

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143341.10 + phase: 0
(633 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
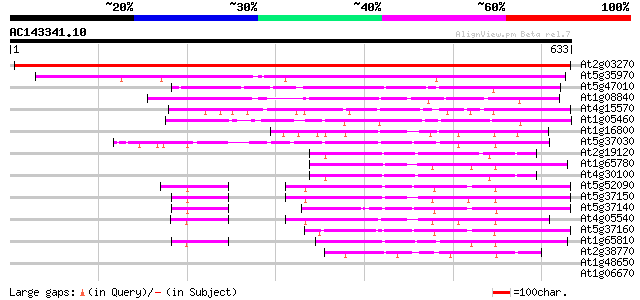
Score E
Sequences producing significant alignments: (bits) Value
At2g03270 helicase like protein 1022 0.0
At5g35970 DNA helicase-like 347 1e-95
At5g47010 UPF1 216 4e-56
At1g08840 hypothetical protein,5' partial 186 4e-47
At4g15570 SEN1 like protein 169 4e-42
At1g05460 unknown protein 150 2e-36
At1g16800 hypothetical protein 141 1e-33
At5g37030 SEN1-like protein 135 5e-32
At2g19120 putative DNA2-NAM7 helicase family protein 134 1e-31
At1g65780 hypothetical protein 134 1e-31
At4g30100 putative protein 131 1e-30
At5g52090 putative protein 124 1e-28
At5g37150 sen1-like protein 124 2e-28
At5g37140 sen1-like protein 120 2e-27
At4g05540 118 1e-26
At5g37160 sen1-like protein 110 2e-24
At1g65810 hypothetical protein 109 4e-24
At2g38770 unknown protein 74 2e-13
At1g48650 hypothetical protein 41 0.002
At1g06670 DEIH-box RNA/DNA helicase 37 0.034
>At2g03270 helicase like protein
Length = 639
Score = 1022 bits (2643), Expect = 0.0
Identities = 519/628 (82%), Positives = 575/628 (90%), Gaps = 1/628 (0%)
Query: 6 SFMILLFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQST 65
S M L ++AEIS S+ +GASRN++TAQK+G+TILNLKCVDVQTGLMGKSLIE QS
Sbjct: 12 STMAPLIDMEKEAEISMSLTSGASRNIETAQKKGTTILNLKCVDVQTGLMGKSLIEFQSN 71
Query: 66 KADVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLNSPLR 125
K DVLPAHKFG HDVVVLKLNK+DLGS L QGVVYRLKDSSITV FD++PE+GLN+ LR
Sbjct: 72 KGDVLPAHKFGNHDVVVLKLNKSDLGSSPLAQGVVYRLKDSSITVVFDEVPEEGLNTSLR 131
Query: 126 LEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVV-FTSINKNLD 184
LEK+ANEVTY RMKD LIQLSKGV +GPASDL+PVLFGERQP+VSKKDV FT NKNLD
Sbjct: 132 LEKLANEVTYRRMKDTLIQLSKGVLRGPASDLVPVLFGERQPSVSKKDVKSFTPFNKNLD 191
Query: 185 YSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVE 244
SQKDAI+KALSSK+VFLLHGPPGTGKTTTVVEI+LQEVKRGSKILACAASNIAVDNIVE
Sbjct: 192 QSQKDAITKALSSKDVFLLHGPPGTGKTTTVVEIVLQEVKRGSKILACAASNIAVDNIVE 251
Query: 245 RLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEK 304
RLVPH+VKLVR+GHPARLLPQV+DSALDAQVL+GDNSGLANDIRKEMK LNGKLLK K+K
Sbjct: 252 RLVPHKVKLVRVGHPARLLPQVLDSALDAQVLKGDNSGLANDIRKEMKALNGKLLKAKDK 311
Query: 305 NTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEA 364
NTRR IQKELRTL +EERKRQQLAV+DVIK +DVILTTL GA ++KL N +FDLVIIDE
Sbjct: 312 NTRRLIQKELRTLGKEERKRQQLAVSDVIKNADVILTTLTGALTRKLDNRTFDLVIIDEG 371
Query: 365 AQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSM 424
AQALEVACWI LLKG+RCILAGDHLQLPPTIQS EAE+KGLGRTLFERLA+LYGDE+ SM
Sbjct: 372 AQALEVACWIALLKGSRCILAGDHLQLPPTIQSAEAERKGLGRTLFERLADLYGDEIKSM 431
Query: 425 LTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDTAGCD 484
LTVQYRMH+LIM+WSSKELY++K+ AH+ VASHML+DLE V K+SSTE TLLL+DTAGCD
Sbjct: 432 LTVQYRMHELIMNWSSKELYDNKITAHSSVASHMLFDLENVTKSSSTEATLLLVDTAGCD 491
Query: 485 MEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSL 544
MEEKKDEE+ST NEGE+EVAMAHAKRL++SGV PSDIGIITPYAAQV+LL++L+ KE L
Sbjct: 492 MEEKKDEEESTYNEGEAEVAMAHAKRLMESGVQPSDIGIITPYAAQVMLLRILRGKEEKL 551
Query: 545 KDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTET 604
KD+EISTVDGFQGREKEAIIISMVRSNSKKEVGFL D+RRMNVAVTR+RRQCCIVCDTET
Sbjct: 552 KDMEISTVDGFQGREKEAIIISMVRSNSKKEVGFLKDQRRMNVAVTRSRRQCCIVCDTET 611
Query: 605 VSSDGFLKRLIEYFEEHGEYQSASEYQN 632
VSSD FLKR+IEYFEEHGEY SASEY N
Sbjct: 612 VSSDAFLKRMIEYFEEHGEYLSASEYTN 639
>At5g35970 DNA helicase-like
Length = 750
Score = 347 bits (890), Expect = 1e-95
Identities = 227/636 (35%), Positives = 350/636 (54%), Gaps = 43/636 (6%)
Query: 30 RNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTKADVLPAHKFGTHDVVVLKL-NKA 88
R+ D Q+ TI NL V TGL G L+ + LP D+V +++ +
Sbjct: 100 RHGDAPQELCDTICNLYAVSTSTGLGGMHLVLFKVGGNHRLPPTTLSPGDMVCIRVCDSR 159
Query: 89 DLGSPALGQGVVYRLKDS--SITVAFDDIPEDGLNSPL--------RLEKVANEVTYHRM 138
G+ A QG V+ L + SI VA + D S L R+ +A+ +TY R
Sbjct: 160 GAGATACTQGFVHNLGEDGCSIGVALESRHGDPTFSKLFGKSVRIDRIHGLADALTYERN 219
Query: 139 KDALIQLSKG--VHKGPASDLIPVLFGERQPTV----------SKKDVVFTSINKNLDYS 186
+AL+ L K K P+ ++ LFG+ + S+ ++ ++K D S
Sbjct: 220 CEALMLLQKNGLQKKNPSISVVATLFGDGEDITWLEQNDYVDWSEAELSDEPVSKLFDSS 279
