

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143341.11 - phase: 0
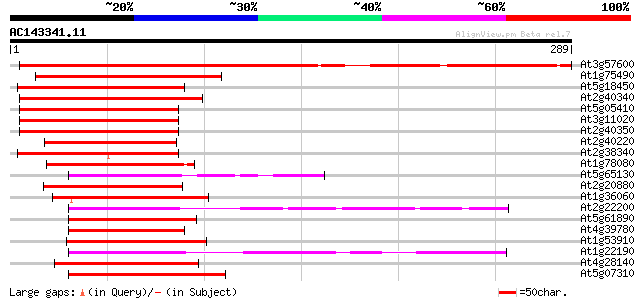
(289 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g57600 AP2 transcription factor - like protein 276 9e-75
At1g75490 transcription factor DREB2A like protein 127 7e-30
At5g18450 AP2-domain DNA-binding protein -like 123 1e-28
At2g40340 AP2 domain transcription factor 121 4e-28
At5g05410 DREB2A (dbj|BAA33794.1) 119 1e-27
At3g11020 DREB2B transcription factor 115 2e-26
At2g40350 AP2 domain transcription factor 114 5e-26
At2g40220 AP2 domain transcription factor (ABI4:abscisic acid-in... 107 6e-24
At2g38340 DREB-like AP2 domain transcription factor 107 1e-23
At1g78080 putative AP2 domain containing protein (At1g78080) 95 5e-20
At5g65130 putative protein 94 7e-20
At2g20880 AP2 domain transcription factor 93 2e-19
At1g36060 putative AP2 domain containing protein RAP2.4 gi|2281633 92 3e-19
At2g22200 pseudogene 91 6e-19
At5g61890 putative protein 91 9e-19
At4g39780 putative protein 90 1e-18
At1g53910 unknown protein 90 1e-18
At1g22190 unknown protein 90 1e-18
At4g28140 putative DNA-binding protein 89 2e-18
At5g07310 putative transcription factor 89 3e-18
>At3g57600 AP2 transcription factor - like protein
Length = 277
Score = 276 bits (706), Expect = 9e-75
Identities = 147/287 (51%), Positives = 193/287 (67%), Gaps = 20/287 (6%)
Query: 6 KKSPLKPWKKGPTRGKGGPQNASCEYRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEE 65
K S +K WKKGP RGKGGPQNA C+YRGVRQRTWGKWVAEIREPKKR RLWLGSFATAEE
Sbjct: 3 KSSSMKQWKKGPARGKGGPQNALCQYRGVRQRTWGKWVAEIREPKKRARLWLGSFATAEE 62
Query: 66 AAMAYDEAARRLYGPDAYLNLPHMQTHSNSTMKTG-KFKWLPSKNFISMFPSCGLLNVNA 124
AAMAYDEAA +LYG DAYLNLPH+Q ++ ++ +FKW+PS+ FISMFPSCG+LNVNA
Sbjct: 63 AAMAYDEAALKLYGHDAYLNLPHLQRNTRPSLSNSQRFKWVPSRKFISMFPSCGMLNVNA 122
Query: 125 QPSVHLIHQRLQEFKQNAVVASQSSFSSSSNDPKAEEIQKVDSKKSHTEDPLPKETIVQT 184
QPSVH+I QRL+E K+ +++ S SSSS + K +D K S E
Sbjct: 123 QPSVHIIQQRLEELKKTGLLSQSYSSSSSSTESKT-NTSFLDEKTSKGE----------- 170
Query: 185 SANKMLGDLQEEKPQIDLNEFLQQMGILKEGSHSEQTESSGSSTVHEVLPRDDNDQLGI- 243
+ M ++KP+IDL EFLQQ+GILK+ + +E +E + H P ++ ++ G
Sbjct: 171 -TDNMFEGGDQKKPEIDLTEFLQQLGILKDENEAEPSE---VAECHSPPPWNEQEETGSP 226
Query: 244 FSDMSVNWEALIEM-HEFAGIEESEATHLEAYDPNDHLNFSTSIWDF 289
F + +W+ LIEM + ++++ +YD D ++F SIWD+
Sbjct: 227 FRTENFSWDTLIEMPRSETTTMQFDSSNFGSYDFEDDVSF-PSIWDY 272
>At1g75490 transcription factor DREB2A like protein
Length = 206
Score = 127 bits (319), Expect = 7e-30
Identities = 59/96 (61%), Positives = 70/96 (72%)
Query: 14 KKGPTRGKGGPQNASCEYRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEA 73
