

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143341.7 + phase: 0
(745 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
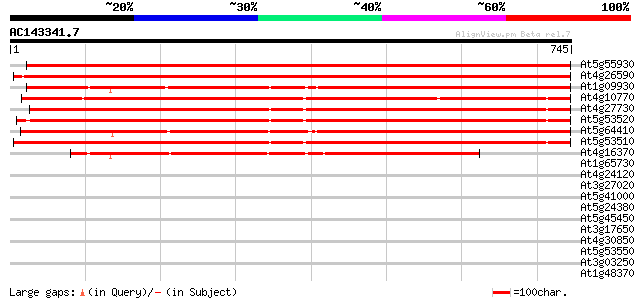
Score E
Sequences producing significant alignments: (bits) Value
At5g55930 sexual differentiation process protein ISP4-like 1067 0.0
At4g26590 isp4 like protein 1060 0.0
At1g09930 unknown protein 844 0.0
At4g10770 oligopeptide transporter like protein 843 0.0
At4g27730 unknown protein 794 0.0
At5g53520 isp4 protein 793 0.0
At5g64410 Isp4-like protein 793 0.0
At5g53510 isp4 protein 790 0.0
At4g16370 isp4 like protein 566 e-161
At1g65730 unknown protein 40 0.004
At4g24120 unknown protein 38 0.024
At3g27020 unknown protein 34 0.26
At5g41000 unknown protein 33 0.44
At5g24380 putative protein 33 0.44
At5g45450 putative protein 32 0.99
At3g17650 unknown protein 32 0.99
At4g30850 unknown protein 31 2.9
At5g53550 EspB-like protein 30 3.8
At3g03250 UDP-glucose pyrophosphorylase like protein 30 3.8
At1g48370 unknown protein 30 4.9
>At5g55930 sexual differentiation process protein ISP4-like
Length = 755
Score = 1067 bits (2759), Expect = 0.0
Identities = 488/723 (67%), Positives = 595/723 (81%), Gaps = 1/723 (0%)
Query: 23 NDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQ 82
ND+PIE+VRLTVPITDDP+ P LTFRTW LGL SCILLAFVNQF G+R+N L ++SV+AQ
Sbjct: 32 NDNPIEEVRLTVPITDDPTLPVLTFRTWTLGLFSCILLAFVNQFFGFRSNQLWVSSVAAQ 91
Query: 83 IIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSGSSGVYAINII 142
I+ LPLGKLMA TLPTK P T WS+S NPGPF++KEHVLITIFA++G+ GVYA +II
Sbjct: 92 IVTLPLGKLMAKTLPTKKFGFPGTNWSWSFNPGPFNMKEHVLITIFANTGAGGVYATSII 151
Query: 143 TIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRAFH 202
TIVKAFY+R ++ AA LL +TQ+LGYGWAGIFR+FLVDSPYMWWP +LVQVSLFRA H
Sbjct: 152 TIVKAFYNRQLNVAAAMLLTQTTQLLGYGWAGIFRKFLVDSPYMWWPSNLVQVSLFRALH 211
Query: 203 EKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQIGSGM 262
EKE +G +R +FF +VF SFAYYIIPGY F +IS +SFVC IWK S+TAQ +GSG+
Sbjct: 212 EKEDLQKGQQTRFRFFIIVFCVSFAYYIIPGYLFPSISAISFVCWIWKSSVTAQIVGSGL 271
Query: 263 KGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWNNLYDAKKFP 322
KGLGIGSFGLDW+TVAGFLGSPLAVP FAI N GFF+++Y+++PI YW N YDA+KFP
Sbjct: 272 KGLGIGSFGLDWSTVAGFLGSPLAVPFFAIANFFGGFFIFLYIVLPIFYWTNAYDAQKFP 331
Query: 323 LISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALTATIS 382
+SHTFD TG TYN+TRILN + FDI++++Y+ YSK+YLSV FA YG F +L ATIS
Sbjct: 332 FYTSHTFDQTGHTYNITRILNEKNFDINLDAYNGYSKLYLSVMFALLYGLSFGSLCATIS 391
Query: 383 HVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMVMMALLAC 442
HV L+ G+ I MWKK T+ K++ GDVH+R+MKKNY+ VP+WWF+ +L++ AL AC
Sbjct: 392 HVALYDGKFIWGMWKKAKTATKDKYGDVHSRLMKKNYQSVPQWWFIAVLVISFAFALYAC 451
Query: 443 EGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYPGKPLAN 502
EGF KQLQLPWWG++L+ IAL FTLPIGVI+ATTN + GLNVITEL+IG++YPGKPLAN
Sbjct: 452 EGFDKQLQLPWWGLILACAIALFFTLPIGVIQATTNQQMGLNVITELIIGYLYPGKPLAN 511
Query: 503 VAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLTSI 562
VAFKTYG+ISM QAL F+GDFKLGHYMKI P+SMFIVQLV TVVAS+V FGT WWL+TS+
Sbjct: 512 VAFKTYGYISMSQALYFVGDFKLGHYMKIPPRSMFIVQLVATVVASTVCFGTTWWLITSV 571
Query: 563 ENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIGLIAPV 622
ENIC+ LLP GSPWTCPGD+VFYNASIIWGV+GP RMFTK+G+YP +NWFFLIGL+APV
Sbjct: 572 ENICNVDLLPVGSPWTCPGDEVFYNASIIWGVIGPGRMFTKEGIYPGMNWFFLIGLLAPV 631
Query: 623 PVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRKFKAWW 682
P W LS KFP +KW++ I++P+I + S +P ++V+Y +W IVG+ FN+Y++R+FK WW
Sbjct: 632 PFWYLSKKFPEKKWLKQIHVPLIFSAVSAMPQAKAVHYWSWAIVGVVFNYYIFRRFKTWW 691
Query: 683 ARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLE-PDHCPLAKCPTAPGVHAEG 741
ARH YILSA LDAG A +G+L++F+ Q+ I P WWGLE DHCPLA CP A GV EG
Sbjct: 692 ARHNYILSAALDAGTAIMGVLIFFAFQNNDISLPDWWGLENSDHCPLAHCPLAKGVVVEG 751
Query: 742 CPV 744
CPV
Sbjct: 752 CPV 754
>At4g26590 isp4 like protein
Length = 753
Score = 1060 bits (2740), Expect = 0.0
Identities = 487/741 (65%), Positives = 597/741 (79%), Gaps = 3/741 (0%)
Query: 5 RVSQNTVIEDAEIDEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVN 64
+V VI D E E NDSPIE+VRLTVPITDDPS P LTFRTW LG+ SC++LAFVN
Sbjct: 14 KVESKIVIADEE--EEDENDSPIEEVRLTVPITDDPSLPVLTFRTWFLGMVSCVVLAFVN 71
Query: 65 QFLGYRTNPLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVL 124
F GYR+NPL ++SV AQII LPLGKLMA TLPT +++P T WS SLNPGPF++KEHVL
Sbjct: 72 NFFGYRSNPLTVSSVVAQIITLPLGKLMATTLPTTKLRLPGTNWSCSLNPGPFNMKEHVL 131
Query: 125 ITIFASSGSSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSP 184
ITIFA++G+ G YA +I+TIVKAFYHR+++P AA LL +TQ+LGYGWAG+FR++LVDSP
Sbjct: 132 ITIFANTGAGGAYATSILTIVKAFYHRNLNPAAAMLLVQTTQLLGYGWAGMFRKYLVDSP 191
Query: 185 YMWWPESLVQVSLFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSF 244
YMWWP +LVQVSLFRA HEKE++ G ++L+FF +VF SF YYI+PGY F +IS +SF
Sbjct: 192 YMWWPANLVQVSLFRALHEKEEKREGKQTKLRFFLIVFFLSFTYYIVPGYLFPSISYLSF 251
Query: 245 VCLIWKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMY 304
VC IW S+TAQQIGSG+ GLGIGSFGLDW+TVAGFLGSPLAVP FAI N GF ++ Y
Sbjct: 252 VCWIWTRSVTAQQIGSGLHGLGIGSFGLDWSTVAGFLGSPLAVPFFAIANSFGGFIIFFY 311
Query: 305 VLIPISYWNNLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSV 364
+++PI YW+N Y+AKKFP +SH FD TG YN TRILN +TF+ID+ +Y +YSK+YLS+
Sbjct: 312 IILPIFYWSNAYEAKKFPFYTSHPFDHTGQRYNTTRILNQKTFNIDLPAYESYSKLYLSI 371
Query: 365 AFAFEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPE 424
FA YG F LTATISHV LF G+ I ++WKK T + K++ GDVHTR+MKKNY++VP+
Sbjct: 372 LFALIYGLSFGTLTATISHVALFDGKFIWELWKKATLTTKDKFGDVHTRLMKKNYKEVPQ 431
Query: 425 WWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLN 484
WWFV +L ++AL ACEGFGKQLQLPWWG+LL+ IA FTLPIGVI ATTN R GLN
Sbjct: 432 WWFVAVLAASFVLALYACEGFGKQLQLPWWGLLLACAIAFTFTLPIGVILATTNQRMGLN 491
Query: 485 VITELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGT 544
VI+EL+IGF+YPGKPLANVAFKTYG +S+ QAL F+GDFKLGHYMKI P+SMFIVQLV T
Sbjct: 492 VISELIIGFLYPGKPLANVAFKTYGSVSIAQALYFVGDFKLGHYMKIPPRSMFIVQLVAT 551
Query: 545 VVASSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKD 604
+VAS+V FGT WWLL+S+ENIC+ +LPK SPWTCPGD VFYNASIIWG++GP RMFT
Sbjct: 552 IVASTVSFGTTWWLLSSVENICNTDMLPKSSPWTCPGDVVFYNASIIWGIIGPGRMFTSK 611
Query: 605 GVYPELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWG 664
G+YP +NWFFLIG +APVPVW + KFP +KWI I+IP+I +GA+ +P ++V+Y +W
Sbjct: 612 GIYPGMNWFFLIGFLAPVPVWFFARKFPEKKWIHQIHIPLIFSGANVMPMAKAVHYWSWF 671
Query: 665 IVGIFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLE-P 723
VGI FN+Y++R++K WWARH YILSA LDAG A +G+L+YF+LQ+ I P WWG E
Sbjct: 672 AVGIVFNYYIFRRYKGWWARHNYILSAALDAGTAVMGVLIYFALQNNNISLPDWWGNENT 731
Query: 724 DHCPLAKCPTAPGVHAEGCPV 744
DHCPLA CPT G+ A+GCPV
Sbjct: 732 DHCPLANCPTEKGIVAKGCPV 752
>At1g09930 unknown protein
Length = 734
Score = 844 bits (2180), Expect = 0.0
Identities = 393/727 (54%), Positives = 533/727 (73%), Gaps = 16/727 (2%)
Query: 23 NDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQ 82
++SP+EQVRLTV DDPS P TFR W LGL SCILL+F+N F GYRT PL IT +S Q
Sbjct: 18 DESPVEQVRLTVSNHDDPSLPVWTFRMWFLGLLSCILLSFLNTFFGYRTQPLMITMISVQ 77
Query: 83 IIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSG----SSGVYA 138
++ LPLGKLMA LP ++ +W FS NPGPF++KEHVLI++FA++G S YA
Sbjct: 78 VVTLPLGKLMARVLPETKYKIG--SWEFSFNPGPFNVKEHVLISMFANAGAGFGSGTAYA 135
Query: 139 INIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLF 198
+ I+ I+ AFY R I +A+++L ++TQ+LGYGWAGI R+ +VD MWWP S++QVSLF
Sbjct: 136 VGIVDIIMAFYKRKISFLASWILVITTQILGYGWAGIMRKLVVDPAQMWWPTSVLQVSLF 195
Query: 199 RAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQI 258
RA HEK+ SR +FF + FV SFA+YI P Y F +S++S+VC + +SITAQQ+
Sbjct: 196 RALHEKDN---ARMSRGKFFVIAFVCSFAWYIFPAYLFLTLSSISWVCWAFPKSITAQQL 252
Query: 259 GSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWN-NLYD 317
GSGM GLGIG+F LDW+ +A +LGSPL P FAI+N++ G+ L MY++IPISYW N+Y+
Sbjct: 253 GSGMSGLGIGAFALDWSVIASYLGSPLVTPFFAIVNVLVGYVLVMYMVIPISYWGMNVYE 312
Query: 318 AKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAAL 377