Query: 187 QKDAISKALSSKN-VFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVER 245
Q+ AI+ ++ K V ++ GPPGTGKT + E+I V++G ++L A +N AVDN+VE+
Sbjct: 280 QRRAIALGVNKKRPVMIVQGPPGTGKTGMLKEVITLAVQQGERVLVTAPTNAAVDNMVEK 339
Query: 246 LVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKN 305
L+ + +VR+G+PAR+ V +L V NS LA+ R E++ L K +
Sbjct: 340 LLHLGLNIVRVGNPARISSAVASKSLGEIV----NSKLAS-FRAELERKKSDLRKDLRQC 394
Query: 306 TRRE-----IQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNT-SFDLV 359
R + I++ L+ L + +K+++ V +++ + V+ T IGA+ + +FDLV
Sbjct: 395 LRDDVLAAGIRQLLKQLGKTLKKKEKETVKEILSNAQVVFATNIGAADPLIRRLETFDLV 454
Query: 360 IIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGD 419
+IDEA Q++E +CWIP+L+G RCIL+GD QL P + S +A + GLG +L ER A L+
Sbjct: 455 VIDEAGQSIEPSCWIPILQGKRCILSGDPCQLAPVVLSRKALEGGLGVSLLERAASLHDG 514
Query: 420 EVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLID 479
+ + LT QYRM+ +I W+SKE+Y +K+ VASH+L D VK T T+ L+L+D
Sbjct: 515 VLATKLTTQYRMNDVIAGWASKEMYGGWLKSAPSVASHLLIDSPFVKATWITQCPLVLLD 574
Query: 480 T--------AGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQV 531
T GC+ S NEGE+++ + H L+ +GV P I + +PY AQV
Sbjct: 575 TRMPYGSLSVGCEERLDPAGTGSLYNEGEADIVVNHVISLIYAGVSPMAIAVQSPYVAQV 634
Query: 532 VLLKMLKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTR 591
LL+ + +E++T+D FQGRE +A+IISMVRSN+ VGFL D RRMNVA+TR
Sbjct: 635 QLLRERLDDFPVADGVEVATIDSFQGREADAVIISMVRSNNLGAVGFLGDSRRMNVAITR 694
Query: 592 ARRQCCIVCDTETVSSDGFLKRLIEYFEEHGEYQSA 627
AR+ +VCD+ T+ + FL RL+ + G + A
Sbjct: 695 ARKHVAVVCDSSTICHNTFLARLLRHIRYFGRVKHA 730
>At5g47010 UPF1
Length = 1254
Score = 216 bits (549), Expect = 4e-56
Identities = 147/446 (32%), Positives = 232/446 (51%), Gaps = 26/446 (5%)
Query: 183 LDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGS-KILACAASNIAVDN 241
L+ SQ +A+ K++ K + L+ GPPGTGKT T I+ K+G ++L CA SN+AVD
Sbjct: 489 LNASQVNAV-KSVLQKPISLIQGPPGTGKTVTSAAIVYHMAKQGQGQVLVCAPSNVAVDQ 547
Query: 242 IVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKT 301
+ E++ +K+VR+ +R + V S ++ L L + E+ L + LK
Sbjct: 548 LAEKISATGLKVVRLCAKSR---EAVSSPVEYLTLHYQVRHLDTSEKSELHKL--QQLKD 602
Query: 302 KEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVII 361
++ +K+ + L R + ++ +++DVI T +GA+ +L N F V+I
Sbjct: 603 EQGELSSSDEKKYKNLKRATER-------EITQSADVICCTCVGAADLRLSNFRFRQVLI 655
Query: 362 DEAAQALEVACWIPLLKGTR-CILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDE 420
DE+ QA E C IPL+ G + +L GDH QL P I +A + GL ++LFERL L
Sbjct: 656 DESTQATEPECLIPLVLGVKQVVLVGDHCQLGPVIMCKKAARAGLAQSLFERLVTLGIKP 715
Query: 421 VTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDT 480
+ L VQYRMH + ++ S Y ++ + ++ + P +
Sbjct: 716 IR--LQVQYRMHPALSEFPSNSFYEGTLQNGVTIIERQTTGIDFPWPVPN-RPMFFYVQL 772
Query: 481 AGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNK 540
EE S LN E+ ++SGV+PS IG+ITPY Q + +
Sbjct: 773 G---QEEISASGTSYLNRTEAANVEKLVTAFLKSGVVPSQIGVITPYEGQRAYIVNYMAR 829
Query: 541 ENSL-----KDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQ 595
SL K+IE+++VD FQGREK+ II+S VRSN + +GFL+D RR+NVA+TRAR
Sbjct: 830 NGSLRQQLYKEIEVASVDSFQGREKDYIILSCVRSNEHQGIGFLNDPRRLNVALTRARYG 889
Query: 596 CCIVCDTETVSSDGFLKRLIEYFEEH 621
I+ + + +S L+ +++EH
Sbjct: 890 IVILGNPKVLSKQPLWNGLLTHYKEH 915
>At1g08840 hypothetical protein,5' partial
Length = 874
Score = 186 bits (471), Expect = 4e-47
Identities = 149/479 (31%), Positives = 231/479 (48%), Gaps = 71/479 (14%)
Query: 156 DLIPVLFGERQPTVSKKDVVFTSINKNLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTV 215
DL P F + + K+L+ Q+ AI K L++K+ L+ G PGTGKT+T+
Sbjct: 439 DLEPPRFDNGSILSQDPAISYIWSEKSLNNDQRQAILKILTAKDYALILGMPGTGKTSTM 498
Query: 216 VEIILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQV 275
V + + RGS IL + +N AVDN++ +L ++ +RIG + +V +S A
Sbjct: 499 VHAVKALLIRGSSILLASYTNSAVDNLLIKLKAQGIEFLRIGRDEAVHEEVRESCFSAM- 557
Query: 276 LRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRREIQKELRTLSREERKRQQLAVTDVIKT 335
N DI+K++ D +K
Sbjct: 558 ----NMCSVEDIKKKL---------------------------------------DQVK- 573
Query: 336 SDVILTTLIGASSKKLGNTSFDLVIIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTI 395
