

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144375.6 - phase: 0
(1419 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
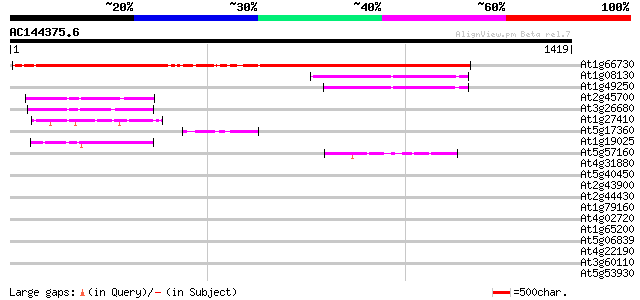
Score E
Sequences producing significant alignments: (bits) Value
At1g66730 hypothetical protein 1217 0.0
At1g08130 DNA ligase like protein 289 7e-78
At1g49250 hypothetical protein 247 4e-65
At2g45700 unknown protein 216 1e-55
At3g26680 unknown protein 211 2e-54
At1g27410 unknown protein (At1g27420) 119 2e-26
At5g17360 unknown protein 94 7e-19
At1g19025 unknown protein 82 2e-15
At5g57160 putative protein 47 6e-05
At4g31880 unknown protein 40 0.007
At5g40450 unknown protein 39 0.028
At2g43900 putative inositol polyphosphate 5'-phosphatase 37 0.081
At2g44430 unknown protein 36 0.18
At1g79160 unknown protein 35 0.31
At4g02720 unknown protein 34 0.53
At1g65200 hypothetical protein 34 0.53
At5g06839 bZip transcription factor AtbZip65 34 0.69
At4g22190 unknown protein 34 0.69
At3g60110 unknown protein 34 0.69
At5g53930 unknown protein 33 0.90
>At1g66730 hypothetical protein
Length = 1417
Score = 1217 bits (3150), Expect = 0.0
Identities = 668/1174 (56%), Positives = 826/1174 (69%), Gaps = 57/1174 (4%)
Query: 7 SQTLESLNSTQLYLNALQSLNLPPPTLPLPPLPSSTIPHSKLIPNTRFLIDSFR--HTTP 64
S+TL +LN+T+LY +A+ S++ P+ P P +IP+SK IPNT F++D FR H +
Sbjct: 20 SETL-NLNTTKLYSSAISSISPQFPS-PKPTSSCPSIPNSKRIPNTNFIVDLFRLPHQS- 76
Query: 65 SSFTYFLSHFHSDHYSPLSSSWSHGIIFCSPITSHLLINILHIPSPFVHPLSLNQSVVID 124
SS +FLSHFHSDHYS LSSSWS GII+CS T+ L+ IL +PS FV L +NQ V ID
Sbjct: 77 SSVAFFLSHFHSDHYSGLSSSWSKGIIYCSHKTARLVAEILQVPSQFVFALPMNQMVKID 136
Query: 125 GSVVTLIDANHCPGAVQFLFKVNETES--PRYVHTGDFRFNREMLLDLNLGEFIGADAVF 182
GS V LI+ANHCPGAVQFLFKV S +YVHTGDFRF EM D L F+G D VF
Sbjct: 137 GSEVVLIEANHCPGAVQFLFKVKLESSGFEKYVHTGDFRFCDEMRFDPFLNGFVGCDGVF 196
Query: 183 LDTTYCHPKFVFPTQNESVDYIVDVVKECDGENVLFLVATYVVGKEKILLEIARRCGKKV 242
LDTTYC+PKFVFP+Q ESV Y+V V+ + E VLFLVATYVVGKEKIL+EIARRC +K+
Sbjct: 197 LDTTYCNPKFVFPSQEESVGYVVSVIDKISEEKVLFLVATYVVGKEKILVEIARRCKRKI 256
Query: 243 CVDGKKMEVLRALGYGESGEFTEDRLESNVHVVGWNVLGETWPYFRPNFVRMKEIMVERG 302
VD +KM +L LG GE G FTED ES+VHVVGWNVLGETWPYFRPNFV+M EIMVE+G
Sbjct: 257 VVDARKMSMLSVLGCGEEGMFTEDENESDVHVVGWNVLGETWPYFRPNFVKMNEIMVEKG 316
Query: 303 YSKVVGFVPTGWTYEVKRDKFKVREKDSCKIHLVPYSEHSNYEELREYVRFLKPKKVVPT 362
Y KVVGFVPTGWTYEVKR+KF VR KDS +IHLVPYSEHSNY+ELRE+++FLKPK+V+PT
Sbjct: 317 YDKVVGFVPTGWTYEVKRNKFAVRFKDSMEIHLVPYSEHSNYDELREFIKFLKPKRVIPT 376
Query: 363 VGLDVEKSDSKHVDKMRKYFAGLVDETANKHEFLKGFKQCDSGRSGFEVGKDVGNDTEPG 422
VG+D+EK D K V+KM+K+F+GLVDE ANK +FL GF R ++ + D
Sbjct: 377 VGVDIEKFDCKEVNKMQKHFSGLVDEMANKKDFLLGFY-----RQSYQKNEKSDVDV-VS 430
Query: 423 HSVEKEVKPSDVGGDKSIDQDVAMSLSSCMGETCIEDPTLLNDEEKEKVVQELSCCLPTW 482
HS E + +K+ +D ++ S G + D T +D +++ +L LP W
Sbjct: 431 HSAEVYEEE-----EKNACEDGGENVPSSRG-PILHDTTPSSD---SRLLIKLRDSLPAW 481
Query: 483 VTRSQMLDLISISGSNVVEAVSNFFERETEFHEQVNSSQTPVPTHRSCSSNDTSPLSKSN 542
VT QMLDLI N V+ VSNF+E E E ++Q + P P S N+ + L +
Sbjct: 482 VTEEQMLDLIKKHAGNPVDIVSNFYEYEAELYKQ---ASLPTP-----SLNNQAVLFDDD 533
Query: 543 LKSFSSNDASPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKALSNQISPSKRKKGSESK 602
+ N K + + K DL R NL K ISP KR K S SK
Sbjct: 534 VTDLQPNPV----KGICPDVQAIQKGFDLPRKM-----NLTK---GTISPGKRGKSSGSK 581