+KG RGKGGP NASC Y+GVRQRTWGKWVAEIREP + RLWLG+F T+ EAA+AYD A
Sbjct: 25 RKGCMRGKGGPDNASCTYKGVRQRTWGKWVAEIREPNRGARLWLGTFDTSREAALAYDSA 84
Query: 74 ARRLYGPDAYLNLPHMQTHSNSTMKTGKFKWLPSKN 109
AR+LYGP+A+LNLP T + + PS N
Sbjct: 85 ARKLYGPEAHLNLPESLRSYPKTASSPASQTTPSSN 120
>At5g18450 AP2-domain DNA-binding protein -like
Length = 307
Score = 123 bits (308), Expect = 1e-28
Identities = 57/86 (66%), Positives = 70/86 (81%)
Query: 5 SKKSPLKPWKKGPTRGKGGPQNASCEYRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAE 64
+KK + +KG +GKGGP+NA+C +RGVRQRTWGKWVAEIREP + TRLWLG+F T+
Sbjct: 8 AKKRNMGRSRKGCMKGKGGPENATCTFRGVRQRTWGKWVAEIREPNRGTRLWLGTFNTSV 67
Query: 65 EAAMAYDEAARRLYGPDAYLNLPHMQ 90
EAAMAYDEAA++LYG +A LNL H Q
Sbjct: 68 EAAMAYDEAAKKLYGHEAKLNLVHPQ 93
>At2g40340 AP2 domain transcription factor
Length = 341
Score = 121 bits (304), Expect = 4e-28
Identities = 56/94 (59%), Positives = 70/94 (73%)
Query: 6 KKSPLKPWKKGPTRGKGGPQNASCEYRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEE 65
+K P K +KG +GKGGP+N C+YRGVRQR WGKWVAEIREP RLWLG+F+++ E
Sbjct: 47 RKPPPKGSRKGCMKGKGGPENGICDYRGVRQRRWGKWVAEIREPDGGARLWLGTFSSSYE 106
Query: 66 AAMAYDEAARRLYGPDAYLNLPHMQTHSNSTMKT 99
AA+AYDEAA+ +YG A LNLP + S+ST T
Sbjct: 107 AALAYDEAAKAIYGQSARLNLPEITNRSSSTAAT 140
>At5g05410 DREB2A (dbj|BAA33794.1)
Length = 335
Score = 119 bits (299), Expect = 1e-27
Identities = 54/82 (65%), Positives = 65/82 (78%)
Query: 6 KKSPLKPWKKGPTRGKGGPQNASCEYRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEE 65
+K P K KKG +GKGGP+N+ C +RGVRQR WGKWVAEIREP + +RLWLG+F TA+E
Sbjct: 54 RKVPAKGSKKGCMKGKGGPENSRCSFRGVRQRIWGKWVAEIREPNRGSRLWLGTFPTAQE 113
Query: 66 AAMAYDEAARRLYGPDAYLNLP 87
AA AYDEAA+ +YGP A LN P
Sbjct: 114 AASAYDEAAKAMYGPLARLNFP 135
>At3g11020 DREB2B transcription factor
Length = 330
Score = 115 bits (289), Expect = 2e-26
Identities = 55/82 (67%), Positives = 62/82 (75%)
Query: 6 KKSPLKPWKKGPTRGKGGPQNASCEYRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEE 65
+K P K KKG +GKGGP N+ C +RGVRQR WGKWVAEIREPK TRLWLG+F TAE+
Sbjct: 53 RKVPAKGSKKGCMKGKGGPDNSHCSFRGVRQRIWGKWVAEIREPKIGTRLWLGTFPTAEK 112
Query: 66 AAMAYDEAARRLYGPDAYLNLP 87
AA AYDEAA +YG A LN P
Sbjct: 113 AASAYDEAATAMYGSLARLNFP 134
>At2g40350 AP2 domain transcription factor
Length = 177
Score = 114 bits (286), Expect = 5e-26
Identities = 50/82 (60%), Positives = 66/82 (79%)
Query: 6 KKSPLKPWKKGPTRGKGGPQNASCEYRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEE 65
+K+P K +KG +GKGGP+N C+Y GVRQRTWGKWVAEIREP + +LWLG+F+++ E
Sbjct: 42 RKAPPKRSRKGCMKGKGGPENGICDYTGVRQRTWGKWVAEIREPGRGAKLWLGTFSSSYE 101