A KFP+ SS FD G YN++ I+N + F++DME+Y ++YLS FA YG FAA+
Sbjct: 313 ANKFPIFSSDLFDKQGQLYNISTIVNNK-FELDMENYQQQGRVYLSTFFAISYGIGFAAI 371
Query: 378 TATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMVMM 437
+T++HV LF+G+ I W++ S K ++ D+HTR+MKK Y+ +P WWF ++L + +++
Sbjct: 372 VSTLTHVALFNGKGI---WQQVRASTKAKM-DIHTRLMKK-YKDIPGWWFYSLLAISLVL 426
Query: 438 ALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYPG 497
+L+ C ++Q+PWWG+LL+ +AL FT+P+ +I ATTN GLN+ITE ++G + PG
Sbjct: 427 SLVLCIFMKDEIQMPWWGLLLASFMALTFTVPVSIITATTNQTPGLNIITEYLMGVLLPG 486
Query: 498 KPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWW 557
+P+ANV FKTYG+ISM QA+ FL DFKLGHYMKI P+SMF+VQ +GTV+A +V+ AW+
Sbjct: 487 RPIANVCFKTYGYISMSQAISFLNDFKLGHYMKIPPRSMFLVQFIGTVIAGTVNISVAWY 546
Query: 558 LLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIG 617
LLTS+ENIC + LLP SPWTCP D VF++AS+IWG+VGPKR+F + G YP LNWFFL G
Sbjct: 547 LLTSVENICQKELLPPNSPWTCPSDRVFFDASVIWGLVGPKRIFGRLGNYPALNWFFLGG 606
Query: 618 LIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRK 677
LI PV VWLL FP + WI IN+P+++ + +PP SVN+ W IVG+ FN++V++
Sbjct: 607 LIGPVLVWLLQKAFPTKTWISQINLPVLLGATAAMPPATSVNFNCWIIVGVIFNYFVFKY 666
Query: 678 FKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKCPTAPGV 737
K WW R+ Y+LSA LDAG+AF+G+LLYFSL GI WWG + ++CPLA CPTAPGV
Sbjct: 667 CKKWWQRYNYVLSAALDAGLAFMGVLLYFSLTMNGISINHWWGAKGENCPLASCPTAPGV 726
Query: 738 HAEGCPV 744
+GCPV
Sbjct: 727 LVDGCPV 733
>At4g10770 oligopeptide transporter like protein
Length = 766
Score = 843 bits (2178), Expect = 0.0
Identities = 395/729 (54%), Positives = 529/729 (72%), Gaps = 10/729 (1%)
Query: 16 EIDEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLK 75
E +E +SPI QV LTVP TDDPS P LTFR WVLG SCILL+F+NQF YRT PL
Sbjct: 46 ENEEEEEENSPIRQVALTVPTTDDPSLPVLTFRMWVLGTLSCILLSFLNQFFWYRTEPLT 105
Query: 76 ITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSGSSG 135
I+++SAQI +PLG+LMAA + T + + W F+LNPGPF++KEHVLITIFA++G+
Sbjct: 106 ISAISAQIAVVPLGRLMAAKI-TDRVFFQGSKWQFTLNPGPFNVKEHVLITIFANAGAGS 164
Query: 136 VYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQV 195
VYAI+++T+VKAFY ++I ++++ ++TQ+LG+GWAGIFR++LV+ MWWP +LVQV
Sbjct: 165 VYAIHVVTVVKAFYMKNITFFVSFIVIVTTQVLGFGWAGIFRKYLVEPAAMWWPANLVQV 224
Query: 196 SLFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITA 255
SLFRA HEKE+R +GG +R QFF + FV SFAYY+ PGY FQ ++++S+VC + S+ A
Sbjct: 225 SLFRALHEKEERTKGGLTRTQFFVIAFVCSFAYYVFPGYLFQIMTSLSWVCWFFPSSVMA 284
Query: 256 QQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWNNL 315
QQIGSG+ GLG+G+ GLDW+T++ +LGSPLA P FA N+ GF L +YVL+PI YW ++
Sbjct: 285 QQIGSGLHGLGVGAIGLDWSTISSYLGSPLASPWFATANVGVGFVLVIYVLVPICYWLDV 344
Query: 316 YDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFA 375
Y AK FP+ SS F S G+ YN+T I+++ F +D+ +Y +YL FA YG FA
Sbjct: 345 YKAKTFPIFSSSLFSSQGSKYNITSIIDS-NFHLDLPAYERQGPLYLCTFFAISYGVGFA 403
Query: 376 ALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMV 435
AL+ATI HV LFHG ++W+++ S K + DVH R+M++ Y+QVPEWWF IL+ V
Sbjct: 404 ALSATIMHVALFHGR---EIWEQSKESFKEKKLDVHARLMQR-YKQVPEWWFWCILVTNV 459
Query: 436 MMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIY 495
+ ACE + QLQLPWWG+LL+ T+A+IFTLPIG+I A TN GLN+ITE +IG+IY
Sbjct: 460 GATIFACEYYNDQLQLPWWGVLLACTVAIIFTLPIGIITAITNQAPGLNIITEYIIGYIY 519
Query: 496 PGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTA 555
PG P+AN+ FK YG+ISM QA+ FL DFKLGHYMKI P++MF+ Q+VGT+++ V+ TA
Sbjct: 520 PGYPVANMCFKVYGYISMQQAITFLQDFKLGHYMKIPPRTMFMAQIVGTLISCFVYLTTA 579
Query: 556 WWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFL 615
WWL+ +I NICD S WTCP D VFY+AS+IWG++GP+R+F G+Y +NWFFL
Sbjct: 580 WWLMETIPNICDS---VTNSVWTCPSDKVFYDASVIWGLIGPRRIFGDLGLYKSVNWFFL 636
Query: 616 IGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVY 675
+G IAP+ VWL S FP Q+WI+ IN+P++I+ S +PP +VNY TW + G F V+
Sbjct: 637 VGAIAPILVWLASRMFPRQEWIKLINMPVLISATSSMPPATAVNYTTWVLAGFLSGFVVF 696
Query: 676 RKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKCPTAP 735
R W R+ Y+LS LDAG+AF+G+LLY L + WWG E D CPLA CPTAP