V+ +T +G +S L N FD+ IIDEA Q PLL + +L GDH QLPP +
Sbjct: 574 --VVASTCLGINSPLLVNRRFDVCIIDEAGQIALPVSIGPLLFASTFVLVGDHYQLPPLV 631
Query: 396 QSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVA 455
QS EA + G+G +LF RL+E + + S+L QYRM + IM+ S+ +Y ++ C
Sbjct: 632 QSTEARENGMGISLFRRLSEAH-PQAISVLQNQYRMCRGIMELSNALIYGDRL----CCG 686
Query: 456 SHMLYDLEGVKKTSST---------EP--TLLLIDTAGCDMEEKKDEEDSTLNEGESEVA 504
S + D V TSS+ EP T++ ++T E +D +++ N E+ +
Sbjct: 687 SAEVADATLVLSTSSSTSPWLKKVLEPTRTVVFVNTDMLRAFEARD-QNAINNPVEASII 745
Query: 505 MAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSLKDIEISTVDGFQGREKEAII 564
+ LV +GV DIGIITPY +Q L++ +EI T+D +QGR+K+ I+
Sbjct: 746 AEIVEELVNNGVDSKDIGIITPYNSQASLIQ----HAIPTTPVEIHTIDKYQGRDKDCIL 801
Query: 565 ISMVRSNSK---KEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKRLIEYFEE 620
+S VRS K L D R+NVA+TRA+++ +V T+S L L+ +E
Sbjct: 802 VSFVRSREKPRSSASSLLGDWHRINVALTRAKKKLIMVGSQRTLSRVPLLMLLLNKVKE 860
>At4g15570 SEN1 like protein
Length = 818
Score = 169 bits (428), Expect = 4e-42
Identities = 166/528 (31%), Positives = 239/528 (44%), Gaps = 96/528 (18%)
Query: 180 NKNLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEII----------------LQEV 223
N+NL+ SQK+AI LS K+ L+ GPPGTGKT T++ I+ EV
Sbjct: 255 NENLNKSQKEAIDVGLSRKSFVLIQGPPGTGKTQTILSILGAIMHATPARVQSKGTDHEV 314
Query: 224 KRGSKILACAASN--------IAVDNIVERLVPH--------------RVKLVRIGHPAR 261
KRG ++ N I N + ++P + ++V R
Sbjct: 315 KRGIQMTIQEKYNHWGRASPWILGVNPRDAIMPEDGDDGFFPTSGNELKPEVVNASRKYR 374
Query: 262 LLPQVV---DSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRREIQKELRTLS 318
L V +SALD VLR +SGL ++ + K+++ K L L
Sbjct: 375 LRVLVCAPSNSALDEIVLRLLSSGLRDE---NAQTYTPKIVRIGLKAHHSVASVSLDHLV 431
Query: 319 REER------KRQQLAVTD-------VIKTSDVILTTLIGASSKKL--GNTSFDLVIIDE 363
++R +Q TD +++ + ++ TL + S L N FD+VIIDE
Sbjct: 432 AQKRGSAIDKPKQGTTGTDIDSIRTAILEEAAIVFATLSFSGSALLAKSNRGFDVVIIDE 491
Query: 364 AAQALEVACWIPLLKGTRC---ILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDE 420
AAQA+E A IPL TRC L GD QLP T+ S A+ G G ++FERL +
Sbjct: 492 AAQAVEPATLIPL--ATRCKQVFLVGDPKQLPATVISTVAQDSGYGTSMFERLQK--AGY 547
Query: 421 VTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDT 480
ML QYRMH I + SK+ Y ++ + + + D K P
Sbjct: 548 PVKMLKTQYRMHPEIRSFPSKQFYEGALEDGSDIEAQTTRDWH---KYRCFGPFCFF--- 601
Query: 481 AGCDMEEKKDEE-----DSTLNEGESEVAMAHAKRLVQSGVLP-----SDIGIITPYAAQ 530
D+ E K+ + S +N E E + RLV + P S + II+PY Q
Sbjct: 602 ---DIHEGKESQHPGATGSRVNLDEVEFVLLIYHRLVT--MYPELKSSSQLAIISPYNYQ 656
Query: 531 VVLLKMLKNKENSL------KDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRR 584
V K K++ + K ++I+TVDGFQGREK+ I S VR+N ++GFLS+ RR
Sbjct: 657 V---KTFKDRFKEMFGTEAEKVVDINTVDGFQGREKDVAIFSCVRANENGQIGFLSNSRR 713
Query: 585 MNVAVTRARRQCCIVCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQN 632
MNV +TRA+ +V T+ SD K LIE E+ S+ N
Sbjct: 714 MNVGITRAKSSVLVVGSAATLKSDPLWKNLIESAEQRNRLFKVSKPLN 761
>At1g05460 unknown protein
Length = 1002
Score = 150 bits (379), Expect = 2e-36
Identities = 142/479 (29%), Positives = 227/479 (46%), Gaps = 62/479 (12%)
Query: 176 FTSINKNLDYSQKDAISKALSSKNV--FLLHGPPGTGKTTTVVEIILQ--EVKRGSKILA 231
F I+ L+ Q +I L K +++HGPPGTGKT T+VE I+Q +R +++L
Sbjct: 390 FVPISPALNAEQICSIEMVLGCKGAPPYVIHGPPGTGKTMTLVEAIVQLYTTQRNARVLV 449
Query: 232 CAASNIAVDNIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEM 291
CA SN A D+I+E+L+ ++ VRI D ++ R N +
Sbjct: 450 CAPSNSAADHILEKLLC--LEGVRIK--------------DNEIFR------LNAATRSY 487
Query: 292 KVLNGKLLKTKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKL 351
+ + ++++ + L+ L+R + + S + +L+ A
Sbjct: 488 EEIKPEIIRFCFFDELIFKCPPLKALTRYK-----------LVVSTYMSASLLNAEGVNR 536
Query: 352 GNTSFDLVIIDEAAQALEVACWIPL----LKGTRCILAGDHLQLPPTIQSVEAEKKGLGR 407
G+ F +++DEA QA E I + L T +LAGD QL P I S +AE GLG+
Sbjct: 537 GH--FTHILLDEAGQASEPENMIAVSNLCLTETVVVLAGDPRQLGPVIYSRDAESLGLGK 594
Query: 408 TLFERLAE----LYGDE-VTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDL 462
+ ERL E GDE + L YR H I+D SK Y+ ++ A +L L
Sbjct: 595 SYLERLFECDYYCEGDENYVTKLVKNYRCHPEILDLPSKLFYDGELVASKEDTDSVLASL 654
Query: 463 EGVKKTSSTEPTLLLIDTAGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSG-VLPSDI 521
