

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144345.7 - phase: 0 /pseudo
(366 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
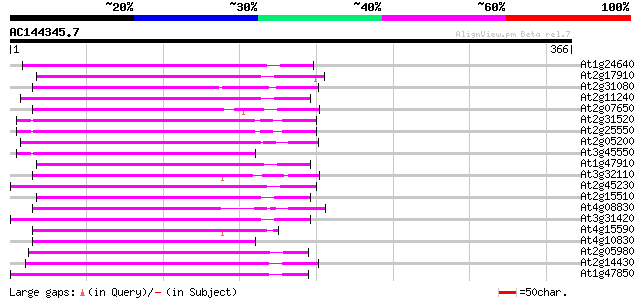
Score E
Sequences producing significant alignments: (bits) Value
At1g24640 hypothetical protein 129 2e-30
At2g17910 putative non-LTR retroelement reverse transcriptase 125 5e-29
At2g31080 putative non-LTR retroelement reverse transcriptase 123 2e-28
At2g11240 pseudogene 122 4e-28
At2g07650 putative non-LTR retrolelement reverse transcriptase 122 4e-28
At2g31520 putative non-LTR retroelement reverse transcriptase 121 5e-28
At2g25550 putative non-LTR retroelement reverse transcriptase 121 5e-28
At2g05200 putative non-LTR retroelement reverse transcriptase 121 7e-28
At3g45550 putative protein 118 4e-27
At1g47910 reverse transcriptase, putative 117 1e-26
At3g32110 non-LTR reverse transcriptase, putative 115 5e-26
At2g45230 putative non-LTR retroelement reverse transcriptase 113 1e-25
At2g15510 putative non-LTR retroelement reverse transcriptase 113 1e-25
At4g08830 putative protein 109 2e-24
At3g31420 hypothetical protein 109 3e-24
At4g15590 reverse transcriptase like protein 108 5e-24
At4g10830 putative protein 108 6e-24
At2g05980 putative non-LTR retroelement reverse transcriptase 108 6e-24
At2g14430 putative non-LTR retroelement reverse transcriptase 107 1e-23
At1g47850 hypothetical protein 106 2e-23
>At1g24640 hypothetical protein
Length = 1270
Score = 129 bits (325), Expect = 2e-30
Identities = 70/190 (36%), Positives = 104/190 (53%), Gaps = 8/190 (4%)
Query: 9 KLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLES 68
K+ + L DR+K +L W +R LS GG+ +LLK V ++PV+ +S FK P LES
Sbjct: 724 KVDMLHYLKDRLKEKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPITTCENLES 783
Query: 69 LFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTL 128
+F+W C+ RKI W WE +C K GGLG R ++ FN ALL K WR+L L
Sbjct: 784 AMASFWWDSCDHSRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLHFPDCL 843
Query: 129 WYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLST 188
R+L +RY + L + S W++IL R+ + + + +RVG+G S
Sbjct: 844 LSRLLKSRYFDATDFLDAALSQRPSFGWRSILFGRELLSK--------GLQKRVGDGASL 895
Query: 189 LFWVDPWLEE 198
W+DPW+++
Sbjct: 896 FVWIDPWIDD 905
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 125 bits (313), Expect = 5e-29
Identities = 65/192 (33%), Positives = 105/192 (53%), Gaps = 12/192 (6%)
Query: 18 DRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFWGG 77
++++ RL+ W ++ LS GG+ +LLK + ++PVY +S FK P + L ++ F+W
Sbjct: 748 EKLQSRLTGWYAKTLSQGGKEVLLKSIALALPVYVMSCFKLPKNLCQKLTTVMMDFWWNS 807
Query: 78 CEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCARY 137
+ RKI W+ W+ + K +GG G + L+ FN ALL K WR+LQE+G+L+ RV +RY
Sbjct: 808 MQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCFNQALLAKQAWRVLQEKGSLFSRVFQSRY 867
Query: 138 GEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPWLE 197
L + S W++IL R+ ++ + +GNG T W D WL
Sbjct: 868 FSNSDFLSATRGSRPSYAWRSILFGRE--------LLMQGLRTVIGNGQKTFVWTDKWLH 919