Sbjct: 697 RYRPNLWQRYNYVLSGALDAGLAFMGVLLYMCLGLENV-SLDWWGNELDGCPLASCPTAP 755
Query: 736 GVHAEGCPV 744
G+ EGCP+
Sbjct: 756 GIIVEGCPL 764
>At4g27730 unknown protein
Length = 736
Score = 794 bits (2051), Expect = 0.0
Identities = 380/721 (52%), Positives = 510/721 (70%), Gaps = 9/721 (1%)
Query: 27 IEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQIIAL 86
+ +V LTVP TDD + P LTFR WVLG+ +CI+L+F+NQF YRT PL IT +SAQI +
Sbjct: 21 VPEVELTVPKTDDSTLPVLTFRMWVLGIGACIVLSFINQFFWYRTMPLSITGISAQIAVV 80
Query: 87 PLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSGSSGVYAINIITIVK 146
PLG LMA LPTK + T + F+LNPG F++KEHVLITIFA+SG+ VYA +I++ +K
Sbjct: 81 PLGHLMARVLPTKRF-LEGTRFQFTLNPGAFNVKEHVLITIFANSGAGSVYATHILSAIK 139
Query: 147 AFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRAFHEKEK 206
+Y RS+ + A+L+ ++TQ+LG+GWAG+FR+ LV+ MWWP +LVQVSLF A HEKEK
Sbjct: 140 LYYKRSLPFLPAFLVMITTQILGFGWAGLFRKHLVEPGEMWWPSNLVQVSLFGALHEKEK 199
Query: 207 RPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQIGSGMKGLG 266
+ RGG SR QFF +V VASFAYYI PGY F ++++S+VC + +SI Q+GSG GLG
Sbjct: 200 KSRGGMSRTQFFLIVLVASFAYYIFPGYLFTMLTSISWVCWLNPKSILVNQLGSGEHGLG 259
Query: 267 IGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWNNLYDAKKFPLISS 326
IGS G DW T++ +LGSPLA P FA +N+ GF L MY++ P+ YW N+YDAK FP+ SS
Sbjct: 260 IGSIGFDWVTISAYLGSPLASPLFASVNVAIGFVLVMYIVTPVCYWLNIYDAKTFPIFSS 319
Query: 327 HTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALTATISHVVL 386
F G+ Y+V I++++ F +D YS I +S FA YG FA L+ATI HV++
Sbjct: 320 QLFMGNGSRYDVLSIIDSK-FHLDRVVYSRTGSINMSTFFAVTYGLGFATLSATIVHVLV 378
Query: 387 FHGEMIVQMWKKTTTSL-KNQLGDVHTRIMKKNYEQVPEWWFVTILILMVMMALLACEGF 445
F+G +WK+T + KN+ D+HTRIMKKNY +VP WWF+ IL+L + + + +
Sbjct: 379 FNGS---DLWKQTRGAFQKNKKMDIHTRIMKKNYREVPLWWFLVILLLNIALIMFISVHY 435
Query: 446 GKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYPGKPLANVAF 505
+QLPWWG+LL+ IA+ FT IGVI ATTN GLN+ITE VIG+IYP +P+AN+ F
Sbjct: 436 NATVQLPWWGVLLACAIAISFTPLIGVIAATTNQAPGLNIITEYVIGYIYPERPVANMCF 495
Query: 506 KTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLTSIENI 565
K YG+ISM QAL F+ DFKLGHYMKI P+SMF+ Q+ GT+VA V+ GTAWWL+ I ++
Sbjct: 496 KVYGYISMTQALTFISDFKLGHYMKIPPRSMFMAQVAGTLVAVVVYTGTAWWLMEEIPHL 555
Query: 566 CDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIGLIAPVPVW 625
CD SLLP S WTCP D VF++AS+IWG+VGP+R+F G Y +NWFFL+G IAP+ VW
Sbjct: 556 CDTSLLPSDSQWTCPMDRVFFDASVIWGLVGPRRVFGDLGEYSNVNWFFLVGAIAPLLVW 615
Query: 626 LLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRKFKAWWARH 685
L + FP Q WI I+IP+++ + +PP +VN+ +W IV F ++++ + WW ++
Sbjct: 616 LATKMFPAQTWIAKIHIPVLVGATAMMPPATAVNFTSWLIVAFIFGHFIFKYRRVWWTKY 675
Query: 686 TYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGL--EPDHCPLAKCPTAPGVHAEGCP 743
Y+LS GLDAG AF+ +LL+ +L GI WWG + D CPLA CPTA GV +GCP
Sbjct: 676 NYVLSGGLDAGSAFMTILLFLALGRKGI-EVQWWGNSGDRDTCPLASCPTAKGVVVKGCP 734
Query: 744 V 744
V
Sbjct: 735 V 735
>At5g53520 isp4 protein
Length = 733
Score = 793 bits (2048), Expect = 0.0
Identities = 377/737 (51%), Positives = 519/737 (70%), Gaps = 11/737 (1%)
Query: 9 NTVIEDAEIDEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLG 68
+T+ E DE SI + QV LTVP TDDP+ P +TFR WVLG+ +C+LL+F+NQF
Sbjct: 6 DTISESECDDEISI----VPQVELTVPKTDDPTSPTVTFRMWVLGITACVLLSFLNQFFW 61
Query: 69 YRTNPLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIF 128
YRTNPL I+SVSAQI +P+G LMA LPT+ T WSF++NPGPFS KEHVLIT+F
Sbjct: 62 YRTNPLTISSVSAQIAVVPIGHLMAKVLPTRRF-FEGTRWSFTMNPGPFSTKEHVLITVF 120
Query: 129 ASSGSSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWW 188
A+SGS VYA +I++ VK +Y R + + A L+ ++TQ+LG+GWAG++R+ LV+ MWW
Sbjct: 121 ANSGSGAVYATHILSAVKLYYKRRLDFLPALLVMITTQVLGFGWAGLYRKHLVEPGEMWW 180
Query: 189 PESLVQVSLFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLI 248
P +LVQVSLFRA HEKE + + G SR QFF + + SF+YY++PGY F ++TVS++C I
Sbjct: 181 PSNLVQVSLFRALHEKENKSKWGISRNQFFVITLITSFSYYLLPGYLFTVLTTVSWLCWI 240
Query: 249 WKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIP 308