Query: 603 SNKKVKVKAKSESSGSKQATITKFFGKAMPVMPGDTQSDQFGSKPGESPEVEELVPTDAG 662
SNKK K KS+ G Q T+ KFF K V+ G + S GS+ E +++V DA
Sbjct: 582 SNKKAKKDPKSKPVGPGQPTLFKFFNK---VLDGGSNSVSVGSETEECNTDKKMVHIDAS 638
Query: 663 NMYKQEIDQFMQIINGDESLKKQAITIIEEAKGDINKALDIYYSNSCNL-----GEREIS 717
YK+ DQF+ I+NG ESL+ A +II+EAKGDI++AL+IYYS + GER +S
Sbjct: 639 EAYKEVTDQFIDIVNGSESLRDYAASIIDEAKGDISRALNIYYSKPREIPGDHAGERGLS 698
Query: 718 VQ----GEC-KVDRPLEKKYVSKELNVIPDISMHRVLRDNVDATHVSLPSDKYNPKEHAC 772
+ +C + E K S+ +I + ++VD +VSLP +KY PKEHAC
Sbjct: 699 SKTIQYPKCSEACSSQEDKKASENSGHAVNICVQTSAEESVDKNYVSLPPEKYQPKEHAC 758
Query: 773 WRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLPAVYLCTNKIAA 832
WR+GQPAPY+HL RTF+ +E EKGKIKA S+LCNMFRSL LSPEDVLPAVYLCTNKIAA
Sbjct: 759 WREGQPAPYIHLVRTFASVESEKGKIKAMSMLCNMFRSLFALSPEDVLPAVYLCTNKIAA 818
Query: 833 DHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQRLLAPPTPLL 892
DHEN+ELNIGGSL+++ALEEACG +R +++MYN LGDLGDVAQ CRQTQ+LL PP PLL
Sbjct: 819 DHENIELNIGGSLISSALEEACGISRSTVRDMYNSLGDLGDVAQLCRQTQKLLVPPPPLL 878
Query: 893 IKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLRIGAMLRTVL 952
++D++S LRKISVQTG GST KK +I+ LMRSCREKE+KFLVRTL RNLRIGAMLRTVL
Sbjct: 879 VRDVFSTLRKISVQTGTGSTRLKKNLIVKLMRSCREKEIKFLVRTLARNLRIGAMLRTVL 938
Query: 953 PALAHAVVMNSRPTVYEEGTAEN-LKAALQVLSVAVVEAYNILPNLDIIVPTLMNKGIEF 1011
PAL A+VMNS + + +E+ + L+ +S AVVEAYNILP+LD++VP+LM+K IEF
Sbjct: 939 PALGRAIVMNSFWNDHNKELSESCFREKLEGVSAAVVEAYNILPSLDVVVPSLMDKDIEF 998
Query: 1012 SVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQRAQIHKLVDGSVLVF 1071
S S+LSMVPGIPIKPMLAKI G+ + L Q KAFTCEYKYDGQRAQIHKL+DG+V +F
Sbjct: 999 STSTLSMVPGIPIKPMLAKIAKGVQEFFNLSQEKAFTCEYKYDGQRAQIHKLLDGTVCIF 1058
Query: 1072 SRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGCRIMSFQELSSRGRGGK 1131
SRNGDE+TSRFPDLVD+IK+ P A TF++DAEVV DR NG ++MSFQELS+R RG K
Sbjct: 1059 SRNGDETTSRFPDLVDVIKQFSCPAAETFMLDAEVVATDRINGNKLMSFQELSTRERGSK 1118
Query: 1132 DTLVTKESIKVGICIFVFDIMFANGEH-LADPLK 1164
D L+T ESIKV +C+FVFDIMF NGE LA PL+
Sbjct: 1119 DALITTESIKVEVCVFVFDIMFVNGEQLLALPLR 1152
>At1g08130 DNA ligase like protein
Length = 790
Score = 289 bits (740), Expect = 7e-78
Identities = 156/399 (39%), Positives = 235/399 (58%), Gaps = 10/399 (2%)
Query: 761 PSDKYNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVL 820
P+D ++P++ +CW G+ P+L +A F L+ +E G+I T ILCNM R+++ +PED++
Sbjct: 158 PND-FDPEKMSCWEKGERVPFLFVALAFDLISNESGRIVITDILCNMLRTVIATTPEDLV 216
Query: 821 PAVYLCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQ 880
VYL N+IA HE VEL IG S + A+ EA G +K+ +LGDLG VA+ R
Sbjct: 217 ATVYLSANEIAPAHEGVELGIGESTIIKAISEAFGRTEDHVKKQNTELGDLGLVAKGSRS 276
Query: 881 TQRLLAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVR 940
TQ ++ P PL + ++ R+I+ ++G S +KK + L+ + + E +L R L
Sbjct: 277 TQTMMFKPEPLTVVKVFDTFRQIAKESGKDSNEKKKNRMKALLVATTDCEPLYLTRLLQA 336
Query: 941 NLRIGAMLRTVLPALAHAVVMNSRPTVYEEGTAENLKAALQVLSVAVVEAYNILPNLDII 1000
LR+G +TVL AL A V N N K+ L+ + V + + +LP DII
Sbjct: 337 KLRLGFSGQTVLAALGQAAVYNEE----HSKPPPNTKSPLEEAAKIVKQVFTVLPVYDII 392
Query: 1001 VPTLMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQRAQI 1060
VP L++ G+ + + G+PI PMLAK T G+ + L FQ+ FTCEYKYDG+RAQI
Sbjct: 393 VPALLSGGVWNLPKTCNFTLGVPIGPMLAKPTKGVAEILNKFQDIVFTCEYKYDGERAQI 452
Query: 1061 HKLVDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGCRIMSF 1120