+ + E ++ GCD E++ S N E + KRL + V DI
Sbjct: 655 NFL---PNKEFPMVFYGIQGCD--EREGNNPSWFNRIEISKVIETIKRLTANDCVQEEDI 709
Query: 522 GIITPYAAQVVLLKMLKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKE------ 575
G+ITPY QV+ +K + ++ + + ++++ +V+ FQG+EK+ IIIS VRS K
Sbjct: 710 GVITPYRQQVMKIKEVLDRLD-MTEVKVGSVEQFQGQEKQVIIISTVRSTIKHNEFDRAY 768
Query: 576 -VGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQNE 633
+GFLS+ RR NVA+TRA I+ + + D +L+ ++ YQ + E
Sbjct: 769 CLGFLSNPRRFNVAITRAISLLVIIGNPHIICKDMNWNKLLWRCVDNNAYQGCGLPEQE 827
>At1g16800 hypothetical protein
Length = 1939
Score = 141 bits (355), Expect = 1e-33
Identities = 119/370 (32%), Positives = 180/370 (48%), Gaps = 72/370 (19%)
Query: 295 NGKLLKTKEKNTR--------REIQKELRTLSREERKR-------QQLAVTDVIKTSDVI 339
+GKL+ E R R+I K+L + +ERK +Q ++K + ++
Sbjct: 1337 DGKLMSDAELGIRLRRLYEQKRKIYKDLSAVQAQERKANYEMRTLKQKLRKSILKEAQIV 1396
Query: 340 LTTLIGA------------SSKKLGNTS----FDLVIIDEAAQALEVACWIPLL----KG 379
+TTL G ++ K G+ S FD V+IDEAAQALE A IPL +G
Sbjct: 1397 VTTLSGCGGDLYSVCAESLAAHKFGSPSEDNLFDAVVIDEAAQALEPATLIPLQLLKSRG 1456
Query: 380 TRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWS 439
T+CI+ GD QLP T+ S A K ++FERL G + MLT QYRMH I +
Sbjct: 1457 TKCIMVGDPKQLPATVLSNVASKFLYECSMFERLQRA-GYPIL-MLTQQYRMHPEICRFP 1514
Query: 440 SKELYNSKVKAHACVASHMLYDLEGVKKTSSTEP--------TLLLIDTAGCDMEEKKDE 491
S Y++K+ L GV +S + P + D E + +
Sbjct: 1515 SMHFYDNKL-------------LNGVDMSSKSAPFHENHHLGPYVFYDIVD-GQEHRSGD 1560
Query: 492 EDSTLNEGESEVA---MAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSLK--- 545
S NE E+E A + K+ S + IGIITPY Q+ +L+ +
Sbjct: 1561 SSSVCNEQEAEAAVQLLRFFKKRYPSEFVAGRIGIITPYKRQLAVLRSRFTGAFGAQVTA 1620
Query: 546 DIEISTVDGFQGREKEAIIISMVRSN-------SKKEVGFLSDRRRMNVAVTRARRQCCI 598
D+E++TVDGFQG+E + +++S VR+ ++ +GF++D RRMNVA+TRA+ +
Sbjct: 1621 DMEMNTVDGFQGKEVDILVLSTVRATHSAPDGVNQSRIGFVADVRRMNVALTRAKLSLWV 1680
Query: 599 VCDTETVSSD 608
+ +T T+ D
Sbjct: 1681 LGNTRTLQRD 1690
Score = 39.3 bits (90), Expect = 0.007
Identities = 21/47 (44%), Positives = 28/47 (58%), Gaps = 6/47 (12%)
Query: 179 INKNLDYSQKDAISKALSSKNVF------LLHGPPGTGKTTTVVEII 219
+ + + SQ AIS A+ S N+ L+ GPPGTGKT T+V II
Sbjct: 1099 LKSSFNESQLQAISVAIGSSNLMKAFDISLIQGPPGTGKTRTIVAII 1145
>At5g37030 SEN1-like protein
Length = 638
Score = 135 bits (341), Expect = 5e-32
Identities = 150/521 (28%), Positives = 239/521 (45%), Gaps = 73/521 (14%)
Query: 118 DGLNSPLRLEKVANEVTYHRMKDALIQL-----SKGVHKGPASDLIPVLFGER----QPT 168
D LN PL L ++ + TY + L +L K + P DLI + +P
Sbjct: 128 DDLN-PLLLGSISVD-TYPNISVILSRLIFHDEKKSLGFAPNFDLIQSVLQPNTAGMEPI 185
Query: 169 VSKK----DVVFTSINKNLDYSQKDAISKALSSKN------VFLLHGPPGTGKTTTVVEI 218
VS + +V+ + L+ SQ+ AI L +++ V L+ GP KT TV +
Sbjct: 186 VSSRTWGQNVLDIIRSSKLNSSQEAAILSCLETRDSNHKNSVKLIWGPL---KTKTVATL 242
Query: 219 ILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRG 278
+L +K K + CA +N A+ + RL+ AL +
Sbjct: 243 LLVLLKLRCKTVVCAPTNTAIVEVTSRLL----------------------ALSNKYSEH 280
Query: 279 DNSGLANDIRKEMKVLNGKLLKTKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDV 338
GL N N K ++ K+ + R + + R + R+ T K S
Sbjct: 281 ATYGLGNIFLAG----NQKRMRIKDTDYLRNVFLDHRI----SKLRKLFLSTCGWKQSLE 332
Query: 339 ILTTLIGASSKKLGNTSFDLVIIDEAAQALEVACWIPL-LKGTR-CILAGDHLQLPPTIQ 396
+ L+ + K GN +++++DEAAQ E L L G R IL GD QLP +
Sbjct: 333 SIIDLLENTETKTGN--IEVLVVDEAAQLKECESVAALQLPGLRHAILIGDEFQLPAMVH 390
Query: 397 SVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVAS 456
+ + EK GR+LFERL L +L VQYRMH I + KE Y ++K A V
Sbjct: 391 NDQCEKAKFGRSLFERLVLL--GHKKHLLDVQYRMHPSISRFPYKEFYGGRIKDAANV-Q 447
Query: 457 HMLYDLEGVKKTSSTEPTLLLIDTAGCDMEEKKDEEDSTLNEGESEVA---MAHAKRLVQ 513
+Y ++ + + + +E+ + DS N E V +++ ++ +
Sbjct: 448 ESIYQKRFLQGNMFGSFSFINVGRG----KEEFGDGDSPKNMVEVAVVSEIISNLFKVSR 503
Query: 514 SGVLPSDIGIITPYAAQV-VLLKMLKNKENSLKD----IEISTVDGFQGREKEAIIISMV 568
+ +G+ITPY QV + + +++K +SL + + +VDGFQG E++ IIIS V
Sbjct: 504 ERKMKMSVGVITPYKGQVRAIQERIRDKYSSLSGELFTVNVRSVDGFQGGEEDIIIISTV 563
Query: 569 RSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDG 609
RSNS +VGFLS+R+R NVA+TRAR ++ + T++ G
Sbjct: 564 RSNSNGKVGFLSNRQRANVALTRARHCLWVIGNESTLARSG 604
>At2g19120 putative DNA2-NAM7 helicase family protein
