

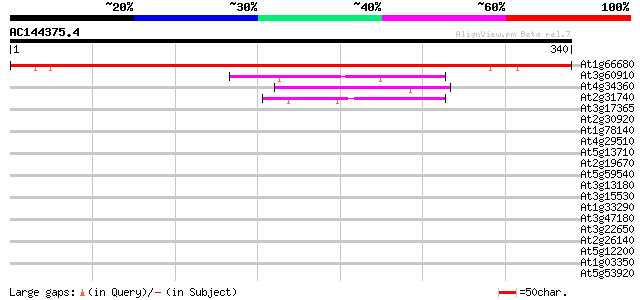
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144375.4 - phase: 0
(340 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g66680 AR401 488 e-138
At3g60910 unknown protein 60 2e-09
At4g34360 unknown protein 59 4e-09
At2g31740 unknown protein 51 8e-07
At3g17365 unknown protein 41 0.001
At2g30920 dihydroxypolyprenylbenzoate methyltransferase 36 0.034
At1g78140 unknown protein 36 0.034
At4g29510 arginine methyltransferase (pam1) 32 0.38
At5g13710 24-sterol C-methyltransferase 32 0.65
At2g19670 putative arginine N-methyltransferase 31 0.85
At5g59540 1-aminocyclopropane-1-carboxylate oxidase - like protein 31 1.1
At3g13180 putative sun (fmu) protein 31 1.1
At3g15530 unknown protein 30 1.4
At1g33290 unknown protein 30 1.4
At3g47180 zinc-finger protein-like protein 30 2.5
At3g22650 hypothetical protein 30 2.5
At2g26140 FtsH like protease 30 2.5
At5g12200 dihydropyrimidinase like protein 29 4.2
At1g03350 unknown protein 29 4.2
At5g53920 ribosomal protein L11 methyltransferase-like protein 28 5.5
>At1g66680 AR401
Length = 358
Score = 488 bits (1257), Expect = e-138
Identities = 250/358 (69%), Positives = 287/358 (79%), Gaps = 18/358 (5%)
Query: 1 MAGIRLQPEDSDVS--QQARAVALV------DLVSDDDRSIAADSWSIKSEYGSTLDDDQ 52
MAGIRL PE+ + + QQARA A V L SDDDRSIAADSWSIKSEYGSTLDDDQ
Sbjct: 1 MAGIRLLPEEPETTPQQQARAAAAVTTTTTDSLASDDDRSIAADSWSIKSEYGSTLDDDQ 60
Query: 53 RHADAAEALSNVNLRAASDYSSDKDEPDAEAVS-SMLGFQSYWDAAYTDELTNFHEHGHA 111
RHADAAEALS+ N R +SDYSSDK+EPDA+ SMLG QSYWDAAY+DELTNF EHGHA
Sbjct: 61 RHADAAEALSSANFRVSSDYSSDKEEPDADGGGQSMLGLQSYWDAAYSDELTNFREHGHA 120
Query: 112 GEVWFGDNVMEVVASWTKTLCIDISQGRLPNHVDDVKADAGELDDKLLSSWNVLDIGTGN 171
GEVWFGD+VME+V SWTK LC++ISQ + +DV + + DK LSSWNVLD+GTGN
Sbjct: 121 GEVWFGDDVMEIVTSWTKDLCVEISQRNMSVSENDVTTEVNDQADKYLSSWNVLDLGTGN 180
Query: 172 GLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDVLETKLEQVFQLVM 231
GLLL +LAK+GFSDLTGTDYS+ A+ LAQ L+ RDGFPNI+F+VDD+L+TKLEQ F+LVM
Sbjct: 181 GLLLHQLAKEGFSDLTGTDYSDGAVELAQHLSQRDGFPNIRFMVDDILDTKLEQQFKLVM 240
Query: 232 DKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCNSTKDELVQEVESFNQRKS- 290
DKGTLDAIGLHPDGPVKR+MYWDSVS+LVAPGGILV+TSCN TKDELV+EVE+FN RKS
Sbjct: 241 DKGTLDAIGLHPDGPVKRVMYWDSVSKLVAPGGILVITSCNHTKDELVEEVENFNIRKSN 300
Query: 291 -----ATAPEPEVAKDDESCR---DPLFQYVSHVRTYPTFMFGGSVGSRVATVAFLRK 340
++ E+ P F+Y+SHVRTYPTFMF GSVGSRVATVAFLRK