H + DG+ ++SRN + +T ++PD+ + KP +FI+D EVV DR+ +I+ F
Sbjct: 453 HFMEDGTFEIYSRNAERNTGKYPDVALALSRLKKPSVKSFILDCEVVAFDREKK-KILPF 511
Query: 1121 QELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHL 1159
Q LS+R R V IKVG+CIF FD+++ NG+ L
Sbjct: 512 QILSTRARKN----VNVNDIKVGVCIFAFDMLYLNGQQL 546
>At1g49250 hypothetical protein
Length = 657
Score = 247 bits (630), Expect = 4e-65
Identities = 136/367 (37%), Positives = 212/367 (57%), Gaps = 12/367 (3%)
Query: 794 EKGKIKATSILCNMFRSLLVLSPEDVLPAVYLCTNKIAADHENVELNIG-GSLVTTALEE 852
E G+I T ILCNM R+++ +P+D+LP VYL N+IA HE ++L +G GS + A+ E
Sbjct: 58 ESGRIVITHILCNMLRTVIATTPDDLLPTVYLAANEIAPAHEGIKLGMGKGSYIIKAISE 117
Query: 853 ACGTNRLKIKEMYNKLGDLGDVAQECRQTQRLLAPPTPLLIKDIYSALRKISVQTGNGST 912
A G +++ Y +LGDLG VA R +Q ++ P PL + + LR I+ ++G GS
Sbjct: 118 AFGRTESHVEQQYTQLGDLGLVANGSRSSQTMIFMPKPLTVVKVADTLRLIAKESGKGSK 177
Query: 913 LRKKGIILHLMRSCREKEMKFLVRTLVRNLRIGAMLRTVLPALAHAVVMNSRPTVYEEGT 972
+KK ++ L+ + + E +L R L NLR+G +TVL AL A V N
Sbjct: 178 DKKKDLMKALLVATTDCEPLYLTRLLQDNLRLGFSRQTVLAALGQAAVYNEE----HSKP 233
Query: 973 AENLKAALQVLSVAVVEAYNILPNLDIIVPTLMNKGIEFSVSSLSMVPGIPIKPMLAKIT 1032
N+K L + V E +++LP DIIV L+ G+ + ++ G+P++PMLAK T
Sbjct: 234 PPNIKNPLDEAATIVKEVFSMLPVYDIIVGALLTSGVWNLPKTCNLTLGVPVRPMLAKAT 293
Query: 1033 NGIPQALKLFQNKAFTCEYKYDGQRAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMIKES 1092
+ L+ F++ FT EYKYDG+RAQI+ + DG+V +FSR+ + +T ++PD+ ++
Sbjct: 294 TRVDLILEKFKDTVFTAEYKYDGERAQIYYMEDGTVEIFSRHAERNTGKYPDVALVLSRL 353
Query: 1093 CKPVASTFIIDAEVVGIDRKNGCRIMSFQELSSRGRGGKDTLVTKESIKVGICIFVFDIM 1152
KP +FI+D EVV +R+ +I+ Q + V IKVG+C+F FDI+
Sbjct: 354 KKPTVKSFILDCEVVTFNREKE-KILPLQSTRAHKN------VNVSDIKVGVCVFAFDIL 406
Query: 1153 FANGEHL 1159
+ NG+ L
Sbjct: 407 YLNGQLL 413
>At2g45700 unknown protein
Length = 723
Score = 216 bits (549), Expect = 1e-55
Identities = 125/335 (37%), Positives = 191/335 (56%), Gaps = 22/335 (6%)
Query: 40 SSTIPHSKLIPNTRFLIDSFRHTTPSSFTYFLSHFHSDHYSPLSSSWSHGIIFCSPITSH 99
S IPH IP T F +D+F++ T +FL+HFH DHY L+ S+SHG I+CS +T+
Sbjct: 389 SKVIPHWNCIPGTPFRVDAFKYLTRDCCHWFLTHFHLDHYQGLTKSFSHGKIYCSLVTAK 448
Query: 100 LLINILHIPSPFVHPLSLNQSVVIDGSVVTLIDANHCPGAVQFLFKVNETESPRYVHTGD 159
L+ + IP + L L Q V I G VT DANHCPG++ LF+ +HTGD
Sbjct: 449 LVNMKIGIPWERLQVLDLGQKVNISGIDVTCFDANHCPGSIMILFE--PANGKAVLHTGD 506
Query: 160 FRFNREMLLDLNLGEFIGADAVFLDTTYCHPKFVFPTQNESVDYIVDVVK-ECDGENVLF 218
FR++ EM L +G I ++ LDTTYC+P++ FP Q + ++V+ ++ E LF
Sbjct: 507 FRYSEEMSNWL-IGSHI--SSLILDTTYCNPQYDFPKQEAVIQFVVEAIQAEAFNPKTLF 563
Query: 219 LVATYVVGKEKILLEIARRCGKKVCVDGKKMEVLRALGYGESG--EFTEDRLESNVHVVG 276
L+ +Y +GKE++ LE+AR +K+ ++ K+++L LG+ + FT ES++HVV
Sbjct: 564 LIGSYTIGKERLFLEVARVLREKIYINPAKLKLLECLGFSKDDIQWFTVKEEESHIHVVP 623
Query: 277 -WNVLGETWPYFRPNFVRMKEI--MVERGYSKVVGFVPTGWTYEVKRDKFKVREKDSCKI 333
W + +F R+K + YS +V F PTGWT + K R I
Sbjct: 624 LWTL---------ASFKRLKHVANRYTNRYSLIVAFSPTGWTSGKTKKKSPGRRLQQGTI 674
Query: 334 --HLVPYSEHSNYEELREYVRFLKPKKVVPTVGLD 366
+ VPYSEHS++ EL+E+V+ + P+ ++P+V D
Sbjct: 675 IRYEVPYSEHSSFTELKEFVQKVSPEVIIPSVNND 709
>At3g26680 unknown protein
Length = 484
Score = 211 bits (537), Expect = 2e-54
Identities = 120/329 (36%), Positives = 180/329 (54%), Gaps = 20/329 (6%)
Query: 44 PHSKLIPNTRFLIDSFRH-TTPSSFTYFLSHFHSDHYSPLSSSWSHGIIFCSPITSHLLI 102