Query: 198 E----KPLS*RR 205
+ +PL+ RR
Sbjct: 920 DGSNRRPLNRRR 931
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 123 bits (308), Expect = 2e-28
Identities = 65/186 (34%), Positives = 103/186 (54%), Gaps = 5/186 (2%)
Query: 16 LVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFW 75
+++R+ RL+ WK R+LS+ GR+ L K VLSSIPV+ +S P + TL+ F W
Sbjct: 631 VLERVSARLAGWKGRSLSLAGRITLTKAVLSSIPVHVMSAILLPVSTLDTLDRYSRTFLW 690
Query: 76 GGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCA 135
G + +K + W +C K EGG+G+R R+ N AL+ K WR+LQ++ +LW RV+
Sbjct: 691 GSTMEKKKQHLLSWRKICKPKAEGGIGLRSARDMNKALVAKVGWRLLQDKESLWARVVRK 750
Query: 136 RYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPW 195
+Y + GG S W T V G+ ++ ++ + G+G + FW+D W
Sbjct: 751 KY-KVGGVQDTSWLKPQPRWSSTWRSVAVGLREV----VVKGVGWVPGDGCTIRFWLDRW 805
Query: 196 LEEKPL 201
L ++PL
Sbjct: 806 LLQEPL 811
>At2g11240 pseudogene
Length = 1044
Score = 122 bits (305), Expect = 4e-28
Identities = 65/189 (34%), Positives = 96/189 (50%), Gaps = 8/189 (4%)
Query: 8 RKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLE 67
+K + +VDRI++R W SR LS G+ +LK VL+S+P Y +S FK + ++
Sbjct: 586 KKRDLFNQIVDRIRQRSLSWSSRFLSTAGKTTMLKSVLASMPTYTMSCFKLLVSLCKRIQ 645
Query: 68 SLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGT 127
S F+W D +K+ WI W + K EGGLG + + FN ALL K WRI+Q
Sbjct: 646 SALTHFWWDSSADKKKMCWIAWSKMAKNKKEGGLGFKDITNFNDALLAKLSWRIVQSPSC 705
Query: 128 LWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLS 187
+ R+L +Y L CS S W+ I +D + + + +G+GL
Sbjct: 706 VLVRILLGKYCRTSSFLDCSVTAASSHGWRGICTGKD--------LIKSQLGKVIGSGLD 757
Query: 188 TLFWVDPWL 196
TL W +PWL
Sbjct: 758 TLVWNEPWL 766
>At2g07650 putative non-LTR retrolelement reverse transcriptase
Length = 732
Score = 122 bits (305), Expect = 4e-28
Identities = 67/196 (34%), Positives = 102/196 (51%), Gaps = 23/196 (11%)
Query: 16 LVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFW 75
+++++ RL+ WK R LS+ GR+ L K VLSSIPV+ +S P + L+ + +F W
Sbjct: 293 ILEKLTTRLAGWKGRFLSLAGRVTLTKAVLSSIPVHTMSTIALPKSTLDGLDKVSRSFLW 352
Query: 76 GGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCA 135
G RK I W+ VC + EGGLG+R+ ++ N ALL K WR++Q+ +LW R++
Sbjct: 353 GSSVTQRKQHLISWKRVCKPRSEGGLGIRKAQDMNKALLSKVGWRLIQDYHSLWARIMRC 412
Query: 136 RYGEEGGRLCCSCRDG---------GSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGL 186
Y + RDG S W L +R+ V + +S +G+G
Sbjct: 413 NYRVQ------DVRDGAWTKVRSVCSSTWRSVALGMREVV--------IPGLSWVIGDGR 458
Query: 187 STLFWVDPWLEEKPLS 202
LFW+D WL PL+
Sbjct: 459 EILFWMDKWLTNIPLA 474
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 121 bits (304), Expect = 5e-28
Identities = 72/196 (36%), Positives = 108/196 (54%), Gaps = 9/196 (4%)
Query: 5 GDPRKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIIS 64
G +K F Y ++DR+K+R S W +R LS G+ I+LK V ++PVY +S FK P GI+S
Sbjct: 914 GRKKKEMFEY-IIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCFKLPKGIVS 972
Query: 65 TLESLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQE 124
+ESL F+W + R I W+ W+ + K EGGLG R L +FN ALL K WR++Q