Query: 66 AAMAYDEAARRLYGPDAYLNLP 87
AA+AYDEA++ +YG A LNLP
Sbjct: 102 AALAYDEASKAIYGQSARLNLP 123
>At2g40220 AP2 domain transcription factor (ABI4:abscisic
acid-insensitive 4 )
Length = 328
Score = 107 bits (268), Expect = 6e-24
Identities = 50/68 (73%), Positives = 56/68 (81%)
Query: 19 RGKGGPQNASCEYRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEAARRLY 78
+GKGGP N+ YRGVRQR+WGKWVAEIREP+KRTR WLG+FATAE+AA AYD AA LY
Sbjct: 43 KGKGGPDNSKFRYRGVRQRSWGKWVAEIREPRKRTRKWLGTFATAEDAARAYDRAAVYLY 102
Query: 79 GPDAYLNL 86
G A LNL
Sbjct: 103 GSRAQLNL 110
>At2g38340 DREB-like AP2 domain transcription factor
Length = 244
Score = 107 bits (266), Expect = 1e-23
Identities = 53/91 (58%), Positives = 62/91 (67%), Gaps = 8/91 (8%)
Query: 5 SKKSPLKPWKKGPTRGKGGPQNASCEYRGVRQRTWGKWVAEIREP--------KKRTRLW 56
+++ K KKG RGKGGP+N C +RGVRQR WGKWVAEIREP + RLW
Sbjct: 44 ARRVQAKGSKKGCMRGKGGPENPVCRFRGVRQRVWGKWVAEIREPVSHRGANSSRSKRLW 103
Query: 57 LGSFATAEEAAMAYDEAARRLYGPDAYLNLP 87
LG+FATA EAA+AYD AA +YGP A LN P
Sbjct: 104 LGTFATAAEAALAYDRAASVMYGPYARLNFP 134
>At1g78080 putative AP2 domain containing protein (At1g78080)
Length = 334
Score = 94.7 bits (234), Expect = 5e-20
Identities = 47/76 (61%), Positives = 57/76 (74%), Gaps = 1/76 (1%)
Query: 20 GKGGPQNASCEYRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEAARRLYG 79
G G P + YRGVRQR WGKWVAEIR P+ RTRLWLG+F TAEEAA+AYD+AA +L G
Sbjct: 141 GSGVPSKPTKLYRGVRQRHWGKWVAEIRLPRNRTRLWLGTFDTAEEAALAYDKAAYKLRG 200
Query: 80 PDAYLNLPHMQTHSNS 95
A LN P+++ H+ S
Sbjct: 201 DFARLNFPNLR-HNGS 215
>At5g65130 putative protein
Length = 277
Score = 94.4 bits (233), Expect = 7e-20
Identities = 57/132 (43%), Positives = 75/132 (56%), Gaps = 16/132 (12%)
Query: 31 YRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEAARRLYGPDAYLNLPHMQ 90
YRGVRQR WGKWVAEIR PK RTRLWLG+F TA+EAA+AYD+AA ++ G +A LN P +
Sbjct: 111 YRGVRQRQWGKWVAEIRLPKNRTRLWLGTFETAQEAALAYDQAAHKIRGDNARLNFPDI- 169
Query: 91 THSNSTMKTGKFKWLPSKNFISMFPSCGLLNVNAQPSVHLIHQRLQEFKQNAVVASQSSF 150
++ G +K + S + + S + N + P Q KQN S F
Sbjct: 170 ------VRQGHYKQILSPSINAKIES--ICNSSDLPLP-------QIEKQNKTEEVLSGF 214
Query: 151 SSSSNDPKAEEI 162
S +P+ EI
Sbjct: 215 SKPEKEPEFGEI 226
>At2g20880 AP2 domain transcription factor
Length = 336
Score = 92.8 bits (229), Expect = 2e-19
Identities = 46/72 (63%), Positives = 53/72 (72%)
Query: 18 TRGKGGPQNASCEYRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEAARRL 77
TR P +A+ YRGVRQR WGKWVAEIR+P+ RTRLWLG+F TAEEAAMAYD A +L
Sbjct: 174 TRPPVQPFSATKLYRGVRQRHWGKWVAEIRKPRNRTRLWLGTFDTAEEAAMAYDREAFKL 233