P K +P T F +D+FR+ YFL+HFH+DHY L+ +WSHG I+CS +TS LL
Sbjct: 146 PFYKKLPGTPFTVDAFRYGCVQGCSAYFLTHFHADHYIGLTKAWSHGPIYCSSLTSRLLR 205
Query: 103 NILHIPSPFVHPLSLNQSVVIDGSVVTLIDANHCPGAVQFLFKVNETESPRYVHTGDFRF 162
L + +HPL L+ I+G VTLI+ANHCPGA F++ + Y+HTGDFR
Sbjct: 206 LSLSVNPSSIHPLELDVEYTINGIKVTLIEANHCPGAALIHFRL--LDGTCYLHTGDFRA 263
Query: 163 NREMLLDLNLGEFIGADAVFLDTTYCHPKFVFPTQNESVDYIVDVVKEC--DGENVLFLV 220
+++M L ++LDTTYC+P++ FP++ + + Y+V + K+ L +V
Sbjct: 264 SKQMQTHPLLFN-QRVHVLYLDTTYCNPRYKFPSKEDVLSYVVRITKDFLRKQPKTLIVV 322
Query: 221 ATYVVGKEKILLEIARRCGKKVCVDGKKMEVLRALGYGESGEFTEDRLESNVHVVGWNVL 280
+Y +GKE + L IA+ G K+ + + +L++ G+ D + N+ G
Sbjct: 323 GSYSIGKECVYLAIAKALGVKIFANASRRRILQSFGW--------DDISKNLSTDGKATC 374
Query: 281 GETWPYFRPNFVRMKEIM--VERGYSKVVGFVPTGWTYEVK----RDKFKVREKDSCKIH 334
P R+ E + Y V+ F PTGWTY K D K + I+
Sbjct: 375 LHVLPMSSLKVERLDEHLKIYREQYGAVLAFRPTGWTYSEKIGEHLDLIKPTSRGKITIY 434
Query: 335 LVPYSEHSNYEELREYVRFLKPKKVVPTV 363
VPYSEHS++ ELRE+V+FL+P K++PTV
Sbjct: 435 GVPYSEHSSFTELREFVQFLRPDKIIPTV 463
>At1g27410 unknown protein (At1g27420)
Length = 422
Score = 119 bits (297), Expect = 2e-26
Identities = 103/365 (28%), Positives = 172/365 (46%), Gaps = 55/365 (15%)
Query: 56 IDSFRHTTPSSFTYFLSHFHSDHYSPLSSSWSHGIIFCSPITSHLL--------INILHI 107
+D +R+ S YFL+H HSDH LS WS G ++CS T+ L +++L +
Sbjct: 8 VDRWRN---GSQAYFLTHIHSDHTRGLSGGWSQGPLYCSRTTASLFPSRFPGFDLSLLRV 64
Query: 108 PSPFVHPLSLNQSVVIDGSVVTL----IDANHCPGAVQFLFKVNETESPRYVHTGDFRFN 163
P SL+ GS V L IDA+HCPG++ FLF+ + +++TGDFR++
Sbjct: 65 -VPLFSWTSLSLRSPSSGSTVRLHLMAIDAHHCPGSIMFLFR---GDFGCFLYTGDFRWD 120
Query: 164 RE------MLLDLNLGEFIGADAVFLDTTYCHPKFVFPTQNESVDYIVDVVKECDGENVL 217
+ L + EF D ++LD TYC+P + FP++ +V + D++ ++
Sbjct: 121 SDASDEARTTLVAAIDEF-PVDILYLDNTYCNPIYSFPSRLVAVQLVADIIASHPSHDI- 178
Query: 218 FLVATYVVGKEKILLEIARRCGKKVCVDGKKMEVLRALGYGESGEFTEDRLESNVHVV-- 275
++A +GKE +L+ ++R K+ V +++ + LG+ + FT D + V V
Sbjct: 179 -IIAVDSLGKEDLLVHVSRILNIKIWVWPERLRTMHLLGFQDI--FTTDTSLTRVRAVPR 235
Query: 276 ---------GWNVLGETWPYFRPNFVRMKEIMVERGYSKVVG-FVPTGWTYEVKRDKFKV 325
G N + T +K +G K+ G F+ E K ++
Sbjct: 236 YSFSIQTLEGLNTMCPTIGIMPSGLPWVKRPF--KGDDKLSGSFLTASMKNETVSAKKEL 293
Query: 326 REKDSCKIH----LVPYSEHSNYEELREYVRFLKPKKVVPTVGLDVEKSDSKHVDKMRKY 381
K H V YS+HS YEE+ E+++ +KPK + V S S +VD + Y
Sbjct: 294 EAAAVHKFHDYMYSVHYSDHSCYEEIGEFIKLVKPKSMKGIV-----VSSSSYVDPL--Y 346
Query: 382 FAGLV 386
+ G +
Sbjct: 347 YFGRI 351
>At5g17360 unknown protein
Length = 175
Score = 93.6 bits (231), Expect = 7e-19
Identities = 66/194 (34%), Positives = 99/194 (51%), Gaps = 35/194 (18%)
Query: 437 DKSIDQDVAMSLSSCMGETCIEDPTLLNDEEKEKVVQELSCCLPTWVTRSQMLDLISISG 496
+K +D+ V+M L TC +E +++EL CLP WVT+ Q+ D+I +
Sbjct: 8 NKPLDEKVSMDL------TC----------GQEAILEELRICLPKWVTQEQVTDMIRGTC 51
Query: 497 SNVVEAVSNFFERETEFHEQVNSSQTPVPTHRSCSSNDTSPLSKSNLKSFSSNDASPFSK 556
N+VE V NF+E ET+F+EQV++ S +S SN +F
Sbjct: 52 RNIVEIVDNFYEHETKFYEQVSA---------VISFTSRDGISSSNHDAF---------V 93
Query: 557 SNLNNTNSTTKKLDLFR-SQESKLTNLRKALSNQISPSKRKKGSESKSNKKVKVKAKSES 615
S L + T++ + + SQ KL ++ + + ISP KR + + K KK K +K ES
Sbjct: 94 SKLEFIHCETERSESCQPSQFHKLKSMSSSQKSSISPVKRNRKIKGKPKKKGKASSKVES 153
Query: 616 SGSKQATITKFFGK 629
G KQA+ITKFF K
Sbjct: 154 GGPKQASITKFFNK 167
>At1g19025 unknown protein
Length = 549