Length = 1090
Score = 134 bits (338), Expect = 1e-31
Identities = 106/268 (39%), Positives = 148/268 (54%), Gaps = 21/268 (7%)
Query: 339 ILTTLIGASSKKLGNT---SFDLVIIDEAAQALEVACWIPLLKGT-RCILAGDHLQLPPT 394
I+ T + +S +KL + FD+V+IDEAAQA EV PL G RC+L GD QLP T
Sbjct: 755 IVFTTVSSSGRKLFSRLTHGFDMVVIDEAAQASEVGVLPPLALGAARCVLVGDPQQLPAT 814
Query: 395 IQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACV 454
+ S A R+LFER +L G T +LTVQYRMH I D+ S+ Y ++K +
Sbjct: 815 VISKAAGTLLYSRSLFERF-QLAGCP-TLLLTVQYRMHPQIRDFPSRYFYQGRLKDSESI 872
Query: 455 ASHMLYDLEGVKKTSSTEPTLLLIDTAGCDMEEKKDEEDSTLNEGESEVAM-AHAKRLVQ 513
+S E K P L + G + ++E V + H ++ ++
Sbjct: 873 SSA---PDEIYYKDPVLRPYLFFNISHGRESHRGGSVSYENVDEARFCVGVYMHLQKTLK 929
Query: 514 S-GVLPSDIGIITPYAAQVVLLKMLKNK------ENSLKDIEISTVDGFQGREKEAIIIS 566
S G +G+ITPY Q LK LK++ ++ LK+I I+TVD FQG+E++ II+S
Sbjct: 930 SLGAGKVSVGVITPYKLQ---LKCLKHEFGNALGQDELKEIYINTVDAFQGQERDVIIMS 986
Query: 567 MVRSNSKKEVGFLSDRRRMNVAVTRARR 594
VR+ S VGF+SD RRMNVA+TRARR
Sbjct: 987 CVRA-SGHGVGFVSDIRRMNVALTRARR 1013
>At1g65780 hypothetical protein
Length = 1065
Score = 134 bits (338), Expect = 1e-31
Identities = 113/311 (36%), Positives = 159/311 (50%), Gaps = 34/311 (10%)
Query: 339 ILTTLIGASSKKLGNTSFDLVIIDEAAQALEVACWIPL-LKGTR-CILAGDHLQLPPTIQ 396
+L + S++ T L++IDEAAQ E IP+ L G R IL GD QLP ++
Sbjct: 547 LLFSTASCSARLYTGTPIQLLVIDEAAQLKECESSIPMQLPGLRHLILVGDERQLPAMVE 606
Query: 397 SVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVAS 456
S A + G GR+LFERLA L + ML +QYRMH I + +KELY K+
Sbjct: 607 SQIALEAGFGRSLFERLALLGHKKY--MLNIQYRMHCSISSFPNKELYGKKI-------- 656
Query: 457 HMLYDLEGVKKTSSTEPTL--------LLIDTAGCDMEEKKDEEDSTLNEGESEVAMAHA 508
D V++ + T+ L I+ A E + E S N E V A
Sbjct: 657 ---LDAPTVRQRNYTKQYLPGEMYGPYSFINIAYGREEYGEGEGRSLKNNVEVVVVAAII 713
Query: 509 KRLVQSGVLPS---DIGIITPYAAQVVLLKMLKNKENSLKD------IEISTVDGFQGRE 559
L+Q ++G+I+PY AQV+ ++ K +E S+ D + I TVDGFQG E
Sbjct: 714 ANLLQVSEKTKTRINVGVISPYKAQVIAIQE-KIQETSIGDAGGLFSLRIRTVDGFQGGE 772
Query: 560 KEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTET-VSSDGFLKRLIEYF 618
++ II+S VRSN VGFL +RRR NV +TRAR I+ + T ++S + LI+
Sbjct: 773 EDIIIVSTVRSNGVGRVGFLGNRRRTNVLLTRARFCLWILGNEATLMNSKSVWRNLIQDA 832
Query: 619 EEHGEYQSASE 629
+E G + SA E
Sbjct: 833 KERGCFHSAGE 843
Score = 41.6 bits (96), Expect = 0.001
Identities = 24/82 (29%), Positives = 42/82 (50%), Gaps = 6/82 (7%)
Query: 183 LDYSQKDAISKALSSKNVF------LLHGPPGTGKTTTVVEIILQEVKRGSKILACAASN 236
L+ SQ+DAI L + + L+ GPPGTGKT T ++ + + L C +N
Sbjct: 238 LNPSQEDAILNCLDVRRCYHANTVRLIWGPPGTGKTKTTSVLLFTLLNAKCRTLTCGPTN 297
Query: 237 IAVDNIVERLVPHRVKLVRIGH 258
++V + R++ ++IG+
Sbjct: 298 VSVLEVASRVLKLVSGSLKIGN 319
>At4g30100 putative protein
Length = 1311
Score = 131 bits (329), Expect = 1e-30
Identities = 102/265 (38%), Positives = 147/265 (54%), Gaps = 15/265 (5%)
Query: 339 ILTTLIGASSKKLGNT---SFDLVIIDEAAQALEVACWIPLLKGT-RCILAGDHLQLPPT 394
I+ T + +S +KL + FD+V+IDEAAQA EV PL G RC+L GD QLP T
Sbjct: 918 IVFTTVSSSGRKLFSRLTHGFDMVVIDEAAQASEVGVLPPLALGAARCVLVGDPQQLPAT 977
Query: 395 IQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACV 454
+ S A R+LFER +L G T +LTVQYRMH I D+ S+ Y ++ V
Sbjct: 978 VISKAAGTLLYSRSLFERF-QLAGCP-TLLLTVQYRMHPQIRDFPSRYFYQGRLTDSESV 1035
Query: 455 ASHMLYDLEGVKKTSSTEPTLLLIDTAGCDMEEKKDEEDSTLNEGESEVAM-AHAKRLVQ 513
++ E K S +P L + G + ++E V + H +R ++
Sbjct: 1036 STA---PDEIYYKDSVLKPYLFFDISHGRESHRGGSVSYENIDEARFCVGVYLHLQRTLK 1092
Query: 514 S-GVLPSDIGIITPYAAQVVLLKMLKNK---ENSLKDIEISTVDGFQGREKEAIIISMVR 569
S G +G+ITPY Q+ LK+ ++ L++I I+TVD FQG+E++ II+S VR
Sbjct: 1093 SLGGGKVSVGVITPYKLQLKCLKIEFGNALSQDELQEIYINTVDAFQGQERDVIIMSCVR 1152
Query: 570 SNSKKEVGFLSDRRRMNVAVTRARR 594
+ S VGF++D RRMNVA+TRA+R
Sbjct: 1153 A-SNHGVGFVADIRRMNVALTRAKR 1176
>At5g52090 putative protein
Length = 676
Score = 124 bits (312), Expect = 1e-28
Identities = 105/338 (31%), Positives = 169/338 (49%), Gaps = 25/338 (7%)
Query: 312 KELRTLSREERKRQQLAVTDV----IKTSDVILTTLIGASSKKLGNT-SFDLVIIDEAAQ 366
K LR L + L D+ ++ +D+IL T GA+ + T + +L+++DEAAQ
Sbjct: 315 KALRLLPKRFEIPDMLENEDIRKFCLQNADIILCTASGAAEMNVERTGNVELLVVDEAAQ 374
Query: 367 ALEVACWIPL-LKGTR-CILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSM 424