Sbjct: 301 LCRGDGNDANNVLSSGSEAASRIDQPPFEYLSHVRTYPTFMFSGSVGSRVATVAFLRK 358
>At3g60910 unknown protein
Length = 252
Score = 60.1 bits (144), Expect = 2e-09
Identities = 42/152 (27%), Positives = 66/152 (42%), Gaps = 23/152 (15%)
Query: 134 DISQGRLPNHVDDVKADAGELDDKLLSSW-------------------NVLDIGTGNGLL 174
D+S N+ D + DA + D L W VL +G GN L+
Sbjct: 4 DVSSCNTYNYGDALYWDARYVQDALSFDWYQCYSSLRPFVRSFVSTSSRVLMVGCGNSLM 63
Query: 175 LQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDVLETKL--EQVFQLVMD 232
+++ K G+ D+ D S AI + Q+ P +K++ DV + + F ++D
Sbjct: 64 SEDMVKDGYEDIMNVDISSVAIEMMQT--KYASVPQLKYMQMDVRDMSYFEDDSFDTIID 121
Query: 233 KGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGG 264
KGTLD++ D + VSRL+ PGG
Sbjct: 122 KGTLDSLMCGSDALLSASRMLGEVSRLIKPGG 153
>At4g34360 unknown protein
Length = 248
Score = 58.9 bits (141), Expect = 4e-09
Identities = 38/116 (32%), Positives = 60/116 (50%), Gaps = 9/116 (7%)
Query: 161 SWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDVLE 220
S +VL++G GN L +EL K G D+T D S A+ QS G+ IK + D+L+
Sbjct: 51 SSSVLELGCGNSQLCEELYKDGIVDITCIDLSSVAVEKMQSRLLPKGYKEIKVVQADMLD 110
Query: 221 TKLE-QVFQLVMDKGTLDAIGL--------HPDGPVKRMMYWDSVSRLVAPGGILV 267
+ + F +V++KGT+D + + P+ K M D V R++ P GI +
Sbjct: 111 LPFDSESFDVVIEKGTMDVLFVDAGDPWNPRPETVSKVMATLDGVHRVLKPDGIFI 166
>At2g31740 unknown protein
Length = 760
Score = 51.2 bits (121), Expect = 8e-07
Identities = 38/117 (32%), Positives = 59/117 (49%), Gaps = 9/117 (7%)
Query: 154 LDDKLLSSWNVLDI---GTGNGLLLQELAKQGFSDLTGTDYSERAIN--LAQSLANRDGF 208
L D SS + L I G GN L + L GF D+T D+S+ I+ L +++ R
Sbjct: 59 LQDSSSSSSDSLQILVPGCGNSRLTEHLYDAGFRDITNVDFSKVVISDMLRRNIRTR--- 115
Query: 209 PNIKFLVDDVLETKL-EQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGG 264
P +++ V D+ + +L ++ F V+DKG LDA+ G Y R++ PGG
Sbjct: 116 PELRWRVMDITKMQLADESFDTVLDKGALDALMEPEVGTKLGNQYLSEAKRVLKPGG 172
>At3g17365 unknown protein
Length = 239
Score = 40.8 bits (94), Expect = 0.001
Identities = 27/79 (34%), Positives = 42/79 (52%), Gaps = 6/79 (7%)
Query: 164 VLDIGTGNGLLLQELAKQGFSDLTGTDYSERAIN-LAQSLANRDGFPNIKFLVDDVLETK 222
VL IG GN + + G+ D+ D S I+ + + ++R P +K+L DV + K
Sbjct: 51 VLVIGCGNSAFSEGMVDDGYEDVVSIDISSVVIDTMIKKYSDR---PQLKYLKMDVRDMK 107
Query: 223 L--EQVFQLVMDKGTLDAI 239
+ F V+DKGTLD+I
Sbjct: 108 AFEDASFDAVIDKGTLDSI 126
>At2g30920 dihydroxypolyprenylbenzoate methyltransferase
Length = 322
