

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143341.1 + phase: 0 /partial
(249 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
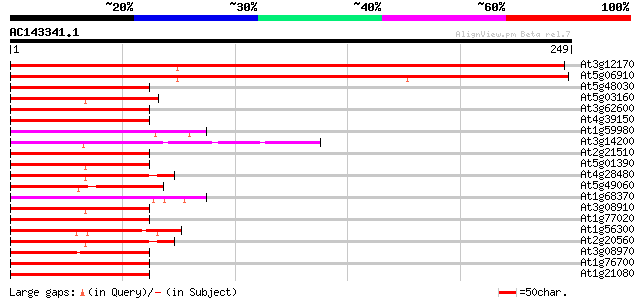
Score E
Sequences producing significant alignments: (bits) Value
At3g12170 hypothetical protein 365 e-101
At5g06910 DnaJ homologue (gb|AAB91418.1|) 354 3e-98
At5g48030 DnaJ protein-like 76 1e-14
At5g03160 unknown protein 71 5e-13
At3g62600 unknown protein 70 1e-12
At4g39150 dnaJ-like protein 70 1e-12
At1g59980 unknown protein 70 1e-12
At3g14200 unknown protein 68 4e-12
At2g21510 putative DnaJ protein 68 4e-12
At5g01390 heat shock protein 40-like 67 7e-12
At4g28480 heat-shock protein 67 7e-12
At5g49060 DNAJ-like protein 66 2e-11
At1g68370 ARG1 protein (Altered Response to Gravity) 66 2e-11
At3g08910 putative heat shock protein 66 2e-11
At1g77020 66 2e-11
At1g56300 unknown protein 66 2e-11
At2g20560 putative heat shock protein 65 3e-11
At3g08970 putative DnaJ protein 64 8e-11
At1g76700 DnaJ like protein 64 8e-11
At1g21080 putative dnaJ protein 64 8e-11
>At3g12170 hypothetical protein
Length = 262
Score = 365 bits (936), Expect = e-101
Identities = 181/247 (73%), Positives = 215/247 (86%), Gaps = 1/247 (0%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+E TAS QEI+KAY+KLALRLHPDKN DE+AKEKFQQLQKVISILGDEEKRA+YDQ
Sbjct: 15 VLGVEATASPQEIRKAYHKLALRLHPDKNKDDEDAKEKFQQLQKVISILGDEEKRAVYDQ 74
Query: 61 TGCIDDDDLAGDV-QNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYKGN 119
TG +DD DL+GDV NL FFK MYKKVTE DIEEFEANYRGS+SEKNDLIELY K+KG
Sbjct: 75 TGSVDDADLSGDVVDNLRDFFKAMYKKVTEEDIEEFEANYRGSESEKNDLIELYNKFKGK 134
Query: 120 MKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEVSETKPPTSPLKRKA 179
M RLFCSMLCS+ KLDSHRFKDI+DEAIAAGE+K+TKAY+KWAKE++E +PPT+P K +
Sbjct: 135 MSRLFCSMLCSNPKLDSHRFKDIIDEAIAAGEVKSTKAYKKWAKEIAEMEPPTNPQKMRR 194
Query: 180 KSNKQSETDLCAIISQRRNERKGQFDSMFASLVSKYGGSDMPEPSEEEFEAAQKKLEKGR 239
K+ K ++ DL A++SQR +ERK +FDSMF+SLVS+YG + EP+EEEFEAAQ+K+E R
Sbjct: 195 KAAKAADKDLYAVMSQRGDERKEKFDSMFSSLVSRYGSNADSEPNEEEFEAAQRKVESRR 254
Query: 240 SSKKSKQ 246
SSKKS++
Sbjct: 255 SSKKSRK 261
>At5g06910 DnaJ homologue (gb|AAB91418.1|)
Length = 284
Score = 354 bits (908), Expect = 3e-98
Identities = 173/252 (68%), Positives = 211/252 (83%), Gaps = 4/252 (1%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+E+ A+ QEI+KAY+KLAL+LHPDKN D+EAK+KFQQLQKVISILGDEEKRA+YDQ
Sbjct: 33 VLGVERRATSQEIRKAYHKLALKLHPDKNQDDKEAKDKFQQLQKVISILGDEEKRAVYDQ 92
Query: 61 TGCIDDDDLAGDV-QNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYKGN 119
TG IDD D+ GD +NL FF+ MYKKV EADIEEFEANYRGS+SEK DL+EL+ K+KG
Sbjct: 93 TGSIDDADIPGDAFENLRDFFRDMYKKVNEADIEEFEANYRGSESEKKDLLELFNKFKGK 152
Query: 120 MKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEVSETKPPTSPL---K 176
M RLFCSMLCSD KLDSHRFKD+LDEAIAAGE+K++KAY+KWA ++SETKPPTSPL K
