
Sbjct: 311 MALRVIAERLSEEEIGGLKELFKMIDTDKSGTITFEELKDSMRRVGSELM-ESEIQELLR 369
Query: 495 TMEPLAYRKMDFEEFCAAATSTYQLEALDGWEDIACAAFEHFESEGNRVISIEELA---R 551
+ +D+ EF AA +LE E+ AAF F+ + + I+IEEL +
Sbjct: 370 AADVDESGTIDYGEFLAATIHLNKLER----EENLVAAFSFFDKDASGYITIEELQQAWK 425
Query: 552 ELNLGPSAY-SVLRDWIRNTDGKL 574
E + S +++D ++ DG++
Sbjct: 426 EFGINDSNLDEMIKDIDQDNDGQI 449
>At3g51850 calcium-dependent like protein kinase
Length = 528
Score = 313 bits (802), Expect = 2e-85
Identities = 177/452 (39%), Positives = 269/452 (59%), Gaps = 18/452 (3%)
Query: 138 KNFGAKYELGKEVGRGHFGHT--CSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRRE 195
+N +Y L +E+GRG FG T C R+ + DL +A K ISK K+ +A+ IEDV+RE
Sbjct: 48 ENIEDRYLLDRELGRGEFGVTYLCIERSSR-DL----LACKSISKRKLRTAVDIEDVKRE 102
Query: 196 VKLLKALSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQ 255
V ++K L ++ +ACED N V++VMELCEGGEL DRI++RG YTE A +
Sbjct: 103 VAIMKHLPKSSSIVTLKEACEDDNAVHLVMELCEGGELFDRIVARG-HYTERAAAGVTKT 161
Query: 256 ILSVAAFCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYY 315
I+ V CH GV+HRDLKPENFLF ++ E++ +K IDFGLS F +P E+ ++IVGS YY
Sbjct: 162 IVEVVQLCHKHGVIHRDLKPENFLFANKKENSPLKAIDFGLSIFFKPGEKFSEIVGSPYY 221
Query: 316 VAPEVLHRSYSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPWPSV 375
+APEVL R+Y E DIWS GVI YILLCG PFWA +E G+ + +LR +F PWP++
Sbjct: 222 MAPEVLKRNYGPEIDIWSAGVILYILLCGVPPFWAESEQGVAQAILRGVIDFKREPWPNI 281
Query: 376 SPEGKDFVKRLLNKDYRKRMTAAQALTHPWLRDESRP--IPLDILIYKLVKSYLHATPFK 433
S K+ V+++L D ++R+TA Q L HPW+++ + +PL ++ +K + FK
Sbjct: 282 SETAKNLVRQMLEPDPKRRLTAKQVLEHPWIQNAKKAPNVPLGDVVKSRLKQFSVMNRFK 341
Query: 434 RAAVKALSKALTDDQLVYLRAQFRLLEPNRDGHVSLDNFKMALARHATDAMRESRVLDII 493
R A++ +++ L+ +++ ++ F ++ + DG VS++ K L +T + ES V +I
Sbjct: 342 RKALRVIAEFLSTEEVEDIKVMFNKMDTDNDGIVSIEELKAGLRDFSTQ-LAESEVQMLI 400
Query: 494 HTMEPLAYRKMDFEEFCAAATSTYQLEALDGWEDIACAAFEHFESEGNRVISIEELAREL 553
++ +D+ EF A + L+ + E + AF +F+ +GN I +EL L
Sbjct: 401 EAVDTKGKGTLDYGEFVAV---SLHLQKVANDEHLR-KAFSYFDKDGNGYILPQELCDAL 456
Query: 554 --NLGPSAYSVLRDWIRNTD-GKLSLLGYTKF 582
+ G V D + D K + Y +F
Sbjct: 457 KEDGGDDCVDVANDIFQEVDTDKDGRISYEEF 488
>At3g57530 calcium-dependent protein kinase
Length = 538
Score = 311 bits (796), Expect = 9e-85
Identities = 175/443 (39%), Positives = 267/443 (59%), Gaps = 20/443 (4%)
Query: 142 AKYELGKEVGRGHFGHTCSARAKKGDLKDQPVAVKIISKSKMTSAIAIEDVRREVKLLKA 201
+KY LG+E+GRG FG T K+ D A K I K K+ +A+ IEDVRREV++++
Sbjct: 61 SKYTLGRELGRGEFGVTYLCTDKE---TDDVFACKSILKKKLRTAVDIEDVRREVEIMRH 117
Query: 202 LSGHKHLIKFHDACEDANNVYIVMELCEGGELLDRILSRGGRYTEEDAKAIVLQILSVAA 261
+ H +++ + ED + V++VMELCEGGEL DRI++RG YTE A A+ I+ V
Sbjct: 118 MPEHPNVVTLKETYEDEHAVHLVMELCEGGELFDRIVARG-HYTERAAAAVTKTIMEVVQ 176
Query: 262 FCHLQGVVHRDLKPENFLFTSRSEDADMKLIDFGLSDFIRPEERLNDIVGSAYYVAPEVL 321
CH GV+HRDLKPENFLF ++ E A +K IDFGLS F +P ER N+IVGS YY+APEVL
Sbjct: 177 VCHKHGVMHRDLKPENFLFGNKKETAPLKAIDFGLSVFFKPGERFNEIVGSPYYMAPEVL 236
Query: 322 HRSYSVEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRADPNFDDLPWPSVSPEGKD 381
R+Y E DIWS GVI YILLCG PFWA TE G+ + ++R+ +F PWP VS KD
Sbjct: 237 KRNYGPEVDIWSAGVILYILLCGVPPFWAETEQGVAQAIIRSVLDFRRDPWPKVSENAKD 296
Query: 382 FVKRLLNKDYRKRMTAAQALTHPWLRDESRP--IPLDILIYKLVKSYLHATPFKRAAVKA 439
++++L+ D ++R+TA Q L HPWL++ + L + +K + K+ A++
Sbjct: 297 LIRKMLDPDQKRRLTAQQVLDHPWLQNAKTAPNVSLGETVRARLKQFTVMNKLKKRALRV 356
Query: 440 LSKALTDDQLVYLRAQFRLLEPNRDGHVSLDNFKMALAR--HATDAMRESRVLDIIHTME 497
+++ L+D++ +R F++++ ++ G +++D K+ L + HA + ++L ++
Sbjct: 357 IAEHLSDEEASGIREGFQIMDTSQRGKINIDELKIGLQKLGHAI-PQDDLQILMDAGDID 415
Query: 498 PLAYRKMDFEEFCAAATSTYQLEALDGWEDIACAAFEHFESEGNRVISIEELARELN--L 555
Y +D +EF A + ++ G ++ AF F+ N I IEEL L+ L
Sbjct: 416 RDGY--LDCDEFIAISVHLRKM----GNDEHLKKAFAFFDQNNNGYIEIEELREALSDEL 469
Query: 556 GPS---AYSVLRDWIRNTDGKLS 575
G S +++RD + DG++S
Sbjct: 470 GTSEEVVDAIIRDVDTDKDGRIS 492
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,100,685
Number of Sequences: 26719
Number of extensions: 678897
Number of successful extensions: 5857
Number of sequences better than 10.0: 995
Number of HSP's better than 10.0 without gapping: 368
Number of HSP's successfully gapped in prelim test: 632
Number of HSP's that attempted gapping in prelim test: 3119
Number of HSP's gapped (non-prelim): 1862
length of query: 596
length of database: 11,318,596
effective HSP length: 105
effective length of query: 491
effective length of database: 8,513,101
effective search space: 4179932591
effective search space used: 4179932591
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144389.13