Score = 35.8 bits (81), Expect = 0.034
Identities = 31/112 (27%), Positives = 56/112 (49%), Gaps = 10/112 (8%)
Query: 165 LDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPN-IKFLVDDVLETKL 223
+DIG G GLL + LA+ G + +TG D ++ + +A+ A+ D + I++L +
Sbjct: 136 IDIGCGGGLLSEPLARMG-ATVTGVDAVDKNVKIARLHADMDPVTSTIEYLCTTAEKLAD 194
Query: 224 E-QVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCNST 274
E + F V+ ++ H P + + S+S L P G V+++ N T
Sbjct: 195 EGRKFDAVLSLEVIE----HVANPAE---FCKSLSALTIPNGATVLSTINRT 239
>At1g78140 unknown protein
Length = 355
Score = 35.8 bits (81), Expect = 0.034
Identities = 32/118 (27%), Positives = 53/118 (44%), Gaps = 21/118 (17%)
Query: 163 NVLDIGTGNGLLLQELAKQG-FSDLTGTDYSERAINLAQSLANRD-GFPNIKFLVDDVLE 220
N++D G+G+ + + FS + DYSE + L N++ FPN + LV
Sbjct: 185 NIIDASCGSGMFSRLFTRSDLFSLVIALDYSENMLRQCYELLNKEENFPNKEKLV----- 239
Query: 221 TKLEQVFQLVMDKGTLDAI----GLH----PDGPVKRMMYWDSVSRLVAPGGILVVTS 270
+ +L G++DA+ LH P V +SR++ PGG+ V T+
Sbjct: 240 LVRADIARLPFLSGSVDAVHAGAALHCWPSPSSAVAE------ISRVLRPGGVFVATT 291
>At4g29510 arginine methyltransferase (pam1)
Length = 390
Score = 32.3 bits (72), Expect = 0.38
Identities = 20/63 (31%), Positives = 36/63 (56%), Gaps = 5/63 (7%)
Query: 164 VLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDVLETKL 223
VLD+G G G+L AK G + + + S+ A ++A+ + +GF + V VL+ K+
Sbjct: 111 VLDVGAGTGILSLFCAKAGAAHVYAVECSQMA-DMAKEIVKANGFSD----VITVLKGKI 165
Query: 224 EQV 226
E++
Sbjct: 166 EEI 168
>At5g13710 24-sterol C-methyltransferase
Length = 336
Score = 31.6 bits (70), Expect = 0.65
Identities = 34/158 (21%), Positives = 59/158 (36%), Gaps = 32/158 (20%)
Query: 98 YTDELTNFHEHG-----HAGEVWFGDNVMEVVASWTKTLCIDISQGRLPNHVDDVKADAG 152
Y D T+F+E+G H + W G+++ E + L + +
Sbjct: 48 YYDLATSFYEYGWGESFHFAQRWKGESLRESIKRHEHFLALQLG---------------- 91
Query: 153 ELDDKLLSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDG----- 207
+ VLD+G G G L+E+A+ S +TG + +E I + L G
Sbjct: 92 -----IQPGQKVLDVGCGIGGPLREIARFSNSVVTGLNNNEYQITRGKELNRLAGVDKTC 146
Query: 208 -FPNIKFLVDDVLETKLEQVFQLVMDKGTLDAIGLHPD 244
F F+ E + V+ + DA G + +
Sbjct: 147 NFVKADFMKMPFPENSFDAVYAIEATCHAPDAYGCYKE 184
>At2g19670 putative arginine N-methyltransferase
Length = 366
Score = 31.2 bits (69), Expect = 0.85
Identities = 20/63 (31%), Positives = 35/63 (54%), Gaps = 5/63 (7%)
Query: 164 VLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDVLETKL 223