+SI Q+GSG GLGIGSFGLDW+T+A +LGSPLA P FA NI AGFFL MYV+ P
Sbjct: 241 SPKSILVNQLGSGSAGLGIGSFGLDWSTIASYLGSPLASPFFASANIAAGFFLVMYVITP 300
Query: 309 ISYWNNLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAF 368
+ Y+ +LY+AK FP+ S F ++G Y VT I++ F +D ++Y+ +++S FA
Sbjct: 301 LCYYLDLYNAKTFPIYSGKLFVASGKEYKVTSIIDAN-FRLDRQAYAETGPVHMSTFFAV 359
Query: 369 EYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSL-KNQLGDVHTRIMKKNYEQVPEWWF 427
YG FA L+A+I HV++F+G+ +W +T + KN+ D+HT+IMK+NY++VP WWF
Sbjct: 360 TYGLGFATLSASIFHVLIFNGK---DLWTQTRGAFGKNKKMDIHTKIMKRNYKEVPLWWF 416
Query: 428 VTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVIT 487
++I + + + + C + Q+QLPWWG L+ IA+ FT +GVI ATTN GLN+IT
Sbjct: 417 LSIFAVNLAVIVFICIYYKTQIQLPWWGAFLACLIAIFFTPLVGVIMATTNQAPGLNIIT 476
Query: 488 ELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVA 547
E +IG+ YP +P+AN+ FKTYG+ISM Q+L FL D KLG YMKI P++MF+ Q+VGT+VA
Sbjct: 477 EYIIGYAYPERPVANICFKTYGYISMSQSLTFLSDLKLGTYMKIPPRTMFMAQVVGTLVA 536
Query: 548 SSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVY 607
+ GTAWWL+ I N+CD +LLP GS WTCP D VF++AS+IWG+VGP+RMF G Y
Sbjct: 537 VIAYAGTAWWLMAEIPNLCDTNLLPPGSQWTCPSDRVFFDASVIWGLVGPRRMFGDLGEY 596
Query: 608 PELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVG 667
+NWFF+ G IAP V+L S FPN+KWI I+IP++I + +PP +VN+ +W ++
Sbjct: 597 SNINWFFVGGAIAPALVYLASRLFPNKKWISDIHIPVLIGATAIMPPASAVNFTSWLVMA 656
Query: 668 IFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCP 727
F +V++ + WW R+ Y+LS G+DAG F+ +LL+ +LQ I WWG + CP
Sbjct: 657 FVFGHFVFKYRREWWQRYNYVLSGGMDAGTGFMSVLLFLALQRSEI-AIDWWGNSGEGCP 715
Query: 728 LAKCPTAPGVHAEGCPV 744
+AKCPTA GV GCPV
Sbjct: 716 VAKCPTAKGVVVHGCPV 732
>At5g64410 Isp4-like protein
Length = 729
Score = 793 bits (2047), Expect = 0.0
Identities = 373/737 (50%), Positives = 517/737 (69%), Gaps = 17/737 (2%)
Query: 15 AEIDEYSIND-SPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNP 73
A DE+S D SPIE+VRLTV TDDP+ P TFR W LGL SC LL+F+NQF YRT P
Sbjct: 2 ATADEFSDEDTSPIEEVRLTVTNTDDPTLPVWTFRMWFLGLISCSLLSFLNQFFSYRTEP 61
Query: 74 LKITSVSAQIIALPLGKLMAATLPTKPIQVPFT-TWSFSLNPGPFSLKEHVLITIFASSG 132
L IT ++ Q+ LP+G +A LP +P + FSLNPGPF++KEHVLI+IFA++G
Sbjct: 62 LVITQITVQVATLPIGHFLAKVLPKTRFGLPGCGSARFSLNPGPFNMKEHVLISIFANAG 121
Query: 133 SS----GVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWW 188
S+ YA+ IITI+KAFY RSI +A +LL ++TQ+LGYGWAG+ R+++V+ +MWW
Sbjct: 122 SAFGSGSAYAVGIITIIKAFYGRSISFIAGWLLIITTQVLGYGWAGLLRKYVVEPAHMWW 181
Query: 189 PESLVQVSLFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLI 248
P +LVQVSLFRA HEK+ + +R +FF + V SF +YI+PGY F ++++S+VC
Sbjct: 182 PSTLVQVSLFRALHEKDDQRM---TRAKFFVIALVCSFGWYIVPGYLFTTLTSISWVCWA 238
Query: 249 WKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIP 308
+ S+TAQQIGSGM+GLG+G+F LDW VA FL SPL P FAI N+ G+ L +Y ++P
Sbjct: 239 FPRSVTAQQIGSGMRGLGLGAFTLDWTAVASFLFSPLISPFFAIANVFIGYVLLIYFVLP 298
Query: 309 ISYWN-NLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFA 367
++YW + Y+A +FP+ SSH F S G TY++ I+N + F++D+ Y +I LS+ FA
Sbjct: 299 LAYWGFDSYNATRFPIFSSHLFTSVGNTYDIPAIVN-DNFELDLAKYEQQGRINLSMFFA 357
Query: 368 FEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWF 427
YG FA + +T++HV LF+G+ I + ++ S K + D+HTR+MK+ Y+ +P WWF
Sbjct: 358 LTYGLGFATIASTLTHVALFYGKEISERFR---VSYKGK-EDIHTRLMKR-YKDIPSWWF 412
Query: 428 VTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVIT 487
++L ++++L C ++Q+PWWG++ + +A +FTLPI +I ATTN GLN+IT
Sbjct: 413 YSMLAATLLISLALCVFLNDEVQMPWWGLVFASAMAFVFTLPISIITATTNQTPGLNIIT 472
Query: 488 ELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVA 547
E +G IYPG+P+ANV FK YG++SM QA+ FL DFKLGHYMKI P+SMF+VQ +GT++A
Sbjct: 473 EYAMGLIYPGRPIANVCFKVYGYMSMAQAVSFLNDFKLGHYMKIPPRSMFLVQFIGTILA 532
Query: 548 SSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVY 607
+++ AWW L SI+NIC E LLP SPWTCPGD VF++AS+IWG+VGPKR+F G Y
Sbjct: 533 GTINITVAWWQLNSIKNICQEELLPPNSPWTCPGDRVFFDASVIWGLVGPKRIFGSQGNY 592
Query: 608 PELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVG 667