Sbjct: 973 EIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRLIQY 1032
Query: 125 RGTLWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGN 184
+L+ RV+ ARY ++ L R S W ++L DG+ + G +G+
Sbjct: 1033 PNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLL---DGIALLKKG-----TRHLIGD 1084
Query: 185 GLSTLFWVDPWLEEKP 200
G + +D ++ P
Sbjct: 1085 GQNIRIGLDNIVDSHP 1100
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 121 bits (304), Expect = 5e-28
Identities = 72/196 (36%), Positives = 108/196 (54%), Gaps = 9/196 (4%)
Query: 5 GDPRKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIIS 64
G +K F Y ++DR+K+R S W +R LS G+ I+LK V ++PVY +S FK P GI+S
Sbjct: 1140 GRKKKEMFEY-IIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCFKLPKGIVS 1198
Query: 65 TLESLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQE 124
+ESL F+W + R I W+ W+ + K EGGLG R L +FN ALL K WR++Q
Sbjct: 1199 EIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRLIQY 1258
Query: 125 RGTLWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGN 184
+L+ RV+ ARY ++ L R S W ++L DG+ + G +G+
Sbjct: 1259 PNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLL---DGIALLKKG-----TRHLIGD 1310
Query: 185 GLSTLFWVDPWLEEKP 200
G + +D ++ P
Sbjct: 1311 GQNIRIGLDNIVDSHP 1326
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 121 bits (303), Expect = 7e-28
Identities = 65/195 (33%), Positives = 103/195 (52%), Gaps = 9/195 (4%)
Query: 8 RKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLE 67
RK + ++D+I+++ W SR LS G+ ++LK VL+S+P+Y +S FK PS + ++
Sbjct: 655 RKRDIFGAIIDKIRQKSHSWASRFLSQAGKQVMLKAVLASMPLYSMSCFKLPSALCRKIQ 714
Query: 68 SLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGT 127
SL F+W D+RK +W+ W + + K GGLG R + N +LL K WR+L +
Sbjct: 715 SLLTRFWWDTKPDVRKTSWVAWSKLTNPKNAGGLGFRDIERCNDSLLAKLGWRLLNSPES 774
Query: 128 LWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLS 187
L R+L +Y + C S W++I+ R+ + + GW+ + NG
Sbjct: 775 LLSRILLGKYCHSSSFMECKLPSQPSHGWRSIIAGRE-ILKEGLGWL-------ITNGEK 826
Query: 188 TLFWVDPWLE-EKPL 201
W DPWL KPL
Sbjct: 827 VSIWNDPWLSISKPL 841
>At3g45550 putative protein
Length = 851
Score = 118 bits (296), Expect = 4e-27
Identities = 64/156 (41%), Positives = 92/156 (58%), Gaps = 1/156 (0%)
Query: 5 GDPRKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIIS 64
G +K F Y ++DR+K R + W ++ LS G+ ILLK V ++PVY +S FK P GI+S
Sbjct: 379 GRKKKEMFNY-IIDRVKERTASWSAKFLSPAGKEILLKSVALAMPVYAMSCFKLPQGIVS 437
Query: 65 TLESLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQE 124
+ESL F+W + R I W+ W+ + K EGGLG R L +FN ALL K WRI+Q
Sbjct: 438 EIESLLMNFWWEKASNKRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRIIQY 497
Query: 125 RGTLWYRVLCARYGEEGGRLCCSCRDGGSVWWQTIL 160
+L+ RV+ ARY ++ + R S W ++L
Sbjct: 498 PNSLFARVMKARYFKDNSIIDAKTRSQQSYGWSSLL 533
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 117 bits (292), Expect = 1e-26
Identities = 59/179 (32%), Positives = 96/179 (52%), Gaps = 8/179 (4%)
Query: 18 DRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFWGG 77
DR++ R++ W ++ LS GG+ +++K V +++P Y +S F+ P I S L S F+W