Query: 78 YGPDAYLNLPHM 89
G A LN P +
Sbjct: 234 RGETARLNFPEL 245
>At1g36060 putative AP2 domain containing protein RAP2.4 gi|2281633
Length = 314
Score = 92.0 bits (227), Expect = 3e-19
Identities = 48/86 (55%), Positives = 59/86 (67%), Gaps = 6/86 (6%)
Query: 23 GPQNASCE-----YRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEAARRL 77
GP + S YRGVRQR WGKWVAEIR P+ RTRLWLG+F TAEEAA+AYD AA +L
Sbjct: 130 GPSSGSVSKPAKLYRGVRQRHWGKWVAEIRLPRNRTRLWLGTFDTAEEAALAYDRAAFKL 189
Query: 78 YGPDAYLNLPHMQTHSNST-MKTGKF 102
G A LN P ++ + S+ TG++
Sbjct: 190 RGDSARLNFPALRYQTGSSPSDTGEY 215
>At2g22200 pseudogene
Length = 261
Score = 91.3 bits (225), Expect = 6e-19
Identities = 73/228 (32%), Positives = 105/228 (46%), Gaps = 41/228 (17%)
Query: 31 YRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEAARRLYGPDAYLNLPHMQ 90
YRGVRQR WGKWVAEIR PK RTRLWLG+F TAE+AA+AYD+AA +L G A LN P+
Sbjct: 71 YRGVRQRHWGKWVAEIRLPKNRTRLWLGTFETAEKAALAYDQAAFQLRGDIAKLNFPN-- 128
Query: 91 THSNSTMKTGKFKWLPSKNFISMFPSCGLLNVNAQPSVHLIHQRLQEFKQNAVVASQSSF 150
L++ + P + +LQ ++ +
Sbjct: 129 ----------------------------LIHEDMNPLPSSVDTKLQAICKS--LRKTEEI 158
Query: 151 SSSSNDPKAEEIQKVDSKKSHTEDPLPK-ETIVQTSANKMLGDLQEEKPQIDLNEFLQQM 209
S S+ K + V K TE LPK E + + +L E P+ D L +
Sbjct: 159 CSVSDQTKEYSVYSVSDK---TELFLPKAELFLPKREHLETNELSNESPRSDETSLLDES 215
Query: 210 GILKEGSHSEQTESSGSSTVHEVLPRDDNDQLGIFSDMSVNWEALIEM 257
E S S++T S T E + + L F + ++W+A+ ++
Sbjct: 216 --QAEYSSSDKTFLDFSDTEFEEI---GSFGLRKFPSVEIDWDAISKL 258
>At5g61890 putative protein
Length = 248
Score = 90.5 bits (223), Expect = 9e-19
Identities = 44/67 (65%), Positives = 50/67 (73%), Gaps = 1/67 (1%)
Query: 31 YRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEAARRLYGPDAYLNLP-HM 89
YRGVRQR WGKW AEIR+PKK R+WLG+F TAE AA+AYDEAA + G A LN P +
Sbjct: 90 YRGVRQRPWGKWAAEIRDPKKAARVWLGTFETAESAALAYDEAALKFKGSKAKLNFPERV 149
Query: 90 QTHSNST 96
Q SNST
Sbjct: 150 QLGSNST 156
>At4g39780 putative protein
Length = 272
Score = 90.1 bits (222), Expect = 1e-18
Identities = 44/60 (73%), Positives = 47/60 (78%)
Query: 31 YRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEAARRLYGPDAYLNLPHMQ 90
YRGVRQR WGKWVAEIR PK RTRLWLG+F TAEEAAMAYD AA +L G A LN P +
Sbjct: 93 YRGVRQRHWGKWVAEIRLPKNRTRLWLGTFDTAEEAAMAYDLAAYKLRGEFARLNFPQFR 152
>At1g53910 unknown protein
Length = 358
Score = 90.1 bits (222), Expect = 1e-18
Identities = 40/72 (55%), Positives = 52/72 (71%)
Query: 30 EYRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEAARRLYGPDAYLNLPHM 89
+YRG+RQR WGKW AEIR+P++ R+WLG+F TAEEAA AYD AARR+ G A +N P
Sbjct: 124 QYRGIRQRPWGKWAAEIRDPREGARIWLGTFKTAEEAARAYDAAARRIRGSKAKVNFPEE 183