Score = 82.0 bits (201), Expect = 2e-15
Identities = 93/343 (27%), Positives = 153/343 (44%), Gaps = 42/343 (12%)
Query: 54 FLIDSFRHTTPSSFT---YFLSHFHSDHYSPLSSSWSHGIIFCSPITSHLLINIL----- 105
F +D+F T + +FL+H H DH +S S I+ ++ L I++L
Sbjct: 11 FAVDTFGPYTETKRRKRHHFLTHAHKDHTVGVSPS---NIVVFPIYSTSLTISLLLQRFP 67
Query: 106 HIPSPFVHPLSLNQSVVIDGS----VVTLIDANHCPGAVQFLFKVNETESPRYVHTGDFR 161
+ + + + QSV++D VT DANHCPGAV FLF E +HTGD R
Sbjct: 68 QLDESYFVRVEIGQSVIVDDPDGEFKVTAFDANHCPGAVMFLF---EGSFGNILHTGDCR 124
Query: 162 FNREMLLDLNLGEFIGAD----------AVFLDTTYCHPKFV--FPTQNESVDYIVDVVK 209
+ L L +++G +FLD T+ FPT++ ++ I++ +
Sbjct: 125 LTLDCLHSLP-EKYVGRSHGMKPKCSLGYIFLDCTFGKSSHSQRFPTKHSAIRQIINCIW 183
Query: 210 ECDGENVLFLVATYVVGKEKILLEIARRCGKKVCVD-GKKMEVLRALGYGESGEFTEDRL 268
V++L A ++G+E +LLE++R G K+ VD +E R+L +ED
Sbjct: 184 NHPDAPVVYL-ACDMLGQEDVLLEVSRTFGSKIYVDKATNLECFRSLMVIVPEIVSEDP- 241
Query: 269 ESNVHVV-GWNVLGETWP---YFRPNFVRMKEIMVERGYSKVVGFVPTGW----TYEVKR 320
S H+ G+ L E + ++ + +++ V W + ++
Sbjct: 242 SSRFHIFSGFPKLYERTSAKLAEARSKLQSEPLIIRPSAQWYVCDDEDDWKSGSIQKQRK 301
Query: 321 DKFKVREKDSCKIHLVPYSEHSNYEELREYVRFLKPKKVVPTV 363
+F KD + V YS HS+ EL ++ L PK VV TV
Sbjct: 302 VRFSEAVKDEFGLWHVCYSMHSSRAELESAMQLLSPKWVVSTV 344
>At5g57160 putative protein
Length = 1184
Score = 47.4 bits (111), Expect = 6e-05
Identities = 77/357 (21%), Positives = 148/357 (40%), Gaps = 62/357 (17%)
Query: 797 KIKATSILCNMFRSLL--VLSPEDVLPAVYLCTNKIAADHENVELNIGGSLVTTALEEAC 854
K K +S + FR L P D AV L + D E + S++ T L +A
Sbjct: 19 KSKTSSQKRSKFRKFLDTYCKPSDYFVAVRLIIPSL--DRERGSYGLKESVLATCLIDAL 76
Query: 855 GTNR-----LKIKEMYN--------KLGDLGDVAQECRQTQRLLAPPTPLLIKDIYSALR 901
G +R +++ G+ +A E Q ++ +A L IK++ L
Sbjct: 77 GISRDAPDAVRLLNWRKGGTAKAGANAGNFSLIAAEVLQRRQGMASGG-LTIKELNDLLD 135
Query: 902 KISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLRIGAMLRTVLPALAHAVVM 961
+++ + + K ++ L++ +EMK+++R ++++L++G
Sbjct: 136 RLA---SSENRAEKTLVLSTLIQKTNAQEMKWVIRIILKDLKLGM--------------- 177
Query: 962 NSRPTVYEE--GTAENLKAALQVLSVAVVEAYNILPNLDIIVPTLMNKGIEFSVSSLSMV 1019
S ++++E AE+L +N+ +L ++ L ++ + +
Sbjct: 178 -SEKSIFQEFHPDAEDL--------------FNVTCDLKLVCEKLRDRHQRHKRQDIEV- 221
Query: 1020 PGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQRAQIHKLVDGS-VLVFSRNGDES 1078
G ++P LA + A K K E K+DG R QIHK +G+ + FSRN +
Sbjct: 222 -GKAVRPQLAMRIGDVNAAWKKLHGKDVVAECKFDGDRIQIHK--NGTDIHYFSRNFLDH 278
Query: 1079 TSRFPDLVDMIKESCKPVASTFIIDAEVVGIDR--KNGCRIMSFQELSSRGRGGKDT 1133
+ + D+I ++ + I+D E++ D S QE++ R G D+
Sbjct: 279 SEYAHAMSDLIVQNI--LVDKCILDGEMLVWDTSLNRFAEFGSNQEIAKAAREGLDS 333
>At4g31880 unknown protein
Length = 873
Score = 40.4 bits (93), Expect = 0.007
Identities = 100/480 (20%), Positives = 183/480 (37%), Gaps = 70/480 (14%)
Query: 303 YSKVVGFVPTGWTYEVKRDKFKVREKDSCKIHLVPYSEHSNYEELREYVRFLKPK----K 358
YS +V + G +++D+ EK+ + H+ +E E+ R PK K
Sbjct: 243 YSNIVASICEGTFSALQQDQVVANEKEDSQGHIKRETEVEKAAEISTPERTDAPKDESGK 302
Query: 359 VVPTVGLDVEKSDSKHVDKMRKYFAGLVDETANKHEFLKGFKQCDSGRSGFEVGKDVGND 418
+ G+ + S D M+K D+T K E +Q D+ R+ D+ N
Sbjct: 303 SGVSNGVAQQNDSSVDTDSMKKQ-----DDTGAKDE----PQQLDNPRN-----TDLNNT 348
Query: 419 TEPGHSVEKEVKPSDVGGDKSIDQDVAMSLSSCMGETCIEDPTLLNDEEKEKVVQELSCC 478
TE VE +++ + S+ Q S ET E LL+ ++
Sbjct: 349 TEEKPDVEHQIEEKE-NESSSVKQADLSKDSDIKEET--EPAELLDSKD----------- 394