+NWFFL G + PV VW L FP + WI +N+P+++ + +PP +VNY +W +VG
Sbjct: 593 AAMNWFFLGGALGPVIVWSLHKAFPKRSWIPLVNLPVLLGATAMMPPATAVNYNSWILVG 652
Query: 668 IFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCP 727
FN +V+R K+WW R+ Y+LSA +DAGVAF+ +LLYFS+ WWG +HC
Sbjct: 653 TIFNLFVFRYRKSWWQRYNYVLSAAMDAGVAFMAVLLYFSV-GMEEKSLDWWGTRGEHCD 711
Query: 728 LAKCPTAPGVHAEGCPV 744
LAKCPTA GV +GCPV
Sbjct: 712 LAKCPTARGVIVDGCPV 728
>At5g53510 isp4 protein
Length = 741
Score = 790 bits (2040), Expect = 0.0
Identities = 378/742 (50%), Positives = 516/742 (68%), Gaps = 9/742 (1%)
Query: 6 VSQNTVIEDAEIDEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQ 65
V ++ + + E ++ +E+V LTVP TDDP+ P LTFR W LGL +CI+L+F+NQ
Sbjct: 5 VEDSSRVMEIEGQNDDLDRCVVEEVELTVPKTDDPTLPVLTFRMWTLGLGACIILSFINQ 64
Query: 66 FLGYRTNPLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLI 125
F YR PL I+ +SAQI +PLG LMA LPT+ + + W FS+NPGPF++KEHVLI
Sbjct: 65 FFWYRQMPLTISGISAQIAVVPLGHLMAKVLPTRMF-LEGSKWEFSMNPGPFNVKEHVLI 123
Query: 126 TIFASSGSSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPY 185
TIFA+SG+ VYA +I++ +K +Y RS+ + A+LL ++TQ LG+GWAG+FR+ LV+
Sbjct: 124 TIFANSGAGTVYATHILSAIKLYYKRSLPFLPAFLLMITTQFLGFGWAGLFRKHLVEPGE 183
Query: 186 MWWPESLVQVSLFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFV 245
MWWP +LVQVSLF A HEKEK+ +GG +R+QFF +V V SFAYYI+PGY F I+++S++
Sbjct: 184 MWWPSNLVQVSLFSALHEKEKKKKGGMTRIQFFLIVLVTSFAYYILPGYLFTMITSISWI 243
Query: 246 CLIWKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYV 305
C + +S+ Q+GSG +GLGIG+ G+DW T++ +LGSPLA P FA IN+ GF + MYV
Sbjct: 244 CWLGPKSVLVHQLGSGEQGLGIGAIGIDWATISSYLGSPLASPLFATINVTIGFVVIMYV 303
Query: 306 LIPISYWNNLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVA 365
PI YW N+Y AK +P+ SS F G++Y+V I++ + F +D + Y+ I +S
Sbjct: 304 ATPICYWLNIYKAKTYPIFSSGLFMGNGSSYDVLSIIDKK-FHLDRDIYAKTGPINMSTF 362
Query: 366 FAFEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSL-KNQLGDVHTRIMKKNYEQVPE 424
FA YG FA L+ATI HV+LF+G +WK+T + +N+ D HTRIMKKNY +VP
Sbjct: 363 FAVTYGLGFATLSATIVHVLLFNGR---DLWKQTRGAFQRNKKMDFHTRIMKKNYREVPM 419
Query: 425 WWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLN 484
WWF IL+L + + + + +QLPWWG+LL+ IA+ FT IGVI ATTN GLN
Sbjct: 420 WWFYVILVLNIALIMFISFYYNATVQLPWWGVLLACAIAVFFTPLIGVIAATTNQEPGLN 479
Query: 485 VITELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGT 544
VITE VIG++YP +P+AN+ FK YG+ISM QAL F+ DFKLG YMKI P+SMF+ Q+VGT
Sbjct: 480 VITEYVIGYLYPERPVANMCFKVYGYISMTQALTFIQDFKLGLYMKIPPRSMFMAQVVGT 539
Query: 545 VVASSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKD 604
+V+ V+ GTAWWL+ I ++CD+SLLP S WTCP D VF++AS+IWG+VGP+RMF
Sbjct: 540 LVSVVVYTGTAWWLMVDIPHLCDKSLLPPDSEWTCPMDRVFFDASVIWGLVGPRRMFGNL 599
Query: 605 GVYPELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWG 664
G Y +NWFFL+G IAP VWL + FP KWI I+ P+I+ S +PP +VN+ +W
Sbjct: 600 GEYAAINWFFLVGAIAPFFVWLATKAFPAHKWISKIHFPVILGATSMMPPAMAVNFTSWC 659
Query: 665 IVGIFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPD 724
IV F ++Y+ + WW ++ Y+LS GLDAG AF+ +L++ S+ GI G WWG D
Sbjct: 660 IVAFVFGHFLYKYKRQWWKKYNYVLSGGLDAGTAFMTILIFLSVGRKGI-GLLWWGNADD 718
Query: 725 --HCPLAKCPTAPGVHAEGCPV 744
+C LA CPTA GV GCPV
Sbjct: 719 STNCSLASCPTAKGVIMHGCPV 740
>At4g16370 isp4 like protein
Length = 637
Score = 566 bits (1458), Expect = e-161
Identities = 276/547 (50%), Positives = 376/547 (68%), Gaps = 13/547 (2%)
Query: 82 QIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSG----SSGVY 137
QI LP+GK MA TLPT + WSFSLNPGPF++KEHV+ITIFA+ G Y
Sbjct: 2 QIAGLPIGKFMARTLPTTSHNL--LGWSFSLNPGPFNIKEHVIITIFANCGVAYGGGDAY 59
Query: 138 AINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSL 197
+I IT++KA+Y +S+ + + L+TQ+LGYGWAGI RR+LVD MWWP +L QVSL
Sbjct: 60 SIGAITVMKAYYKQSLSFICGLFIVLTTQILGYGWAGILRRYLVDPVDMWWPSNLAQVSL 119
Query: 198 FRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQ 257
FRA HEKE + +G T R++FF V ASF YY +PGY F ++ S+VC W SITAQQ
Sbjct: 120 FRALHEKENKSKGLT-RMKFFLVALGASFIYYALPGYLFPILTFSSWVCWAWPNSITAQQ 178