E L L G R IL GD QLP + + EK GR+LFERL L ++ +
Sbjct: 375 LKECESVAALQLPGLRHAILIGDEFQLPAMVHNEMCEKAKFGRSLFERLVLLGHNK--HL 432
Query: 425 LTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLI----DT 480
L VQYRMH I + +KE Y ++K V +Y +K + + + +
Sbjct: 433 LDVQYRMHPSISRFPNKEFYGGRIKDAENV-KESIYQKRFLKGNMFDSFSFINVGRGKEE 491
Query: 481 AGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQV-VLLKMLKN 539
G K E + ++E S + +R ++ V G+++PY Q+ + + + +
Sbjct: 492 FGDGHSPKNMVEVAVISEIISNLYKVSCERRMKVSV-----GVVSPYKGQMRAIQEKIGD 546
Query: 540 KENSLKD----IEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQ 595
K +SL + + +VDGFQG E++ IIIS VRSN +VGFL++R+R NVA+TRAR
Sbjct: 547 KYSSLSGQQFTLNVRSVDGFQGGEEDIIIISTVRSNGNGKVGFLNNRQRANVALTRARHC 606
Query: 596 CCIVCDTETVSSDGFL-KRLIEYFEEHGEYQSASEYQN 632
++ + T++ G + LI G + A++ N
Sbjct: 607 LWVIGNETTLALSGSIWATLISESRTRGCFHDATDEMN 644
Score = 48.1 bits (113), Expect = 1e-05
Identities = 30/84 (35%), Positives = 45/84 (52%), Gaps = 7/84 (8%)
Query: 171 KKDVVFTSINKN-LDYSQKDAISKALSSKN------VFLLHGPPGTGKTTTVVEIILQEV 223
+ D+V I L+ SQ+DAI L ++N V L+ GPP TGKT TV ++ +
Sbjct: 45 RSDLVLDIIRSTKLNSSQEDAILGCLETRNCTHKNSVKLIWGPPRTGKTKTVATLLFALL 104
Query: 224 KRGSKILACAASNIAVDNIVERLV 247
K K + CA +N A+ + RL+
Sbjct: 105 KLRCKTVVCAPTNTAIVQVTSRLL 128
>At5g37150 sen1-like protein
Length = 839
Score = 124 bits (310), Expect = 2e-28
Identities = 110/344 (31%), Positives = 167/344 (47%), Gaps = 37/344 (10%)
Query: 312 KELRTLSREERKRQQLAVTDV----IKTSDVILTTLIGASSKKLGNT-SFDLVIIDEAAQ 366
K LR L + L D+ ++ +D+IL T GA+ + T + +L+++DEAAQ
Sbjct: 478 KALRLLPKRFEIPDMLENEDIRKFCLQNADIILCTASGAAEMNVERTGNVELLVVDEAAQ 537
Query: 367 ALEVACWIPL-LKGTR-CILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSM 424
E L L G R IL GD QLP + + EK GR+LFERL L ++ +
Sbjct: 538 LKECESVAALQLPGLRHAILIGDEFQLPAMVHNEMCEKAKFGRSLFERLVLLGHNK--HL 595
Query: 425 LTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTL-------LL 477
L VQYRMH I + +KE Y ++K D E VK++ + L
Sbjct: 596 LDVQYRMHPSISRFPNKEFYGGRIK-----------DAENVKESIYQKRFLQGNMFGSFS 644
Query: 478 IDTAGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGV---LPSDIGIITPYAAQV-VL 533
G EE D S N E V L + + +G+++PY Q+ +
Sbjct: 645 FINVGRGKEEFGDGH-SPKNMVEVAVVSEIISNLFKVSCERRMKVSVGVVSPYKGQMRAI 703
Query: 534 LKMLKNKENSLKD----IEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAV 589
+ + +K +SL + + +VDGFQG E++ IIIS VRSNS +VGFL++R+R NVA+
Sbjct: 704 QEKIGDKYSSLSGQQFTLNVRSVDGFQGGEEDIIIISTVRSNSNGKVGFLNNRQRANVAL 763
Query: 590 TRARRQCCIVCDTETVSSDGFL-KRLIEYFEEHGEYQSASEYQN 632
TRAR ++ + T++ G + LI G + A++ N
Sbjct: 764 TRARHCLWVIGNETTLALSGSIWATLISESRTRGCFYDATDEMN 807
Score = 50.4 bits (119), Expect = 3e-06
Identities = 28/71 (39%), Positives = 41/71 (57%), Gaps = 6/71 (8%)
Query: 183 LDYSQKDAISKALSSKN------VFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASN 236
L+ SQ+DAI L ++N V L+ GPPGTGKT TV ++ +K K + CA +N
Sbjct: 221 LNSSQEDAILGCLETRNCTHKNSVKLIWGPPGTGKTKTVATLLFALLKLRCKTVVCAPTN 280
Query: 237 IAVDNIVERLV 247
A+ + RL+
Sbjct: 281 TAIVQVASRLL 291
>At5g37140 sen1-like protein
Length = 692
Score = 120 bits (301), Expect = 2e-27
Identities = 103/315 (32%), Positives = 164/315 (51%), Gaps = 24/315 (7%)
Query: 330 TDVIKTSDVILTTLIGASSKKLGNT-SFDLVIIDEAAQALEVACWIPL-LKGTR-CILAG 386
T ++ +D+I T GA+ T S DL+++DEAAQ E L L G R +L G
Sbjct: 358 TFCLQNADIIFCTASGAADMNPIRTGSVDLLVVDEAAQLKECESVAALQLSGLRHAVLIG 417
Query: 387 DHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNS 446
D LQLP + + EA+ GR+LFERL L ++ +L VQYRMH I + +KE Y
Sbjct: 418 DELQLPAMVHN-EAK---FGRSLFERLVLLGHNK--HLLNVQYRMHPSISRFPNKEFYGG 471
Query: 447 KVKAHACVASHMLYDLEGVKKTSSTEPTLLLI----DTAGCDMEEKKDEEDSTLNEGESE 502
++K A V +Y ++ + + + + G K E + ++E S
Sbjct: 472 RIKDAANV-QESIYQKRFLQGNMFGSFSFINVGRGEEEFGDGHSPKNMVEVAVISEIISN 530
Query: 503 VAMAHAKRLVQSGVLPSDIGIITPYAAQV-VLLKMLKNKENSLKDI---EISTVDGFQGR 558
+ ++R ++ V G+++PY QV + + NK +SL + + +VDGFQG
Sbjct: 531 LFKVSSERRIKMSV-----GVVSPYKGQVRAIQERTTNKYSSLSGLFTLNVRSVDGFQGG 585
Query: 559 EKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDG-FLKRLIEY 617
E++ IIIS VRSN +VGFL++R+R NVA+TRAR ++ + T++ G +LI
Sbjct: 586 EEDIIIISTVRSNGNGKVGFLNNRQRANVALTRARHCLWVIGNETTLALSGSSWAKLISE 645