Sbjct: 153 MNRLFCSMLCSDPKLDSHRFKDMLDEAIAAGEVKSSKAYEKWANKISETKPPTSPLRKRK 212
Query: 177 RKAKSNKQSETDLCAIISQRRNERKGQFDSMFASLVSKYGGSDMPEPSEEEFEAAQKKLE 236
+K + K SETDLC +I++R+ ERKG+ DSMF+SL+S+YGG EP+EEEFEAAQ+++E
Sbjct: 213 KKKSAAKDSETDLCLMIAKRQEERKGKVDSMFSSLISRYGGDAEAEPTEEEFEAAQRRIE 272
Query: 237 KGRSSKKSKQSK 248
R K + K
Sbjct: 273 SKRKPSKKSRGK 284
>At5g48030 DnaJ protein-like
Length = 456
Score = 76.3 bits (186), Expect = 1e-14
Identities = 37/62 (59%), Positives = 44/62 (70%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+ K A + EIKKAYY LA +LHPD N D EA+ KFQ++ K IL D+EKR LYDQ
Sbjct: 98 VLGVSKNAQEGEIKKAYYGLAKKLHPDMNKDDPEAETKFQEVSKAYEILKDKEKRDLYDQ 157
Query: 61 TG 62
G
Sbjct: 158 VG 159
>At5g03160 unknown protein
Length = 482
Score = 71.2 bits (173), Expect = 5e-13
Identities = 35/67 (52%), Positives = 50/67 (74%), Gaps = 1/67 (1%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGD-EEAKEKFQQLQKVISILGDEEKRALYD 59
+LG+ +TAS EIKKAY KLAL+ HPDKN G+ EEA+ KF+++ ILGD++KRA +D
Sbjct: 374 ILGISRTASISEIKKAYKKLALQWHPDKNVGNREEAENKFREIAAAYEILGDDDKRARFD 433
Query: 60 QTGCIDD 66
+ ++D
Sbjct: 434 RGEDLED 440
>At3g62600 unknown protein
Length = 346
Score = 70.1 bits (170), Expect = 1e-12
Identities = 32/62 (51%), Positives = 44/62 (70%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VL + K AS ++IK+AY KLAL+ HPDKN G+EEA KF ++ +L DEEKR +Y++
Sbjct: 30 VLQVPKGASDEQIKRAYRKLALKYHPDKNQGNEEATRKFAEINNAYEVLSDEEKREIYNK 89
Query: 61 TG 62
G
Sbjct: 90 YG 91
>At4g39150 dnaJ-like protein
Length = 345
Score = 69.7 bits (169), Expect = 1e-12
Identities = 33/62 (53%), Positives = 43/62 (69%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
+LG++ AS EIKKAYY A ++HPDKNPGD +A + FQ L + +LGD EKR YD+
Sbjct: 10 ILGVKIDASGAEIKKAYYVQARQVHPDKNPGDPQAAKNFQILGEAYQVLGDPEKRTAYDK 69
Query: 61 TG 62
G
Sbjct: 70 YG 71
>At1g59980 unknown protein
Length = 414
Score = 69.7 bits (169), Expect = 1e-12
Identities = 38/94 (40%), Positives = 55/94 (58%), Gaps = 7/94 (7%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+ ++ QEIK AY ++ALR HPDKNP D A E F+++ +L D E R LYD
Sbjct: 27 VLGIPSNSTDQEIKSAYRRMALRYHPDKNPDDPVAAEMFKEVTFAYEVLSDPENRRLYDT 86
Query: 61 TGC----IDDDDLAGDVQNLHA---FFKTMYKKV 87
TG +++DL D+ +L A F ++ K+
Sbjct: 87 TGSEAVGPENEDLELDLSSLGAVNTIFAALFNKL 120
>At3g14200 unknown protein
Length = 230
Score = 68.2 bits (165), Expect = 4e-12
Identities = 49/141 (34%), Positives = 70/141 (48%), Gaps = 9/141 (6%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPG---DEEAKEKFQQLQKVISILGDEEKRAL 57
VLGL+K S+ E++ AY KLALR HPD+ EEAK+KFQ +Q+ S+L D KR L
Sbjct: 16 VLGLKKECSKTELRSAYKKLALRWHPDRCSSMEFVEEAKKKFQAIQEAYSVLSDSNKRFL 75
Query: 58 YDQTGCIDDDDLAGDVQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYK 117
YD DDD D + F M + ++ + + N S + DL + ++
Sbjct: 76 YDVGAYNTDDD--DDQNGMGDFLNEMATMMNQS--KPSDNNTGDSFEQLQDLFN--EMFQ 129