Sbjct: 554 DRLQSRINGWSAKFLSKGGKEVMIKSVAATLPRYVMSCFRLPKAITSKLTSAVAKFWWSS 613
Query: 78 CEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCARY 137
D R + W+ W+ +CS K +GGLG R + +FN ALL K WR++ +L+ +V RY
Sbjct: 614 NGDSRGMHWMAWDKLCSSKSDGGLGFRNVDDFNSALLAKQLWRLITAPDSLFAKVFKGRY 673
Query: 138 GEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPWL 196
+ L S W++++ R V + +RVG+G S W DPW+
Sbjct: 674 FRKSNPLDSIKSYSPSYGWRSMISARSLV--------YKGLIKRVGSGASISVWNDPWI 724
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 115 bits (287), Expect = 5e-26
Identities = 65/189 (34%), Positives = 97/189 (50%), Gaps = 9/189 (4%)
Query: 16 LVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFW 75
++ R RL+ WK R LS GRL L K VL+SI V+ +S K P + L+ + AF W
Sbjct: 1307 VIKRFSSRLAGWKGRMLSFAGRLTLTKAVLTSILVHTMSTIKLPQSTLDGLDKVSRAFLW 1366
Query: 76 GGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCA 135
G + +K + W VC + EGGLG+R N AL+ K WR+L + +LW +V+ +
Sbjct: 1367 GSTLEKKKQHLVAWTRVCLPRREGGLGIRSATAMNKALIAKVGWRVLNDGSSLWAQVVRS 1426
Query: 136 RY--GEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVD 193
+Y G+ R + S W+++ GVG D W + +G+G FW D
Sbjct: 1427 KYKVGDVHDRNWTVAKSNWSSTWRSV-----GVGLRDVIWREQHWV--IGDGRQIRFWTD 1479
Query: 194 PWLEEKPLS 202
WL E P++
Sbjct: 1480 RWLSETPIA 1488
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 113 bits (283), Expect = 1e-25
Identities = 66/200 (33%), Positives = 101/200 (50%), Gaps = 8/200 (4%)
Query: 1 LPIGGDPRKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPS 60
LP K+A L DR+ +++ W+S LS GG+ ILLK V ++P Y +S FK P
Sbjct: 753 LPESFQGSKVATLSYLKDRLGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPK 812
Query: 61 GIISTLESLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWR 120
I +ES+ F+W ++ R + W W + K GGLG + + FN+ALLGK WR
Sbjct: 813 TICQQIESVMAEFWWKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWR 872
Query: 121 ILQERGTLWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISR 180
++ E+ +L +V +RY + L S W++I + + + Q I
Sbjct: 873 MITEKDSLMAKVFKSRYFSKSDPLNAPLGSRPSFAWKSIYEAQVLIKQ--------GIRA 924
Query: 181 RVGNGLSTLFWVDPWLEEKP 200
+GNG + W DPW+ KP
Sbjct: 925 VIGNGETINVWTDPWIGAKP 944
>At2g15510 putative non-LTR retroelement reverse transcriptase
Length = 1138
Score = 113 bits (283), Expect = 1e-25
Identities = 67/179 (37%), Positives = 91/179 (50%), Gaps = 8/179 (4%)
Query: 18 DRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFWGG 77
D++K RLS W +R LS+GG+ ILLK V ++ VY +S FK L S + F+W
Sbjct: 546 DKLKSRLSGWFARTLSLGGKEILLKAVAMAMQVYAMSCFKLTKTTCKNLTSAMSDFWWNA 605
Query: 78 CEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCARY 137
E RK W+ WE +C K GGLG R + FN ALL K WRILQ L+ R +RY
Sbjct: 606 LEHKRKTHWVSWEKLCLAKESGGLGFRDIESFNQALLAKQSWRILQYPSFLFARFFKSRY 665
Query: 138 GEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPWL 196
++ L S +IL R+ + + + VGNG S W+DPW+
Sbjct: 666 FDDEEFLEADLGVRPSYACCSILFGRE--------LLAKGLRKEVGNGKSLNVWMDPWI 716
>At4g08830 putative protein
Length = 947
Score = 109 bits (273), Expect = 2e-24