Query: 479 LPTWVTRSQMLDLISISGSNVVEAVSNFFERETEFHEQVNSSQTPVPTHRSCSSNDTSPL 538
V S +D S+V A S+ E E Q+ S+T S ++T+ +
Sbjct: 395 ----VLTSPPVD------SSVTAATSS--ENEKNKSVQILPSKT--------SGDETANV 434
Query: 539 SKSNLKSFSSNDASPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKALSN-QISPSKRKK 597
S ++ + P +N S+T+++ S ++ + S Q++ KK
Sbjct: 435 SSPSMAEELPEQSVPKKTANQKKKESSTEEVKPSASIATEEVSEEPNTSEPQVTKKSGKK 494
Query: 598 GSESKSNKKVKVKAKSESSGSKQATITKFFGKAMPVMPGDTQSDQFGSKPGESPEV-EEL 656
+ S K +K +S +K A ++ T+ + KPG + EE
Sbjct: 495 VASSSKTKPTVPPSKKSTSETKVAKQSEKKVVGSDNAQESTKPKEEKKKPGRGKAIDEES 554
Query: 657 VPTDAGNMYKQEIDQFMQIINGDESLKKQAITIIEEAKGDINKALDIYYSNSCNLGEREI 716
+ T +G+ K + S K A +EAK + ++ + +LG+ +
Sbjct: 555 LHTSSGDNEKPAV-----------SSGKLASKSKKEAKQTVEESPNSNTKRKRSLGQGKA 603
Query: 717 S----VQGECKVDRPLEKKYVSKELNVIPDI-SMHRVLRDNVDATHVSLPSDKYNPKEHA 771
S V KV P+++ Y + H V+ D+ D + L + K++P + +
Sbjct: 604 SGESLVGSRIKVWWPMDQAYYKGVVESYDAAKKKHLVIYDDGDQEILYLKNQKWSPLDES 663
Score = 30.4 bits (67), Expect = 7.6
Identities = 25/101 (24%), Positives = 39/101 (37%), Gaps = 2/101 (1%)
Query: 519 SSQTPVPTHRSCSSNDTSPLSKSNLKSFSSNDASPFSKSNLNNTNSTTKKLDLFRSQESK 578
SS+T + S D+ S+ S + + S+ K D+ S SK
Sbjct: 722 SSKTSQDDKTASKSKDSKEASREEEASSEEESEEEEPPKTVGKSGSSRSKKDI--SSVSK 779
Query: 579 LTNLRKALSNQISPSKRKKGSESKSNKKVKVKAKSESSGSK 619
+ + + PSK S+SKS V AKS++ K
Sbjct: 780 SGKSKASSKKKEEPSKATTSSKSKSGPVKSVPAKSKTGKGK 820
>At5g40450 unknown protein
Length = 2910
Score = 38.5 bits (88), Expect = 0.028
Identities = 65/325 (20%), Positives = 125/325 (38%), Gaps = 52/325 (16%)
Query: 387 DETANKHEFLKGFKQCDSGRSGFEVGKDVGNDTEPGHSVEKEVKPSDVGGDKSIDQDVAM 446
+ TA++ E LKG + E +++ + + +EV D + D+
Sbjct: 1238 ETTAHESESLKGDNHQEKNAEPVEATQNL----DDAEQISREVTV-----DTEREADITE 1288
Query: 447 SLSSCM-GETCIEDPTLLNDEEKEKVVQELSCCLPTWVTRSQMLDLISISGSNVVEAVSN 505
+ G T IE PT+ + E + E S L V +S
Sbjct: 1289 KIEKVQEGPTVIETPTI----QGEDIESETSLELKEEVDQSS------------------ 1326
Query: 506 FFERETEFHEQVNSSQTPVPTHRSCSSNDTSPLSKSN-LKSFSSNDASPFSKSNLNNTNS 564
++TE HE V P + DTS + ++ LK+ +N + P + +++ S
Sbjct: 1327 ---KDTEEHEHVLERDIPQCETLKAEAVDTSTVEEAAILKTLETNISEPEA---MHSETS 1380
Query: 565 TTKKLDLFRSQESKLTNLRKALSNQISPSKRKKGSESKSNKKVKVKAKSESSGSKQA--- 621
K+D + Q+ T SN++ S + + + + K ES GS+++
Sbjct: 1381 LDLKVD--KEQKEAETVKTVIFSNEVGTSDAQAEEFGEHTEPCSSEIKDESQGSEESVEV 1438
Query: 622 ----TITKFFGKAMPVMPGDTQSDQFGSKPGESPEVEELVPTDAGNMYKQEIDQFMQIIN 677
T+ + V D QS + P++ + T+ G+ + +EI ++
Sbjct: 1439 KSKETVQGESSEEKDVNMLDVQSGESEKYQENEPDISLVSKTENGDKF-EEIPSVVEGAG 1497
Query: 678 GDESLKKQAITIIEEAKGDINKALD 702
DE+ Q + +E + ++LD
Sbjct: 1498 LDETTHNQTLLDVESV---VKQSLD 1519
>At2g43900 putative inositol polyphosphate 5'-phosphatase
Length = 1305
Score = 37.0 bits (84), Expect = 0.081
Identities = 34/123 (27%), Positives = 52/123 (41%), Gaps = 5/123 (4%)
Query: 528 RSCSSNDTSPLSKSNLKSFSSNDASPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKALS 587
+S N+ SKS+ KS +++ KS+ ++ + ++KK D SK + S
Sbjct: 1111 QSLKKNEGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSD--GDSNSKSSKKSDGDS 1168
Query: 588 NQISPSKRKKGSESKSNKKVKVKAKSESSGSKQATITKFFGKAMPVMPGDTQSDQFGSKP 647
N S K S SKS+KK + S+SS K+ GDT S
Sbjct: 1169 NSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSC---SKSQKKSDGDTNSKSQKKGD 1225
Query: 648 GES 650
