

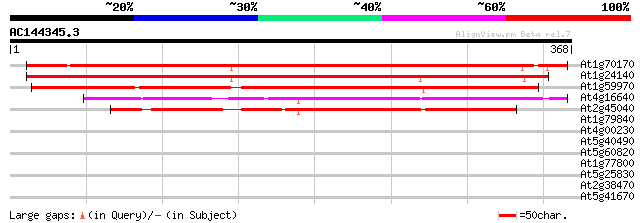
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144345.3 + phase: 0
(368 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g70170 predicted GPI-anchored protein 383 e-106
At1g24140 metalloproteinase like protein, predicted GPI-anchor 377 e-105
At1g59970 predicted GPI-anchored protein 355 3e-98
At4g16640 proteinase-like protein, predicted GPI-anchored protei... 273 1e-73
At2g45040 metalloproteinase like protein ,predicted GPI-anchored... 251 3e-67
At1g79840 homeobox protein (GLABRA2) 32 0.55
At4g00230 subtilisin-type serine endopeptidase XSP1 32 0.72
At5g40490 ribonucleoprotein -like 31 0.94
At5g60820 ring finger protein - like 31 1.2
At1g77800 putative phorbol ester / diacylglycerol binding protein 29 3.6
At5g25830 GATA transcription factor - like 29 4.7
At2g38470 putative WRKY-type DNA binding protein 29 4.7
At5g41670 6-phosphogluconate dehydrogenase 28 8.0
>At1g70170 predicted GPI-anchored protein
Length = 378
Score = 383 bits (983), Expect = e-106
Identities = 197/376 (52%), Positives = 251/376 (66%), Gaps = 24/376 (6%)
Query: 12 IFYITLISITLTSVSARFFPDPSSMPAWNSSNAPPGAWDGYKNFTGCSRGKTYDGLSKLK 71
+F + + ++ SA FFP+ +++P + N WD + NFTGC G+ DGL ++K
Sbjct: 5 VFGFLSLFLIVSPASAWFFPNSTAVPP-SLRNTTRVFWDAFSNFTGCHHGQNVDGLYRIK 63
Query: 72 NYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCGVA 131
YF FGYIP NFTDDFD+ L++AV YQ NFNLN+TGELD T+ +IV PRCG
Sbjct: 64 KYFQRFGYIPETFSGNFTDDFDDILKAAVELYQTNFNLNVTGELDALTIQHIVIPRCGNP 123
Query: 132 DIINGTTSMNSGK-----FNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENLDD 186
D++NGT+ M+ G+ N S T+ H V Y+ FPG+PRWP + LTYAFDP L +
Sbjct: 124 DVVNGTSLMHGGRRKTFEVNFSRTHLHAVKRYTLFPGEPRWPRNRRDLTYAFDPKNPLTE 183
Query: 187 ATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAFSP 246
K VF+ AF +WS VT + FT + S+S+SDI IGFY+GDHGDGE FDGVLGTLAHAFSP
Sbjct: 184 EVKSVFSRAFGRWSDVTALNFTLSESFSTSDITIGFYTGDHGDGEPFDGVLGTLAHAFSP 243
Query: 247 TDGRLHLDKAEDWVVNGDVTE-SSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTIS 305
G+ HLD E+WVV+GD+ S++ AVDLESV VHEIGHLLGLGHSS+EE+IMYPTI+
Sbjct: 244 PSGKFHLDADENWVVSGDLDSFLSVTAAVDLESVAVHEIGHLLGLGHSSVEESIMYPTIT 303
Query: 306 SRTKKVELESDDIEGIQMLYGSNPNFTGTT---TATSRDRDSSFGGRHG----------- 351
+ +KV+L +DD+EGIQ LYG+NPNF GTT + T RD+ GG
Sbjct: 304 TGKRKVDLTNDDVEGIQYLYGANPNFNGTTSPPSTTKHQRDT--GGFSAAWRIDGSSRST 361
Query: 352 -VSLFLSLVFLGFGFL 366
VSL LS V L FL
Sbjct: 362 IVSLLLSTVGLVLWFL 377
>At1g24140 metalloproteinase like protein, predicted GPI-anchor
Length = 384
Score = 377 bits (967), Expect = e-105
Identities = 184/353 (52%), Positives = 241/353 (68%), Gaps = 11/353 (3%)
Query: 12 IFYITLISITLTSVSARFFPDPSSMPAWNSSNAPPGAWDGYKNFTGCSRGKTYDGLSKLK 71
+F + L+ + VSA F+ + S++P NA W+ + NFTGC GK YDGL LK
Sbjct: 6 VFMVFLLFFAPSPVSAGFYTNSSAIPPQLLRNATGNPWNSFLNFTGCHAGKKYDGLYMLK 65
Query: 72 NYFNHFGYIPNGPPS-NFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCGV 130
YF HFGYI S NFTDDFD+ L++AV YQ+NF LN+TG LD+ T+ ++V PRCG