Identities = 63/191 (32%), Positives = 96/191 (49%), Gaps = 29/191 (15%)
Query: 16 LVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFW 75
+++R+ RL+ WK R+LS GRL L K VLS IP++ +S P + L+ L F
Sbjct: 445 VLERVSSRLAGWKGRSLSFAGRLTLTKSVLSLIPIHTMSTISLPQSTLEGLDKLARVFLL 504
Query: 76 GGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCA 135
G + +K+ + W+ VC K EGGLG+R + N AL+ K WR++ +R +LW R+L +
Sbjct: 505 GSSAEKKKLHLVAWDRVCLPKSEGGLGIRTSKCMNKALVSKVGWRLINDRYSLWARILRS 564
Query: 136 RYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPW 195
+Y + +R+ V + S W+ VGNG LFW D W
Sbjct: 565 KYR---------------------VGLREVVSR-GSRWV-------VGNGRDILFWSDNW 595
Query: 196 LEEKPLS*RRI 206
L + L R +
Sbjct: 596 LSHEALINRAV 606
>At3g31420 hypothetical protein
Length = 1491
Score = 109 bits (272), Expect = 3e-24
Identities = 62/196 (31%), Positives = 97/196 (48%), Gaps = 8/196 (4%)
Query: 1 LPIGGDPRKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPS 60
LP + +K + +++++K + W + LS GG+ +LLK V ++PVY ++ FK
Sbjct: 911 LPEQFNKKKKELFNYIIEKVKDKTQGWSKKFLSQGGKEVLLKAVALAMPVYSMNIFKLTK 970
Query: 61 GIISTLESLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWR 120
+ ++SL F+W + + + W W+ + K EGGLG + L FNLALLGK W
Sbjct: 971 EVCEEIDSLLARFWWSSGNETKGMHWFTWKRMSIPKKEGGLGFKELENFNLALLGKQTWH 1030
Query: 121 ILQERGTLWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISR 180
+LQ L RVL RY E + S W++IL R+ + +
Sbjct: 1031 LLQHPNCLMARVLRGRYFPETNVMNAVQGRRASFVWKSILHGRN--------LLKKGLRC 1082
Query: 181 RVGNGLSTLFWVDPWL 196
VG+G W+DPWL
Sbjct: 1083 CVGDGSLINAWLDPWL 1098
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 108 bits (270), Expect = 5e-24
Identities = 58/163 (35%), Positives = 89/163 (54%), Gaps = 6/163 (3%)
Query: 16 LVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFW 75
+++R+ RLS WKSR+LS+ GR+ L K VL SIP++ +S P+ ++ L+ + F W
Sbjct: 566 VLERVSSRLSGWKSRSLSLAGRITLTKAVLMSIPIHTMSSILLPASLLEQLDKVSRNFLW 625
Query: 76 GGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCA 135
G + RK + W+ VC K GGLG+R ++ N ALL K WR+L ++ +LW RVL
Sbjct: 626 GSTVEKRKQHLLSWKKVCRPKAAGGLGLRASKDMNRALLAKVGWRLLNDKVSLWARVLRR 685
Query: 136 RY---GEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWML 175
+Y S W + +R+GV + GW+L
Sbjct: 686 KYKVTDVHDSSWLVPKATWSSTWRSIGVGLREGVAK---GWIL 725
>At4g10830 putative protein
Length = 1294
Score = 108 bits (269), Expect = 6e-24
Identities = 57/145 (39%), Positives = 85/145 (58%)
Query: 16 LVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFW 75
+++R+K+R S W ++ LS G+ I+LK V S+PVY +S FK P I+S +E+L F+W
Sbjct: 1130 IIERVKKRTSSWSAKYLSPAGKEIMLKSVAMSMPVYAMSCFKLPLNIVSEIEALLMNFWW 1189
Query: 76 GGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCA 135
R+I WI W+ + K EGGLG R L +FN ALL K WR++ +L+ R++ A
Sbjct: 1190 EKNAKKREIPWIAWKRLQYSKKEGGLGFRDLAKFNDALLAKQVWRMINNPNSLFARIMKA 1249
Query: 136 RYGEEGGRLCCSCRDGGSVWWQTIL 160
RY E L + S W ++L
Sbjct: 1250 RYFREDSILDAKRQRYQSYGWTSML 1274
>At2g05980 putative non-LTR retroelement reverse transcriptase
Length = 1352
Score = 108 bits (269), Expect = 6e-24