G+S
Sbjct: 1226 GDS 1228
Score = 33.1 bits (74), Expect = 1.2
Identities = 29/123 (23%), Positives = 50/123 (40%), Gaps = 5/123 (4%)
Query: 528 RSCSSNDTSPLSKSNLKSFSSNDASPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKALS 587
+S +D SKS+ KS +++ KS+ ++ + ++KK D +S+ K
Sbjct: 1159 KSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSCSKSQ-----KKSD 1213
Query: 588 NQISPSKRKKGSESKSNKKVKVKAKSESSGSKQATITKFFGKAMPVMPGDTQSDQFGSKP 647
+ +KKG S+K K SS S + K+ GD+ S
Sbjct: 1214 GDTNSKSQKKGDGDSSSKSHKKNDGDSSSKSHKKNDGDSSSKSHKKSDGDSSSKSHKKSE 1273
Query: 648 GES 650
G+S
Sbjct: 1274 GDS 1276
Score = 31.6 bits (70), Expect = 3.4
Identities = 25/85 (29%), Positives = 42/85 (49%), Gaps = 2/85 (2%)
Query: 532 SNDTSPLSKSNLKSFSSNDASPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKALSNQIS 591
SN ++ S+S K+ +++ KS+ ++ + ++KK D SK + SN S
Sbjct: 1103 SNPSNSKSQSLKKNEGDSNSKSSKKSDGDSNSKSSKKSD--GDSNSKSSKKSDGDSNSKS 1160
Query: 592 PSKRKKGSESKSNKKVKVKAKSESS 616
K S SKS+KK + S+SS
Sbjct: 1161 SKKSDGDSNSKSSKKSDGDSNSKSS 1185
>At2g44430 unknown protein
Length = 646
Score = 35.8 bits (81), Expect = 0.18
Identities = 30/107 (28%), Positives = 45/107 (42%), Gaps = 3/107 (2%)
Query: 544 KSFSSNDASPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKALSNQISPSKRKKGSESKS 603
K+ SSND S SK + T T D +S L L+K + + + K G K
Sbjct: 521 KTNSSNDNS--SKQDTGKTEKKTVSADKKKSVADFLKRLKKNSPQKEAKDQNKSGGNVKK 578
Query: 604 NKKVKVK-AKSESSGSKQATITKFFGKAMPVMPGDTQSDQFGSKPGE 649
+ K K + +S S G K+A + K P P ++ S G+
Sbjct: 579 DSKTKPRELRSSSVGKKKAEVENTPVKRAPGRPQKKTAEATASASGK 625
>At1g79160 unknown protein
Length = 239
Score = 35.0 bits (79), Expect = 0.31
Identities = 19/52 (36%), Positives = 28/52 (53%), Gaps = 3/52 (5%)
Query: 5 SKSQTLESLNSTQLYLNALQSLNLPPPTL---PLPPLPSSTIPHSKLIPNTR 53
+ S ++ + + T + Q LN PPPTL PL L T P KL+P++R
Sbjct: 87 TSSSSITNADQTNQTVPTFQDLNFPPPTLNNSPLLNLFDDTTPELKLLPSSR 138
>At4g02720 unknown protein
Length = 422
Score = 34.3 bits (77), Expect = 0.53
Identities = 44/184 (23%), Positives = 76/184 (40%), Gaps = 22/184 (11%)
Query: 528 RSCSSNDTSPLSKSNLK-SFSSNDASPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKAL 586
RS S SK K S+ S+ S S+S+ + + R SK R +
Sbjct: 112 RSRKRKSKSSRSKRRRKRSYDSDSESEGSESDSEEEDRRRR-----RKSSSKRKKSRSSR 166
Query: 587 SNQISPSKRKKG----SESKSNKKVKVKAKSESSGSKQATITK---------FFGKAMPV 633
S + S R+K S+ S++ K + + SSG ++ T +K K
Sbjct: 167 SFRKKRSHRRKTKYSDSDESSDEDSKAEISASSSGEEEDTKSKSKRRKKSSDSSSKRSKG 226
Query: 634 MPGDTQSDQFGSKPGESPEVEELVPTDAGNMYKQEIDQFMQIINGDESLKKQAITIIEEA 693
+ SD G++ +V+E V + ++E+ +F ++I E KK + EE
Sbjct: 227 EKTKSGSDSDGTEEDSKMQVDETVKNTELELDEEELKKFKEMI---ELKKKSSAVDEEEE 283
Query: 694 KGDI 697
+GD+
Sbjct: 284 EGDV 287
>At1g65200 hypothetical protein
Length = 1101
Score = 34.3 bits (77), Expect = 0.53
Identities = 34/142 (23%), Positives = 58/142 (39%), Gaps = 24/142 (16%)
Query: 576 ESKLTNLRKALSNQISPSKRKKGSESKSNKKVKVKAKSESSGSKQATITKFFGKAMPVMP 635
E+ L + K PSK+KK +K+NKK S + Q +
Sbjct: 697 EADLISEEKQEEGMTLPSKKKK---NKNNKKNSTSMSSHLDKTVQHEHS----------- 742
Query: 636 GDTQSDQFGSKPGESPEVEELVPTDAG------NMYKQEIDQFMQIINGDESLKKQAITI 689
+ + P P E+ + +++G N+ QE + + ++G++SL K ++
Sbjct: 743 --ANLELDSTSPSLKPVEEDTLTSESGLLEMKSNINNQE--ETTKALHGEDSLSKHPVSA 798
Query: 690 IEEAKGDINKALDIYYSNSCNL 711
EEA N LD+ CNL