Query: 618 FEEHGEYQSASEYQN 632
G + A++ +N
Sbjct: 646 SRTLGCFYDAADEKN 660
Score = 47.0 bits (110), Expect = 3e-05
Identities = 27/71 (38%), Positives = 39/71 (54%), Gaps = 6/71 (8%)
Query: 183 LDYSQKDAISKALSSKN------VFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASN 236
L+ SQ+ AI L +N V L+ GPPGTGKT TV ++ +K K + CA +N
Sbjct: 74 LNSSQEAAILSCLKIRNCNHKHSVKLIWGPPGTGKTKTVATLLFCLLKLSCKTVVCAPTN 133
Query: 237 IAVDNIVERLV 247
A+ + RL+
Sbjct: 134 TAIVEVTSRLL 144
>At4g05540
Length = 660
Score = 118 bits (295), Expect = 1e-26
Identities = 104/320 (32%), Positives = 153/320 (47%), Gaps = 36/320 (11%)
Query: 312 KELRTLSREERKRQQLAVTDV----IKTSDVILTTLIGASSKKLGNT-SFDLVIIDEAAQ 366
+ LR LS+ L D+ ++ + +I T GA+ S D++++DEAAQ
Sbjct: 353 RTLRLLSKRFEIPALLMKEDIRTFCLQNAHIIFCTASGAAEMTAERAGSIDMLVVDEAAQ 412
Query: 367 ALEVACWIPL-LKGTR-CILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSM 424
E L L+G +L GD LQLP +QS EK R+LFERL L +
Sbjct: 413 LKECESVAALQLQGLHHAVLIGDELQLPAMVQSEVCEKAKFVRSLFERLDSL--GHKKHL 470
Query: 425 LTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTL-------LL 477
L VQYRMH I + + E Y K+ D E VK+++ + L
Sbjct: 471 LNVQYRMHPSISLFPNMEFYGGKIS-----------DAEIVKESTYQKRFLQGNMFGSFS 519
Query: 478 IDTAGCDMEEKKDEEDSTLNEGESEVA---MAHAKRLVQSGVLPSDIGIITPYAAQV-VL 533
G EE D S N E V + + ++ +G+I+PY AQV +
Sbjct: 520 FINVGLGKEEFGDGH-SPKNMVEIAVVSEILTNLLKVSSETKTKMSVGVISPYKAQVSAI 578
Query: 534 LKMLKNKENSLKD----IEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAV 589
+ + +K S+ D + + +VDGFQG E++ IIIS VRSN +GFLS+R+R NVA+
Sbjct: 579 QERIGDKYTSVSDNLFTLNVRSVDGFQGGEEDIIIISTVRSNCNGNIGFLSNRQRANVAL 638
Query: 590 TRARRQCCIVCDTETVSSDG 609
TRAR ++ + T+S G
Sbjct: 639 TRARHCLWVIGNERTLSLSG 658
Score = 43.9 bits (102), Expect = 3e-04
Identities = 25/72 (34%), Positives = 38/72 (52%), Gaps = 6/72 (8%)
Query: 182 NLDYSQKDAISKALSSK------NVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAAS 235
NL+ SQ+ AI L ++ +V L+ GPPGTG T TV ++ + K + C +
Sbjct: 79 NLNSSQESAILACLETREIRDKTSVKLIWGPPGTGNTKTVATLLFALLSLSCKTVVCTPT 138
Query: 236 NIAVDNIVERLV 247
N AV + RL+
Sbjct: 139 NTAVVAVASRLL 150
>At5g37160 sen1-like protein
Length = 871
Score = 110 bits (275), Expect = 2e-24
Identities = 96/316 (30%), Positives = 155/316 (48%), Gaps = 26/316 (8%)
Query: 333 IKTSDVILTTLIGASS---KKLGNTSFDLVIIDEAAQALEVACWIPLLKGTRC--ILAGD 387
++ +D+I T ++ ++G S DL+++DE AQ E L C +L GD
Sbjct: 536 LQNADIIFCTASSVANINPARIG--SVDLLVVDETAQLKECESVAALQLPGLCHALLIGD 593
Query: 388 HLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSK 447
QLP + + E +K GR+LFERL L G +L VQYRMH I + +KE Y +
Sbjct: 594 EYQLPAMVHNEECDKAKFGRSLFERLV-LIGHS-KHLLNVQYRMHPSISRFPNKEFYGGR 651
Query: 448 VKAHACVASHMLYDLEGVKKTSSTEPTLLLI----DTAGCDMEEKKDEEDSTLNEGESEV 503
+ A V +Y+ ++ + + + + G K E + +++ S +
Sbjct: 652 ITDAANV-QESIYEKRFLQGNMFGTFSFINVGRGKEEFGDGHSPKNMVEVAVISKIISNL 710
Query: 504 AMAHAKRLVQSGVLPSDIGIITPYAAQV-VLLKMLKNKENSLK-----DIEISTVDGFQG 557
++R + V G+I+PY QV + + + +K NSL + + +VDGFQG
Sbjct: 711 FKVSSQRKQKMSV-----GVISPYKGQVRAIQERVGDKYNSLSVDQLFTLNVQSVDGFQG 765
Query: 558 REKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFL-KRLIE 616
E + IIIS VR N VGFLS+R+R NVA+TRAR ++ + T++ G + LI
Sbjct: 766 GEVDVIIISTVRCNVNGNVGFLSNRQRANVALTRARHCLWVIGNGTTLALSGSIWAELIS 825
Query: 617 YFEEHGEYQSASEYQN 632
G + A + +N
Sbjct: 826 ESRTRGCFYDAVDDKN 841
Score = 42.0 bits (97), Expect = 0.001
Identities = 25/71 (35%), Positives = 39/71 (54%), Gaps = 6/71 (8%)
Query: 183 LDYSQKDAISKALSSKN------VFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASN 236
L+ SQ+ AI L ++N V L+ GPPGTGKT TV ++ ++ K + CA +N
Sbjct: 244 LNSSQEAAILGFLKTRNCKHKESVKLIWGPPGTGKTKTVATLLSTLMQLKCKTVVCAPTN 303
Query: 237 IAVDNIVERLV 247
+ + RL+
Sbjct: 304 TTIVAVASRLL 314
>At1g65810 hypothetical protein
Length = 1050
Score = 109 bits (273), Expect = 4e-24
Identities = 95/294 (32%), Positives = 155/294 (52%), Gaps = 17/294 (5%)
Query: 346 ASSKKLGNTS-FDLVIIDEAAQALEVACWIPL-LKGTR-CILAGDHLQLPPTIQSVEAEK 402
+SS +L +S L++IDEAAQ E IPL L+G + IL GD QLP I+S A +
Sbjct: 542 SSSARLHMSSPIQLLVIDEAAQLKECESAIPLQLRGLQHAILIGDEKQLPAMIKSNIASE 601
Query: 403 KGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDL 462