Query: 258 IGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWN-NLY 316
+GSG GLG+G+F LDW ++ + GSPL P +I+N+ GF +++Y+++P+ YW N +
Sbjct: 179 VGSGYHGLGVGAFTLDWAGISAYHGSPLVAPWSSILNVGVGFIMFIYIIVPVCYWKFNTF 238
Query: 317 DAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAA 376
DA+KFP+ S+ F ++G Y+ T+IL T FD+D+ +Y+NY K+YLS FA G FA
Sbjct: 239 DARKFPISSNQLFTTSGQKYDTTKIL-TPQFDLDIGAYNNYGKLYLSPLFALSIGSGFAR 297
Query: 377 LTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMVM 436
TAT++HV LF+G I WK+T +++ D+H ++M+ +Y++VPEWWF +L V
Sbjct: 298 FTATLTHVALFNGRDI---WKQTWSAVNTTKLDIHGKLMQ-SYKKVPEWWFYILLAGSVA 353
Query: 437 MALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYP 496
M+LL + + +QLPWWG+L + +A I TLPIGVI+ATTN + G ++I + +IG+I P
Sbjct: 354 MSLLMSFVWKESVQLPWWGMLFAFALAFIVTLPIGVIQATTNQQPGYDIIGQFIIGYILP 413
Query: 497 GKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAW 556
GKP+AN+ FK YG IS V AL FL D KLGHYMKI P M+ QLVGTVVA V+ G AW
Sbjct: 414 GKPIANLIFKIYGRISTVHALSFLADLKLGHYMKIPPPCMYTAQLVGTVVAGVVNLGVAW 473
Query: 557 WLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLI 616
W+L SI++ICD SPWTCP V ++AS+IWG++GP+R+F G+Y L FFLI
Sbjct: 474 WMLESIQDICDIEGDHPNSPWTCPKYRVTFDASVIWGLIGPRRLFGPGGMYRNLVGFFLI 533
Query: 617 GLIAPVP 623
G + PVP
Sbjct: 534 GAVLPVP 540
>At1g65730 unknown protein
Length = 688
Score = 40.4 bits (93), Expect = 0.004
Identities = 43/182 (23%), Positives = 77/182 (41%), Gaps = 13/182 (7%)
Query: 420 EQVPEWWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIA--LIFTLPIGVIEATT 477
+++P W+ VT +++ +++++ QL+ W+ IL+ IA L F G
Sbjct: 386 DRIPSWFAVTGYVVLAIVSIITVPHIFHQLK--WYHILIMYIIAPVLAFCNAYGCGLTDW 443
Query: 478 NIRSGLNVITELVIGF---IYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPK 534
++ S + IG G LA +A + A + DFK G+ +P+
Sbjct: 444 SLASTYGKLAIFTIGAWAGASNGGVLAGLAACGVMMNIVSTASDLMQDFKTGYMTLASPR 503
Query: 535 SMFIVQLVGTVVASSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGV 594
SMF+ Q +GT + + W + + G+ + P V+ N SI+ GV
Sbjct: 504 SMFLSQAIGTAMGCVISPCVFWLFYKAFPDFGQ-----PGTAYPAPYALVYRNMSIL-GV 557
Query: 595 VG 596
G
Sbjct: 558 EG 559
>At4g24120 unknown protein
Length = 665
Score = 37.7 bits (86), Expect = 0.024
Identities = 51/215 (23%), Positives = 87/215 (39%), Gaps = 29/215 (13%)
Query: 350 DMESYSNYSKIYLSVAFAFEYG-FCFAA-LTATISHVVLFHGEMIVQMWKKTTTSLKNQL 407
+M+S Y K++LSVA G + F L TI++V + N L
Sbjct: 311 NMKSIYGY-KVFLSVALILGDGLYTFVKILFVTIANV------------NARLKNKPNDL 357
Query: 408 GDVHTRIMKKNYEQ--------VPEWWFVTILILMVMMALLACEGFGKQLQLPWWGILLS 459
DV + +K+ ++ +P W+ V+ + ++ + QL+ W+ ++++
Sbjct: 358 DDVGHKKQRKDLKEDENFLRDKIPMWFAVSGYLTFAAVSTVVVPLIFPQLK--WYYVIVA 415
Query: 460 LTIA--LIFTLPIGVIEATTNIRSGLNVITELVIGFIYPGKPLANVAFKTYGHISMVQAL 517
A L F G N+ I VI + + G I V ++
Sbjct: 416 YIFAPSLAFCNAYGAGLTDINMAYNYGKIGLFVIAAVTGRENGVVAGLAGCGLIKSVVSV 475
Query: 518 G--FLGDFKLGHYMKIAPKSMFIVQLVGTVVASSV 550
+ DFK HY +PK+MF Q++GTVV V
Sbjct: 476 SCILMQDFKTAHYTMTSPKAMFASQMIGTVVGCIV 510
>At3g27020 unknown protein
Length = 676
Score = 34.3 bits (77), Expect = 0.26
Identities = 42/150 (28%), Positives = 64/150 (42%), Gaps = 11/150 (7%)
Query: 451 LPWWGILLSLTIA--LIFTLPIGVIEATTNIRSGLNVITELVIGFIY--PGKPLANVAFK 506
L W+ +L S IA L F G ++ S I +I + G +A +A
Sbjct: 415 LKWYFVLCSYFIAPALAFCNSYGTGLTDWSLASTYGKIGLFIIASVVGSDGGVIAGLAAC 474
Query: 507 TYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLTSIENIC 566
+ A + DFK G+ + KSMF+ QLVGT + + T W T+ + I
Sbjct: 475 GVMMSIVSTAADLMQDFKTGYLTLSSAKSMFVSQLVGTAMGCVIAPLTFWLFWTAFD-IG 533
Query: 567 DESLLPKGSPWTCPGDDVFYNASIIWGVVG 596
D P G P+ P +F +I+ G+ G
Sbjct: 534 D----PNG-PYKAPYAVIFREMAIL-GIEG 557
>At5g41000 unknown protein
Length = 670
Score = 33.5 bits (75), Expect = 0.44
Identities = 27/81 (33%), Positives = 38/81 (46%), Gaps = 7/81 (8%)
Query: 516 ALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLTSIENICDESLLPKGS 575
A + DFK G+ + KSMF+ QL+GT + + T W T+ + I D L K +
Sbjct: 481 AADLMQDFKTGYLTLSSAKSMFVTQLLGTAMGCIIAPLTFWLFWTAFD-IGDPDGLYK-A 538