Identities = 67/184 (36%), Positives = 92/184 (49%), Gaps = 9/184 (4%)
Query: 13 WYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNA 72
+ PLV++I+ R++ W +R LS GRL L+K VLSSI ++LS F+ P + +E +F+A
Sbjct: 933 YLPLVEKIRARITSWTNRFLSFAGRLQLIKSVLSSITNFWLSVFRLPKACLQEIEKMFSA 992
Query: 73 FFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRV 132
F W G + K I W VC K EGGLG++ L+E N L K WRIL R +LW +
Sbjct: 993 FLWSGPDLNTKKAKIAWSEVCKLKEEGGLGLKPLKEANEVSLLKLIWRILSARDSLWVKW 1052
Query: 133 LCARYGEEGGRLCCSCRDG-GSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFW 191
+ + G GS W+ IL RD V +G T FW
Sbjct: 1053 VNKHLIRKETFWSVKENTGLGSWLWRKILKQRDKARLFH--------RMEVRSGTFTSFW 1104
Query: 192 VDPW 195
D W
Sbjct: 1105 HDHW 1108
>At2g14430 putative non-LTR retroelement reverse transcriptase
Length = 1277
Score = 107 bits (266), Expect = 1e-23
Identities = 60/192 (31%), Positives = 99/192 (51%), Gaps = 9/192 (4%)
Query: 11 AFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLF 70
A + PL+D+++ ++S W +R+LS GRL L+ V+ S+ +++S ++ P+G I +E L
Sbjct: 926 ADYSPLLDKVRSKISSWTARSLSYAGRLALINSVIVSLSNFWMSAYRLPAGCIKEIEKLC 985
Query: 71 NAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWY 130
+AF W G E K I W ++C K EGGLG++ L E N K WR++ + +LW
Sbjct: 986 SAFLWSGPELNPKKAKITWTSLCKLKQEGGLGIKSLLEANKVSCLKLIWRLVSRQSSLWV 1045
Query: 131 RVLCARYGEEGGRLCCSCRDG-GSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTL 189
+ +G + R GS W+ +L+ RD + + +G ST
Sbjct: 1046 NWVWTYIIRKGSFWSANDRSSLGSWMWKKLLNYRDVAKSM--------CKVEIKSGSSTS 1097
Query: 190 FWVDPWLEEKPL 201
FW D W + + L
Sbjct: 1098 FWYDNWSQLRQL 1109
>At1g47850 hypothetical protein
Length = 799
Score = 106 bits (264), Expect = 2e-23
Identities = 61/196 (31%), Positives = 98/196 (49%), Gaps = 9/196 (4%)
Query: 1 LPIGGDPRKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPS 60
LP+ A + PL+D+++ ++S W +R+LS GRL L+ V+ S+ +++S ++ P+
Sbjct: 347 LPLLTKQMTTADYSPLLDKVRSKISSWTARSLSYAGRLALINSVIVSLSNFWMSAYRLPA 406
Query: 61 GIISTLESLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWR 120
G I +E L +AF W G E K I W ++C K EGGLG++ L E N K WR
Sbjct: 407 GCIKEIEKLCSAFLWSGPELNPKKAKITWTSLCKLKQEGGLGIKSLLEANKVSCLKLIWR 466
Query: 121 ILQERGTLWYRVLCARYGEEGGRLCCSCRDG-GSVWWQTILDVRDGVGQIDSGWMLDNIS 179
++ + +LW + +G + R GS W+ +L RD +
Sbjct: 467 LVSRQSSLWVNWVWTYIIRKGSFWSANDRSSLGSWMWKKLLKYRDVAKSM--------CK 518
Query: 180 RRVGNGLSTLFWVDPW 195
+ +G ST FW D W
Sbjct: 519 VEIKSGSSTSFWYDNW 534
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.350 0.159 0.603
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,224,423
Number of Sequences: 26719
Number of extensions: 352740
Number of successful extensions: 1399
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 1265
Number of HSP's gapped (non-prelim): 81
length of query: 366
length of database: 11,318,596
effective HSP length: 101
effective length of query: 265
effective length of database: 8,619,977
effective search space: 2284293905
effective search space used: 2284293905
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144345.7