Sbjct: 66 QYFQHFGYITETNLSGNFTDDFDDILKNAVEMYQRNFQLNVTGVLDELTLKHVVIPRCGN 125
Query: 131 ADIINGTTSMNSGK------FNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENL 184
D++NGT++M+SG+ F FH V HYSFFPG+PRWP + LTYAFDP L
Sbjct: 126 PDVVNGTSTMHSGRKTFEVSFAGRGQRFHAVKHYSFFPGEPRWPRNRRDLTYAFDPRNAL 185
Query: 185 DDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAF 244
+ K VF+ AF +W +VT +TFT +S+SDI IGFYSG+HGDGE FDG + TLAHAF
Sbjct: 186 TEEVKSVFSRAFTRWEEVTPLTFTRVERFSTSDISIGFYSGEHGDGEPFDGPMRTLAHAF 245
Query: 245 SPTDGRLHLDKAEDWVVNGDVTES--SLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYP 302
SP G HLD E+W+V+G+ + S+S AVDLESV VHEIGHLLGLGHSS+E +IMYP
Sbjct: 246 SPPTGHFHLDGEENWIVSGEGGDGFISVSEAVDLESVAVHEIGHLLGLGHSSVEGSIMYP 305
Query: 303 TISSRTKKVELESDDIEGIQMLYGSNPNFTGTTT--ATSRDRDSSFGGRHGVS 353
TI + +KV+L +DD+EG+Q LYG+NPNF G+ + +++ RD+ G G S
Sbjct: 306 TIRTGRRKVDLTTDDVEGVQYLYGANPNFNGSRSPPPSTQQRDTGDSGAPGRS 358
>At1g59970 predicted GPI-anchored protein
Length = 360
Score = 355 bits (910), Expect = 3e-98
Identities = 177/336 (52%), Positives = 225/336 (66%), Gaps = 11/336 (3%)
Query: 15 ITLISITLTSVSARFFPDPSSMPAWNSSNAPPGAWDGYKNFTGCSRGKTYDGLSKLKNYF 74
I + T+ +SA+F+ + SS+P NA AW+ + GC G+ +GLSKLK YF
Sbjct: 8 ILIFFFTVNPISAKFYTNVSSIPPLQFLNATQNAWETFSKLAGCHIGENINGLSKLKQYF 67
Query: 75 NHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCGVADII 134
FGYI N TDDFD+ L+SA+ TYQKNFNL +TG+LD +T+ IVKPRCG D+I
Sbjct: 68 RRFGYITT--TGNCTDDFDDVLQSAINTYQKNFNLKVTGKLDSSTLRQIVKPRCGNPDLI 125
Query: 135 NGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENLDDATKQVFAN 194
+G + MN GK T YSFFPG+PRWP+ + LTYAF P NL D K+VF+
Sbjct: 126 DGVSEMNGGKI------LRTTEKYSFFPGKPRWPKRKRDLTYAFAPQNNLTDEVKRVFSR 179
Query: 195 AFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAFSPTDGRLHLD 254
AF +W++VT + FT + S +DI IGF+SG+HGDGE FDG +GTLAHA SP G LHLD
Sbjct: 180 AFTRWAEVTPLNFTRSESILRADIVIGFFSGEHGDGEPFDGAMGTLAHASSPPTGMLHLD 239
Query: 255 KAEDWVV-NGDVTESSL--SNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTKKV 311
EDW++ NG+++ L + VDLESV VHEIGHLLGLGHSS+E+AIM+P IS +KV
Sbjct: 240 GDEDWLISNGEISRRILPVTTVVDLESVAVHEIGHLLGLGHSSVEDAIMFPAISGGDRKV 299
Query: 312 ELESDDIEGIQMLYGSNPNFTGTTTATSRDRDSSFG 347
EL DDIEGIQ LYG NPN G + SR+ S+ G
Sbjct: 300 ELAKDDIEGIQHLYGGNPNGDGGGSKPSRESQSTGG 335
>At4g16640 proteinase-like protein, predicted GPI-anchored protein
(by homology)
Length = 364
Score = 273 bits (697), Expect = 1e-73
Identities = 149/322 (46%), Positives = 193/322 (59%), Gaps = 21/322 (6%)
Query: 49 WDGYKNFTGCSRGKTYDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFN 108
W + G G+S+LK Y + FGY+ +G F+D FD LESA+ YQ+N
Sbjct: 52 WHDFSRLVDVQIGSHVSGVSELKRYLHRFGYVNDGSEI-FSDVFDGPLESAISLYQENLG 110
Query: 109 LNITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWP 168
L ITG LD +T+ + PRCGV+D ++ HT AHY++F G+P+W
Sbjct: 111 LPITGRLDTSTVTLMSLPRCGVSDT----------HMTINNDFLHTTAHYTYFNGKPKW- 159
Query: 169 EGTQTLTYAFDPSENLDDAT----KQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYS 224
TLTYA + LD T K VF AF+QWS V ++F E ++++D+KIGFY+