Query: 576 PWTCPGDDVFYNASIIWGVVG 596
P+ V Y I GV G
Sbjct: 539 PYA-----VIYREMAILGVEG 554
>At5g24380 putative protein
Length = 652
Score = 33.5 bits (75), Expect = 0.44
Identities = 36/148 (24%), Positives = 66/148 (44%), Gaps = 32/148 (21%)
Query: 420 EQVPEWWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIA--LIFTLPIGVIEATT 477
E +P W + +++++A QL+ W+ +L++ +A L F G
Sbjct: 364 ESIPLWMACVGYLFFSLVSIIAIPLMFPQLK--WYFVLVAYLLAPSLSFCNAYG------ 415
Query: 478 NIRSGLNVITELVIGFIYPGKP----LANVAFKTYGHISMVQALGFLG-----------D 522
+GL T++ + + Y GK +A +A K G ++ + A G + D
Sbjct: 416 ---AGL---TDMNMAYNY-GKAALFVMAALAGKNDGVVAGMVACGLIKSIVSVSADLMHD 468
Query: 523 FKLGHYMKIAPKSMFIVQLVGTVVASSV 550
FK GH + +P+SM + Q +GT + V
Sbjct: 469 FKTGHLTQTSPRSMLVAQAIGTAIGCVV 496
>At5g45450 putative protein
Length = 216
Score = 32.3 bits (72), Expect = 0.99
Identities = 14/44 (31%), Positives = 24/44 (53%)
Query: 522 DFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLTSIENI 565
DFK G+ +PKSMF+ Q++GT + V W + +++
Sbjct: 24 DFKTGYLTLSSPKSMFVSQVIGTAMGCVVSPCVFWLFYKAFDDL 67
>At3g17650 unknown protein
Length = 714
Score = 32.3 bits (72), Expect = 0.99
Identities = 14/44 (31%), Positives = 24/44 (53%)
Query: 522 DFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLTSIENI 565
DFK G+ +PKSMF+ Q++GT + V W + +++
Sbjct: 516 DFKTGYLTLSSPKSMFVSQVIGTAMGCVVSPCVFWLFYKAFDDL 559
>At4g30850 unknown protein
Length = 358
Score = 30.8 bits (68), Expect = 2.9
Identities = 28/81 (34%), Positives = 36/81 (43%), Gaps = 19/81 (23%)
Query: 400 TTSLKNQLGDVHTRIMKKNYEQVPEW-WFVTI------LILMVMMALLACEGFGKQLQLP 452
T SL N GDV NYE +P+W W V + LI M L AC ++ L
Sbjct: 142 TRSLLNSQGDV-------NYEAIPKWPWLVFLTGAMGCLICSSMSHLFACH--SRRFNLF 192
Query: 453 WWGI---LLSLTIALIFTLPI 470
+W + +SL I F PI
Sbjct: 193 FWRLDYAGISLMIVCSFFAPI 213
>At5g53550 EspB-like protein
Length = 669
Score = 30.4 bits (67), Expect = 3.8
Identities = 12/29 (41%), Positives = 17/29 (58%)
Query: 522 DFKLGHYMKIAPKSMFIVQLVGTVVASSV 550
DFK GH +P+SM + Q +GT + V
Sbjct: 482 DFKTGHLTLTSPRSMLVSQAIGTAIGCVV 510
Score = 29.3 bits (64), Expect = 8.4
Identities = 30/126 (23%), Positives = 51/126 (39%), Gaps = 20/126 (15%)
Query: 493 FIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHF 552
F G + F T+G ++ + ++G I P + I L G V++ +
Sbjct: 238 FFSGGTECGFIQFPTFGLEALKNTMTYVGAGM------ICPHIVNISLLFGAVLS----W 287
Query: 553 GTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNW 612
G W L+ ++ S LP+ S + G VF + S+I G DG+Y +
Sbjct: 288 GIMWPLIKGLKGDWFPSTLPENSMKSLNGYKVFISISLILG----------DGLYQFIKI 337
Query: 613 FFLIGL 618
F G+
Sbjct: 338 LFKTGI 343
>At3g03250 UDP-glucose pyrophosphorylase like protein
Length = 469
Score = 30.4 bits (67), Expect = 3.8
Identities = 20/83 (24%), Positives = 37/83 (44%), Gaps = 2/83 (2%)
Query: 279 GFLGSPLAVPG-FAIINIMAGFFLYMYVLIPISYWNNLYDAKKFPLISSHTFDSTGATYN 337
G LG+ + G ++I + G ++I I NN Y K PL+ ++F++ T+
Sbjct: 87 GGLGTTMGCTGPKSVIEVRDGLTFLDLIVIQIENLNNKYGCK-VPLVLMNSFNTHDDTHK 145
Query: 338 VTRILNTETFDIDMESYSNYSKI 360
+ DI + S Y ++
Sbjct: 146 IVEKYTNSNVDIHTFNQSKYPRV 168
>At1g48370 unknown protein
Length = 724
Score = 30.0 bits (66), Expect = 4.9
Identities = 12/44 (27%), Positives = 24/44 (54%)
Query: 522 DFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLTSIENI 565
DFK G+ +P++MF+ Q++GT + V W + +++
Sbjct: 526 DFKTGYLTLSSPRAMFVSQVIGTAMGCLVSPCVFWLFYKAFDDL 569
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.141 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,559,648
Number of Sequences: 26719
Number of extensions: 715133
Number of successful extensions: 1955
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1879
Number of HSP's gapped (non-prelim): 34
length of query: 745
length of database: 11,318,596
effective HSP length: 107
effective length of query: 638
effective length of database: 8,459,663
effective search space: 5397264994
effective search space used: 5397264994
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC143341.7