VLD+G G G+L AK G + + + S+ A + A+ + +GF + V VL+ K+
Sbjct: 87 VLDVGAGTGILSLFCAKAGAAHVYAVECSQMA-DTAKEIVKSNGFSD----VITVLKGKI 141
Query: 224 EQV 226
E++
Sbjct: 142 EEI 144
>At5g59540 1-aminocyclopropane-1-carboxylate oxidase - like protein
Length = 366
Score = 30.8 bits (68), Expect = 1.1
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 4/52 (7%)
Query: 277 ELVQEVESFNQRKSAT---APEPEVAKD-DESCRDPLFQYVSHVRTYPTFMF 324
+L S N R S + AP+P ++ E+CRD +F+Y HV ++ +F
Sbjct: 143 DLYSSSPSVNWRDSFSCYIAPDPPAPEEIPETCRDAMFEYSKHVLSFGGLLF 194
>At3g13180 putative sun (fmu) protein
Length = 523
Score = 30.8 bits (68), Expect = 1.1
Identities = 16/43 (37%), Positives = 24/43 (55%)
Query: 254 DSVSRLVAPGGILVVTSCNSTKDELVQEVESFNQRKSATAPEP 296
DS S+LV GG+LV ++C+ +E VE+F R +P
Sbjct: 444 DSASKLVKHGGVLVYSTCSIDPEENEGRVEAFLLRHPEFTIDP 486
>At3g15530 unknown protein
Length = 288
Score = 30.4 bits (67), Expect = 1.4
Identities = 34/135 (25%), Positives = 58/135 (42%), Gaps = 19/135 (14%)
Query: 153 ELDDKLLSS---WN----VLDIGTGNGLLLQ----ELAKQGFSD-LTGTDYSERAINLAQ 200
E+ +++SS W+ LD+G G G+LL +L K G S + G D S+R
Sbjct: 97 EMAQRMVSSVGDWSCVKTALDLGCGRGILLNAVATQLKKTGSSGRVVGLDRSKRTTLSTL 156
Query: 201 SLANRDGFPNIKFLVDDVLETKL--EQVFQLVMDKGTLDAIGLH-----PDGPVKRMMYW 253
A +G + ++T + F +V+ + +G + +RM
Sbjct: 157 RTAKLEGVQEYVTCREGDVKTLPFGDNYFDVVVSSVFVHTVGKEHGQKSVEAAAERMRVL 216
Query: 254 DSVSRLVAPGGILVV 268
+ R+V PGG+ VV
Sbjct: 217 GEIVRVVKPGGLCVV 231
>At1g33290 unknown protein
Length = 379
Score = 30.4 bits (67), Expect = 1.4
Identities = 22/81 (27%), Positives = 36/81 (44%), Gaps = 8/81 (9%)
Query: 144 VDDVKADAGELDDKLLSSWNVLDIGTGNGLLLQELAKQ--------GFSDLTGTDYSERA 195
VD++ +A L + ++ V+ IGT +G LQ + K G +T D RA
Sbjct: 248 VDEIGTEAEALACRSIAERGVMLIGTAHGEQLQNIIKNPTLSDLIGGIETVTLGDEEARA 307
Query: 196 INLAQSLANRDGFPNIKFLVD 216
+S+ R P FL++
Sbjct: 308 RRSQKSILERKAPPTFYFLIE 328
>At3g47180 zinc-finger protein-like protein
Length = 210
Score = 29.6 bits (65), Expect = 2.5
Identities = 16/62 (25%), Positives = 30/62 (47%)
Query: 24 DLVSDDDRSIAADSWSIKSEYGSTLDDDQRHADAAEALSNVNLRAASDYSSDKDEPDAEA 83
D S+D+ I + + G +D+D+ + + SN NL D+ ++DE D +
Sbjct: 57 DEESEDEEEINENYYEYFDSNGFGVDEDEINEFLEDQESNSNLEEEDDFLEEEDEIDPDQ 116
Query: 84 VS 85
+S
Sbjct: 117 LS 118
>At3g22650 hypothetical protein
Length = 372
Score = 29.6 bits (65), Expect = 2.5
Identities = 22/69 (31%), Positives = 32/69 (45%), Gaps = 15/69 (21%)