Sbjct: 160 -NRDTLTYAISKTHKLDYLTSEDVKTVFRRAFSQWSSVIPVSFEEVDDFTTADLKIGFYA 218
Query: 225 GDHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEI 284
GDHGDG FDGVLGTLAHAF+P +GRLHLD AE W+V+ D+ SS AVDLESV HEI
Sbjct: 219 GDHGDGLPFDGVLGTLAHAFAPENGRLHLDAAETWIVDDDLKGSS-EVAVDLESVATHEI 277
Query: 285 GHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDS 344
GHLLGLGHSS E A+MYP++ RTKKV+L DD+ G+ LYG NP + S D
Sbjct: 278 GHLLGLGHSSQESAVMYPSLRPRTKKVDLTVDDVAGVLKLYGPNPKLRLDSLTQSEDSIK 337
Query: 345 SFGGRHGVSLFLSLVFLGFGFL 366
+ H FLS F+G+ L
Sbjct: 338 NGTVSH---RFLSGNFIGYVLL 356
>At2g45040 metalloproteinase like protein ,predicted GPI-anchored
protein (by homology)
Length = 342
Score = 251 bits (642), Expect = 3e-67
Identities = 133/272 (48%), Positives = 172/272 (62%), Gaps = 25/272 (9%)
Query: 67 LSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKP 126
+ ++K + +GY+P S+ D + E A+ YQKN L ITG+ D T++ I+ P
Sbjct: 50 IPEIKRHLQQYGYLPQNKESD-----DVSFEQALVRYQKNLGLPITGKPDSDTLSQILLP 104
Query: 127 RCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQT-LTYAFDPSENLD 185
RCG D + T+ FHT Y +FPG+PRW LTYAF ENL
Sbjct: 105 RCGFPDDVEPKTAP-----------FHTGKKYVYFPGRPRWTRDVPLKLTYAFS-QENLT 152
Query: 186 DAT-----KQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTL 240
++VF AF +W+ V ++F E Y +DIKIGF++GDHGDGE FDGVLG L
Sbjct: 153 PYLAPTDIRRVFRRAFGKWASVIPVSFIETEDYVIADIKIGFFNGDHGDGEPFDGVLGVL 212
Query: 241 AHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIM 300
AH FSP +GRLHLDKAE W V+ D +SS+ AVDLESV VHEIGH+LGLGHSS+++A M
Sbjct: 213 AHTFSPENGRLHLDKAETWAVDFDEEKSSV--AVDLESVAVHEIGHVLGLGHSSVKDAAM 270
Query: 301 YPTISSRTKKVELESDDIEGIQMLYGSNPNFT 332
YPT+ R+KKV L DD+ G+Q LYG+NPNFT
Sbjct: 271 YPTLKPRSKKVNLNMDDVVGVQSLYGTNPNFT 302
>At1g79840 homeobox protein (GLABRA2)
Length = 747
Score = 32.0 bits (71), Expect = 0.55
Identities = 33/154 (21%), Positives = 63/154 (40%), Gaps = 26/154 (16%)
Query: 189 KQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAFSPTD 248
+ + A++++QW+K+TT T D+++ H GE ++ + + P
Sbjct: 525 RAIAASSYHQWTKITTKT--------GQDMRVSSRKNLHDPGEPTGVIVCASSSLWLPVS 576
Query: 249 GRLHLD------KAEDW--VVNGDVTES--------SLSNAVDLESVVVHE--IGHLLGL 290
L D + +W + NG +S N+V +++V E I L
Sbjct: 577 PALLFDFFRDEARRHEWDALSNGAHVQSIANLSKGQDRGNSVAIQTVKSREKSIWVLQDS 636
Query: 291 GHSSIEEAIMYPTISSRTKKVELESDDIEGIQML 324
+S E ++Y + T ++ L D IQ+L
Sbjct: 637 STNSYESVVVYAPVDINTTQLVLAGHDPSNIQIL 670
>At4g00230 subtilisin-type serine endopeptidase XSP1
Length = 749
Score = 31.6 bits (70), Expect = 0.72
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 14/45 (31%)
Query: 45 PPGAWDG----YKNFTGCSRGKTYDGLSKL--KNYFNHFGYIPNG 83
PP W G YKNFTGC+ +K+ YF H G +P G
Sbjct: 163 PPAKWKGSCGPYKNFTGCN--------NKIIGAKYFKHDGNVPAG 199
>At5g40490 ribonucleoprotein -like
Length = 423
Score = 31.2 bits (69), Expect = 0.94
Identities = 19/70 (27%), Positives = 30/70 (42%), Gaps = 1/70 (1%)
Query: 282 HEIGHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRD 341