LGR+LFERL L ++ +L +QYRMH I + ++E Y+ K+ V Y+
Sbjct: 602 ADLGRSLFERLVLLGHNK--QLLNMQYRMHPSISIFPNREFYDMKILDAPSVRLRS-YEK 658
Query: 463 EGVKKTSSTEPTLLLIDTAGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPS--- 519
+ + + + + I E+ E S+ N E V +L
Sbjct: 659 KFLPEKMYGPYSFINIAYG----REQFGEGYSSKNLVEVSVVAEIVSKLYSVSRKTGRTI 714
Query: 520 DIGIITPYAAQVVLLKMLKNKENSLKD---IEISTVDGFQGREKEAIIISMVRSNSKKEV 576
+G+I+PY AQV ++ ++ + + + + +VDGFQG E++ IIIS VRSN +
Sbjct: 715 SVGVISPYKAQVFAIQERIGEKYNTEGTFTVSVRSVDGFQGGEEDIIIISTVRSNGNGAI 774
Query: 577 GFLSDRRRMNVAVTRARRQCCIVCDTETVSSD-GFLKRLIEYFEEHGEYQSASE 629
GFLS+++R NVA+TRAR I+ + T++++ ++L++ + + +A E
Sbjct: 775 GFLSNQQRTNVALTRARYCLWILGNEATLTNNRSVWRQLVDDAKARNCFHNAEE 828
Score = 55.1 bits (131), Expect = 1e-07
Identities = 31/71 (43%), Positives = 43/71 (59%), Gaps = 6/71 (8%)
Query: 183 LDYSQKDAISKALSSK------NVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASN 236
L+ SQ+DAI + L +K N+ L+ GPPGTGKT T ++L +K + L CA +N
Sbjct: 267 LNSSQEDAILRCLEAKSCNHSNNIKLIWGPPGTGKTKTTSVLLLNFLKMRCRTLTCAPTN 326
Query: 237 IAVDNIVERLV 247
IAV + RLV
Sbjct: 327 IAVLEVCSRLV 337
>At2g38770 unknown protein
Length = 1444
Score = 73.9 bits (180), Expect = 2e-13
Identities = 71/266 (26%), Positives = 129/266 (47%), Gaps = 37/266 (13%)
Query: 356 FDLVIIDEAAQALEVACWIPLL---------KGTRCILAGDHLQLPPTIQSVEAEKKG-L 405
+D ++++E+AQ LE+ +IP+L + RCIL GDH QLPP ++++ +K +
Sbjct: 1117 YDNLLMEESAQILEIETFIPMLLQRQEDGHARLKRCILIGDHHQLPPVVKNMAFQKYSHM 1176
Query: 406 GRTLFERLAELYGDEVTSMLTVQYRMHQLIM---DWSSKELYNSKVKAHACVASHMLYDL 462
++LF R L + L Q R + +W ++L + + A +
Sbjct: 1177 DQSLFTRFVRLGIPYIE--LNAQGRARPSLAKLYNWRYRDLGDLSIVKEAPI-------F 1227
Query: 463 EGVKKTSSTEPTLLLIDTAGCDMEEKKDEEDSTL---NEGESEVAMAHAKRLVQSGVLPS 519
+ S E L+ + D E + + S N+GE+E ++ + G +
Sbjct: 1228 QRANAGFSYEYQLVNVP----DYEGRGESTPSPWFYQNQGEAEYIVSVYIYMRLLGYPAN 1283
Query: 520 DIGIITPYAAQVVLLKMLKNKENSLKDI-----EISTVDGFQGREKEAIIISMVRSNSKK 574
I I+T Y Q +L++ + N+ +++TVD FQG++ + I++S+VR+ +
Sbjct: 1284 KISILTTYNGQKLLIRDVINRRCVPYPFIGPPSKVTTVDKFQGQQNDFILLSLVRT---R 1340
Query: 575 EVGFLSDRRRMNVAVTRARRQCCIVC 600
VG L D RR+ VA++RAR + C
Sbjct: 1341 FVGHLRDVRRLVVAMSRARLGLYVFC 1366
Score = 35.0 bits (79), Expect = 0.13
Identities = 28/77 (36%), Positives = 40/77 (51%), Gaps = 7/77 (9%)
Query: 205 GPPGTGKTTTVVEI--ILQEVKRGSKILACAASNIAV----DNIVERLVPHRVKLVRIGH 258
GPPGTGKT T V+I +L + L SN A+ + I+ER VP R L+R+G
Sbjct: 887 GPPGTGKTDTAVQILNVLYHNCPSQRTLIITHSNQALNDLFEKIMERDVPARY-LLRLGQ 945
Query: 259 PARLLPQVVDSALDAQV 275
+ L +D + +V
Sbjct: 946 GEQELATDLDFSRQGRV 962
>At1g48650 hypothetical protein
Length = 1167
Score = 40.8 bits (94), Expect = 0.002
Identities = 26/85 (30%), Positives = 46/85 (53%), Gaps = 7/85 (8%)
Query: 185 YSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQ---EVKRGS--KILACAASNIAV 239
Y +KDA+ KA+++ V ++ G G GKTT + + IL+ E RG+ I+ I+
Sbjct: 272 YKEKDALLKAIAANQVVVVSGETGCGKTTQLPQYILESEIEAARGATCSIICTQPRRISA 331
Query: 240 DNIVERLVPHRVKLV--RIGHPARL 262
++ ER+ R + + +G+ RL
Sbjct: 332 ISVSERVAAERGEQIGESVGYKVRL 356
>At1g06670 DEIH-box RNA/DNA helicase
Length = 1576
Score = 37.0 bits (84), Expect = 0.034
Identities = 27/94 (28%), Positives = 46/94 (48%), Gaps = 6/94 (6%)
Query: 175 VFTSINKNLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEV----KRGSKIL 230
+F + +K S +DAI A+ S V L+ G G GKTT V + +L + K KI+
Sbjct: 210 IFEARSKLPIASFRDAIISAVESNQVVLIAGETGCGKTTQVPQYLLDHMWHSKKEACKII 269
Query: 231 ACAASNIAVDNIVERLVPHRVKLV--RIGHPARL 262
I+ ++ +R+ R + + +G+ RL
Sbjct: 270 CTQPRRISAISVSDRISWERGETIGRTVGYKVRL 303
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,869,871
Number of Sequences: 26719
Number of extensions: 535303
Number of successful extensions: 2256
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 32
Number of HSP's that attempted gapping in prelim test: 2104
Number of HSP's gapped (non-prelim): 123
length of query: 633
length of database: 11,318,596
effective HSP length: 105
effective length of query: 528
effective length of database: 8,513,101
effective search space: 4494917328
effective search space used: 4494917328
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC143341.10