Query: 80 DAEAVSSMLGFQSYWDAAYTDELTNFHEHGHAG--EVWFGDNVMEVVASWTKTLCIDISQ 137
D A+SS+ G D L+ H+ G EVW + + + V SWTK +D++
Sbjct: 238 DTAALSSLRG----------DRLSLLHQSGETMKIEVWITNKLSDEVVSWTK--YVDVTS 285
Query: 138 GRLPN-HVD 145
LP H D
Sbjct: 286 PDLPTLHTD 294
>At2g26140 FtsH like protease
Length = 717
Score = 29.6 bits (65), Expect = 2.5
Identities = 24/62 (38%), Positives = 30/62 (47%), Gaps = 6/62 (9%)
Query: 55 ADAAEALSNVNLRAASDYSSDKDEPDAEAVSS--MLGFQSYWDAAYTDE---LTNFHEHG 109
AD A ++ L+AA D S D D E M+G + A +DE LT FHE G
Sbjct: 431 ADLANLVNVAALKAAMDGSKDVTMSDLEFAKDRIMMGSERK-SAVISDESRKLTAFHEGG 489
Query: 110 HA 111
HA
Sbjct: 490 HA 491
>At5g12200 dihydropyrimidinase like protein
Length = 531
Score = 28.9 bits (63), Expect = 4.2
Identities = 20/69 (28%), Positives = 31/69 (43%), Gaps = 3/69 (4%)
Query: 170 GNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDVLETKLEQVFQL 229
G+G LQ+ G L GTD+ N Q D F I V+ LE ++ ++
Sbjct: 339 GHGKALQDALSTGILQLVGTDHC--TFNSTQKALGLDDFRRIPNGVNG-LEERMHLIWDT 395
Query: 230 VMDKGTLDA 238
+++ G L A
Sbjct: 396 MVESGQLSA 404
>At1g03350 unknown protein
Length = 470
Score = 28.9 bits (63), Expect = 4.2
Identities = 27/92 (29%), Positives = 42/92 (45%), Gaps = 11/92 (11%)
Query: 13 VSQQARAVALVDLVSDDDRSIAADSWSIKSEYGSTLD-------DDQRHADAAEALSNVN 65
V + + V+ + L +D S ++K E ST D+ AD+ +SNV
Sbjct: 267 VVEATKDVSRLKLEGNDGMGGGDVSETVKDEVESTYSVAKVSTQDEVTSADSVTEVSNVG 326
Query: 66 LRAASDYSSDKDEPDAEAVSSMLGFQSYWDAA 97
L+ D S +K E D+E V +S+ DAA
Sbjct: 327 LKTDKD-SEEKKETDSEEVPEE---KSFVDAA 354
>At5g53920 ribosomal protein L11 methyltransferase-like protein
Length = 371
Score = 28.5 bits (62), Expect = 5.5
Identities = 17/45 (37%), Positives = 21/45 (45%)
Query: 165 LDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFP 209
LD GTG+G+L K G + G D AIN A A + P
Sbjct: 225 LDYGTGSGILAIAALKFGAASSVGVDIDPLAINSAIHNAALNNIP 269
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,805,220
Number of Sequences: 26719
Number of extensions: 335542
Number of successful extensions: 713
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 697
Number of HSP's gapped (non-prelim): 27
length of query: 340
length of database: 11,318,596
effective HSP length: 100
effective length of query: 240
effective length of database: 8,646,696
effective search space: 2075207040
effective search space used: 2075207040
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144375.4