H G G G + E M + ++ ++EL +E I+ PN T + D
Sbjct: 165 HSTGRSRGFGFVTYESEDMVDHLLAKGNRIELSGTQVE-IKKAEPKKPNSVTTPSKRFGD 223
Query: 342 RDSSFGGRHG 351
S+FGG +G
Sbjct: 224 SRSNFGGGYG 233
>At5g60820 ring finger protein - like
Length = 419
Score = 30.8 bits (68), Expect = 1.2
Identities = 25/108 (23%), Positives = 50/108 (46%), Gaps = 8/108 (7%)
Query: 226 DHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIG 285
DH ++F +L L H DG L++ + + +GDV+ S L D ++
Sbjct: 28 DHLRDQSFSQILYPLPHWIQSDDGDLYISETD--FSSGDVSVSDLVFTTDDGLDLLDRRS 85
Query: 286 HLLGLGHSSIEEAIMYPTISSRT---KKVELESDDIE---GIQMLYGS 327
++ L H +E++ + P + + E+ D++E G++M G+
Sbjct: 86 FVMDLFHQRVEQSQVTPLDNDGIDDFENYEMREDNVELDFGLEMESGN 133
>At1g77800 putative phorbol ester / diacylglycerol binding protein
Length = 1506
Score = 29.3 bits (64), Expect = 3.6
Identities = 26/93 (27%), Positives = 41/93 (43%), Gaps = 9/93 (9%)
Query: 233 FDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGDVTE---SSLSNAVDLESVVVHEIGHLLG 289
FDG L TL H +G HL AE ++ + LS +LE +++ LLG
Sbjct: 837 FDGHLVTLTHLAGSEEGNKHLQGAETFLQLSKARKLGILDLSPEDELEGELLYYQLQLLG 896
Query: 290 --LGHSSIEEAIMYPTISSRTKKVELESDDIEG 320
+ + + ++Y KK+ LE D+ G
Sbjct: 897 TAVSRKQLSDNLVYEV----AKKLPLEIDEQHG 925
>At5g25830 GATA transcription factor - like
Length = 331
Score = 28.9 bits (63), Expect = 4.7
Identities = 18/55 (32%), Positives = 30/55 (53%), Gaps = 8/55 (14%)
Query: 183 NLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVL 237
N DD V A++ TT T T+++++S++D+ + GD DG +F G L
Sbjct: 25 NDDDEENDVVADS------TTTTTITDSSNFSAADLPS--FHGDVQDGTSFSGDL 71
>At2g38470 putative WRKY-type DNA binding protein
Length = 512
Score = 28.9 bits (63), Expect = 4.7
Identities = 22/88 (25%), Positives = 35/88 (39%), Gaps = 5/88 (5%)
Query: 134 INGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENLDDATKQVFA 193
IN N+ FN +FHT + P T T T + ++ + +Q
Sbjct: 94 INEGDKSNNNNFNLFDFSFHTQSSGVSAPTTT-----TTTTTTTTTTNSSIFQSQEQQKK 148
Query: 194 NAFNQWSKVTTITFTEATSYSSSDIKIG 221
N QWS+ T +A SY+ + + G
Sbjct: 149 NQSEQWSQTETRPNNQAVSYNGREQRKG 176
>At5g41670 6-phosphogluconate dehydrogenase
Length = 487
Score = 28.1 bits (61), Expect = 8.0
Identities = 19/67 (28%), Positives = 31/67 (45%), Gaps = 7/67 (10%)
Query: 172 QTLTYAFDPSENLDDATKQVFANAFNQWSKVTTITF-TEATSYSSSDIKIGFYSGDHGDG 230
Q ++ A+D +N+ + A F +W++ +F E TS I D+GDG
Sbjct: 200 QLISEAYDVLKNVGGLSNDELAEIFTEWNRGELESFLVEITS------DIFRVKDDYGDG 253
Query: 231 EAFDGVL 237
E D +L
Sbjct: 254 ELVDKIL 260
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,004,596
Number of Sequences: 26719
Number of extensions: 415947
Number of successful extensions: 1042
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1022
Number of HSP's gapped (non-prelim): 14
length of query: 368
length of database: 11,318,596
effective HSP length: 101
effective length of query: 267
effective length of database: 8,619,977
effective search space: 2301533859
effective search space used: 2301533859
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144345.3