Query: 118 GNMKRLFCSMLCSDAKLDSHR 138
G+ S CS + S R
Sbjct: 130 GDAAAFPSSSSCSTSNFTSSR 150
>At2g21510 putative DnaJ protein
Length = 346
Score = 68.2 bits (165), Expect = 4e-12
Identities = 31/62 (50%), Positives = 43/62 (69%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
+LG++ AS EIKKAYY A ++HPDKNPGD +A + FQ L + +L + +KRA YD+
Sbjct: 10 ILGVKTDASDAEIKKAYYLKARKVHPDKNPGDPQAAKNFQVLGEAYQVLSNPDKRAAYDK 69
Query: 61 TG 62
G
Sbjct: 70 YG 71
>At5g01390 heat shock protein 40-like
Length = 335
Score = 67.4 bits (163), Expect = 7e-12
Identities = 30/63 (47%), Positives = 48/63 (75%), Gaps = 1/63 (1%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGD-EEAKEKFQQLQKVISILGDEEKRALYD 59
VL ++++A+ E+KKAY KLA++ HPDKNP + +EA+ KF+Q+ + +L D +KRA+Y+
Sbjct: 8 VLEVDRSANDDELKKAYRKLAMKWHPDKNPNNKKEAEAKFKQISEAYDVLSDPQKRAIYE 67
Query: 60 QTG 62
Q G
Sbjct: 68 QYG 70
>At4g28480 heat-shock protein
Length = 348
Score = 67.4 bits (163), Expect = 7e-12
Identities = 32/74 (43%), Positives = 54/74 (72%), Gaps = 4/74 (5%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGD-EEAKEKFQQLQKVISILGDEEKRALYD 59
VL ++++A+ ++KKAY KLA++ HPDKNP + ++A+ KF+Q+ + +L D +KRA+YD
Sbjct: 8 VLQVDRSANDDDLKKAYRKLAMKWHPDKNPNNKKDAEAKFKQISEAYDVLSDPQKRAVYD 67
Query: 60 QTGCIDDDDLAGDV 73
Q G ++ L G+V
Sbjct: 68 QYG---EEGLKGNV 78
>At5g49060 DNAJ-like protein
Length = 354
Score = 66.2 bits (160), Expect = 2e-11
Identities = 34/70 (48%), Positives = 46/70 (65%), Gaps = 5/70 (7%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKN--PGDEEAKEKFQQLQKVISILGDEEKRALY 58
+LGLEK S EI+KAY KL+L++HPDKN PG EEA F+++ K + L D R +
Sbjct: 103 ILGLEKNCSVDEIRKAYRKLSLKVHPDKNKAPGSEEA---FKKVSKAFTCLSDGNSRRQF 159
Query: 59 DQTGCIDDDD 68
DQ G +D+ D
Sbjct: 160 DQVGIVDEFD 169
>At1g68370 ARG1 protein (Altered Response to Gravity)
Length = 410
Score = 66.2 bits (160), Expect = 2e-11
Identities = 40/94 (42%), Positives = 55/94 (57%), Gaps = 7/94 (7%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VL + K A+ QEIK AY KLAL+ HPDKN + +A E F+++ SIL D EKR YD
Sbjct: 21 VLCVSKDANDQEIKSAYRKLALKYHPDKNANNPDASELFKEVAFSYSILSDPEKRRHYDN 80
Query: 61 TG--CIDDD--DLAGDVQNL---HAFFKTMYKKV 87
G +D D D+ D+ NL + F ++ K+
Sbjct: 81 AGFEALDADGMDMEIDLSNLGTVNTMFAALFSKL 114
>At3g08910 putative heat shock protein
Length = 323
Score = 65.9 bits (159), Expect = 2e-11
Identities = 29/63 (46%), Positives = 46/63 (72%), Gaps = 1/63 (1%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGD-EEAKEKFQQLQKVISILGDEEKRALYD 59
VL +++ A ++KKAY KLA++ HPDKNP + ++A+ KF+Q+ + +L D +KRA+YD
Sbjct: 8 VLQVDRNAKDDDLKKAYRKLAMKWHPDKNPNNKKDAEAKFKQISEAYDVLSDPQKRAIYD 67
Query: 60 QTG 62
Q G
Sbjct: 68 QYG 70
>At1g77020
Length = 358
Score = 65.9 bits (159), Expect = 2e-11
Identities = 32/62 (51%), Positives = 42/62 (67%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+ +AS++EI+KAYY A ++HPDKN GD A EKFQ L + +L D R YD+