Query: 90 QTHSNSTMKTGK 101
+NS ++ K
Sbjct: 184 NMKANSQKRSVK 195
>At1g22190 unknown protein
Length = 261
Score = 90.1 bits (222), Expect = 1e-18
Identities = 65/226 (28%), Positives = 96/226 (41%), Gaps = 53/226 (23%)
Query: 31 YRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEAARRLYGPDAYLNLPHMQ 90
YRGVRQR WGKWVAEIR P+ RTRLWLG+F TAEEAA+AYD+AA +L G A LN P ++
Sbjct: 83 YRGVRQRHWGKWVAEIRLPRNRTRLWLGTFDTAEEAALAYDKAAYKLRGDFARLNFPDLR 142
Query: 91 THSNSTMKTGKFKWLPSKNFISMFPSCGLLNVNAQPSVHLIHQRLQEFKQNAVVASQSSF 150
N QP + +L+ QN +Q
Sbjct: 143 H-----------------------------NDEYQPLQSSVDAKLEAICQNLAETTQKQV 173
Query: 151 SSSSNDPKAEEIQKVDSKKSHTEDPLPKETIVQTSANKMLGDLQEEKPQIDLNEFLQQMG 210
S+ + V K LP+E ++ +L +
Sbjct: 174 RSTKKSSSRKRSSTVAVK-------LPEEDYSSAGSSPLLTE-----------------S 209
Query: 211 ILKEGSHSEQTESSGSSTVHEVLPRDDNDQLGIFSDMSVNWEALIE 256
GS S +E + T E+ P + + L + ++W+++++
Sbjct: 210 YGSGGSSSPLSELTFGDTEEEIQPPWNENALEKYPSYEIDWDSILQ 255
>At4g28140 putative DNA-binding protein
Length = 292
Score = 89.4 bits (220), Expect = 2e-18
Identities = 44/74 (59%), Positives = 51/74 (68%)
Query: 24 PQNASCEYRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEAARRLYGPDAY 83
P A+ YRGVRQR WGKWVAEIR+P+ R RLWLG+F TAEEAAMAYD A +L G A
Sbjct: 136 PYIATKLYRGVRQRQWGKWVAEIRKPRSRARLWLGTFDTAEEAAMAYDRQAFKLRGHSAT 195
Query: 84 LNLPHMQTHSNSTM 97
LN P + S +
Sbjct: 196 LNFPEHFVNKESEL 209
>At5g07310 putative transcription factor
Length = 263
Score = 89.0 bits (219), Expect = 3e-18
Identities = 45/82 (54%), Positives = 54/82 (64%), Gaps = 1/82 (1%)
Query: 31 YRGVRQRTWGKWVAEIREPKKRTRLWLGSFATAEEAAMAYDEAARRLYGPDAYLNLP-HM 89
YRGVRQR WGKW AEIR+P+K R+WLG+F TAE AA+AYD AA + G A LN P
Sbjct: 92 YRGVRQRPWGKWAAEIRDPQKAARVWLGTFETAEAAALAYDNAALKFKGSKAKLNFPERA 151
Query: 90 QTHSNSTMKTGKFKWLPSKNFI 111
Q SN++ TG + S N I
Sbjct: 152 QLASNTSTTTGPPNYYSSNNQI 173
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.128 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,832,895
Number of Sequences: 26719
Number of extensions: 285829
Number of successful extensions: 1004
Number of sequences better than 10.0: 183
Number of HSP's better than 10.0 without gapping: 157
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 784
Number of HSP's gapped (non-prelim): 214
length of query: 289
length of database: 11,318,596
effective HSP length: 98
effective length of query: 191
effective length of database: 8,700,134
effective search space: 1661725594
effective search space used: 1661725594
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC143341.11