Sbjct: 799 HEEATTRYNSILDMVLKALCNL 820
>At5g06839 bZip transcription factor AtbZip65
Length = 418
Score = 33.9 bits (76), Expect = 0.69
Identities = 21/58 (36%), Positives = 33/58 (56%), Gaps = 2/58 (3%)
Query: 2 APKSKSQTLESLNSTQLYLNALQSLNLPPPTLPLPPLPSSTIPHSKLIPNTRFLIDSF 59
A ++ SQ LE+LN Q +++ S +LPP + PLPP S+ + H L N ++ F
Sbjct: 310 AEEALSQGLEALN--QSLSDSIVSDSLPPASAPLPPHLSNFMSHMSLALNKLSALEGF 365
>At4g22190 unknown protein
Length = 387
Score = 33.9 bits (76), Expect = 0.69
Identities = 35/135 (25%), Positives = 53/135 (38%), Gaps = 24/135 (17%)
Query: 494 ISGSNVVEAVSNFFERETEFHE---------QVNSSQTPVPTHRSCSSNDTSPLSKSNLK 544
++ S+ +A S+ + FH+ V S P C D SP S +L
Sbjct: 88 LTNSSFPQADSDLLSADELFHDGVLLPLDLLSVKSELQSDPNIAECDP-DPSP-STGSLI 145
Query: 545 SFSSNDASPFSKSNLNNTNSTTKKL-DLFRSQESKLTNLRKALSNQISPSKRKKGSESKS 603
+ +D P S L + +K+ D+FR E+K P K++K E+K
Sbjct: 146 TEQKSDLEPGLGSELTRETTVSKRWRDIFRKSETK------------PPGKKEKVKENKK 193
Query: 604 NKKVKVKAKSESSGS 618
KK S SGS
Sbjct: 194 EKKKTGSGPSSGSGS 208
>At3g60110 unknown protein
Length = 644
Score = 33.9 bits (76), Expect = 0.69
Identities = 43/209 (20%), Positives = 75/209 (35%), Gaps = 20/209 (9%)
Query: 422 GHSVEKEVKPSDVGGDKS--IDQDVAMSLSSCMGETCIEDPTLLNDEEKEKVVQELSCCL 479
GH V K S V KS + SS + +T + DE+K + V E
Sbjct: 407 GHCVIKSEAESSVSRQKSSVLSLVPCKKKSSALKKTSPSSSSRQKDEKKSQEVSEEKIVT 466
Query: 480 PTWVT-----RSQMLDLISISGSNVVEAVSNFFERETEFHEQVNSSQTPVPTHRSCSSND 534
T T R ++ ++ N +++T+ + + S +
Sbjct: 467 TTATTSARSSRRTSKEIAVVAKDTKTGRAKNNIKKQTDTKTESSDDDDDEKEENSKTEKK 526
Query: 535 TSPLSKSNLKSFSSNDASPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKALSNQISPSK 594
T K ++ F K++ TT K +Q+ N++K +Q K
Sbjct: 527 TVADKKKSVADFLKR----IKKNSPQKGKETTSK-----NQKKNDGNVKKENDHQ----K 573
Query: 595 RKKGSESKSNKKVKVKAKSESSGSKQATI 623
+ G+ K N KVK + S+G K+ +
Sbjct: 574 KSDGNVKKENSKVKPRELRSSTGKKKVEV 602
>At5g53930 unknown protein
Length = 529
Score = 33.5 bits (75), Expect = 0.90
Identities = 17/44 (38%), Positives = 27/44 (60%)
Query: 575 QESKLTNLRKALSNQISPSKRKKGSESKSNKKVKVKAKSESSGS 618
+ S + + K SN++ K+KK +KS K ++K +SESSGS
Sbjct: 3 KSSSSSKIIKDSSNKLRSVKKKKSKRNKSKKIRRIKDESESSGS 46
Score = 30.4 bits (67), Expect = 7.6
Identities = 29/129 (22%), Positives = 50/129 (38%), Gaps = 10/129 (7%)
Query: 534 DTSPLSKSNLKSFSSNDASPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKALSNQISPS 593
D+S +S K S + S + + + S+ L+ S E +K S
Sbjct: 13 DSSNKLRSVKKKKSKRNKSKKIRRIKDESESSGSDSSLYSSSEDDYRRKKKRRSKLSKKR 72
Query: 594 KRKKGSESKSN----------KKVKVKAKSESSGSKQATITKFFGKAMPVMPGDTQSDQF 643
RK+ S S+S+ KK + K K E+ G K+ + + + T S+Q
Sbjct: 73 SRKRYSSSESDDDSDDDRLLKKKKRSKRKDENVGKKKKKVVSRKRRKRDLSSSSTSSEQS 132
Query: 644 GSKPGESPE 652
+ ES +
Sbjct: 133 DNDGSESDD 141
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,616,889
Number of Sequences: 26719
Number of extensions: 1410589
Number of successful extensions: 6222
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 58
Number of HSP's that attempted gapping in prelim test: 6097
Number of HSP's gapped (non-prelim): 138
length of query: 1419
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1307
effective length of database: 8,326,068
effective search space: 10882170876
effective search space used: 10882170876
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144375.6