Sbjct: 10 VLGVTPSASEEEIRKAYYIKARQVHPDKNQGDPLAAEKFQVLGEAYQVLSDPVHREAYDR 69
Query: 61 TG 62
TG
Sbjct: 70 TG 71
>At1g56300 unknown protein
Length = 156
Score = 65.9 bits (159), Expect = 2e-11
Identities = 38/85 (44%), Positives = 53/85 (61%), Gaps = 10/85 (11%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDK---NPGDE-EAKEKFQQLQKVISILGDEEKRA 56
+LG+ K AS +I+ AY KLA++ HPD+ NPG EAK +FQQ+Q+ S+L DE KR+
Sbjct: 17 ILGIRKDASVSDIRTAYRKLAMKWHPDRYARNPGVAGEAKRRFQQIQEAYSVLNDENKRS 76
Query: 57 LYDQTGCI-----DDDDLAGDVQNL 76
+YD G DDDD +Q +
Sbjct: 77 MYD-VGLYDPHEDDDDDFCDFMQEM 100
>At2g20560 putative heat shock protein
Length = 337
Score = 65.1 bits (157), Expect = 3e-11
Identities = 31/74 (41%), Positives = 53/74 (70%), Gaps = 4/74 (5%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGD-EEAKEKFQQLQKVISILGDEEKRALYD 59
VL ++++AS ++KKAY KLA++ HPDKNP + ++A+ F+Q+ + +L D +K+A+YD
Sbjct: 8 VLQVDRSASDDDLKKAYRKLAMKWHPDKNPNNKKDAEAMFKQISEAYEVLSDPQKKAVYD 67
Query: 60 QTGCIDDDDLAGDV 73
Q G ++ L G+V
Sbjct: 68 QYG---EEGLKGNV 78
>At3g08970 putative DnaJ protein
Length = 572
Score = 63.9 bits (154), Expect = 8e-11
Identities = 32/62 (51%), Positives = 43/62 (68%), Gaps = 1/62 (1%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+ K A Q+EI+KA++K +L+ HPDKN D+ A+EKF ++ IL DEEKR YD
Sbjct: 31 VLGVSKDAKQREIQKAFHKQSLKYHPDKNK-DKGAQEKFAEINNAYEILSDEEKRKNYDL 89
Query: 61 TG 62
G
Sbjct: 90 YG 91
>At1g76700 DnaJ like protein
Length = 398
Score = 63.9 bits (154), Expect = 8e-11
Identities = 30/62 (48%), Positives = 39/62 (62%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+ TA++ EIKKAYY A ++HPDKNP D +A FQ L + +L D +R YD
Sbjct: 10 VLGVSPTATESEIKKAYYIKARQVHPDKNPNDPQAAHNFQVLGEAYQVLSDSGQRQAYDA 69
Query: 61 TG 62
G
Sbjct: 70 CG 71
>At1g21080 putative dnaJ protein
Length = 391
Score = 63.9 bits (154), Expect = 8e-11
Identities = 30/62 (48%), Positives = 40/62 (64%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+ TA++ EIKKAYY A ++HPDKNP D +A FQ L + +L D +R YD
Sbjct: 10 VLGVSPTATEAEIKKAYYIKARQVHPDKNPNDPQAAHNFQVLGEAYQVLSDPGQRQAYDT 69
Query: 61 TG 62
+G
Sbjct: 70 SG 71
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.128 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,770,944
Number of Sequences: 26719
Number of extensions: 253644
Number of successful extensions: 1080
Number of sequences better than 10.0: 125
Number of HSP's better than 10.0 without gapping: 91
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 925
Number of HSP's gapped (non-prelim): 150
length of query: 249
length of database: 11,318,596
effective HSP length: 97
effective length of query: 152
effective length of database: 8,726,853
effective search space: 1326481656
effective search space used: 1326481656
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC143341.1


