

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143339.10 + phase: 0 /pseudo
(349 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
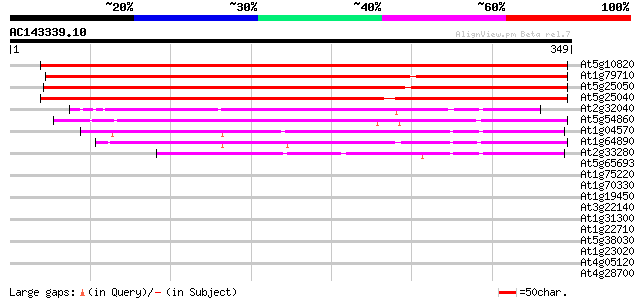
Score E
Sequences producing significant alignments: (bits) Value
At5g10820 unknown protein 456 e-128
At1g79710 hypothetical protein 321 3e-88
At5g25050 putative membrane protein 308 4e-84
At5g25040 putative membrane protein 282 2e-76
At2g32040 unknown protein 137 7e-33
At5g54860 unknown protein 103 2e-22
At1g04570 unknown protein 86 4e-17
At1g64890 unknown protein 68 8e-12
At2g33280 hypothetical protein 64 9e-11
At5g65693 unknown protein 38 0.007
At1g75220 integral membrane protein, putative 32 0.67
At1g70330 unknown protein (At1g70330) 31 0.88
At1g19450 integral membrane protein, putative 31 0.88
At3g22140 hypothetical protein 31 1.2
At1g31300 unknown protein 30 1.5
At1g22710 putative sucrose transport protein, SUC2 30 1.5
At5g38030 putative transmembrane protein 29 4.4
At1g23020 putative superoxide-generating NADPH oxidase flavocyto... 29 4.4
At4g05120 putative protein 28 7.5
At4g28700 ammonium transporter - like protein 28 9.7
>At5g10820 unknown protein
Length = 503
Score = 456 bits (1172), Expect = e-128
Identities = 203/328 (61%), Positives = 267/328 (80%)
Query: 20 LLSILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPST 79
++S+L +P +WL ML++ LN +F+ G+ LVYG+ QGF+GS+FK+VTDYYWKDVQ++QPS
Sbjct: 35 VVSVLIQPFQWLQMLSSRLNLSFVLGVVLVYGVNQGFSGSIFKVVTDYYWKDVQQVQPSV 94
Query: 80 VQLYVGIYFIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVA 139
VQLY+G+Y+IPW+++PIWG+ TD FPI GY+R+PYFVV+GV+G VSA + V G++
Sbjct: 95 VQLYMGLYYIPWVMRPIWGLFTDVFPIKGYKRKPYFVVSGVLGLVSAIAIVVLGKLPAAL 154
Query: 140 ALMCFVGVSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFV 199
AL C +GVSA++AIADV IDACIA NSI + LAPD+QSLC CS +GAL+GY SG FV
Sbjct: 155 ALSCLLGVSAAMAIADVVIDACIATNSINIRSLAPDIQSLCMVCSSAGALVGYATSGVFV 214
Query: 200 HHLGPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYP 259
H LGPQG+LG++A PA ++LGF IYEKR++ + +K K + +G ++ M KT+KYP
Sbjct: 215 HQLGPQGALGVLAFSPATIVILGFFIYEKRSSTVPTQKTKKDTDGLGVAVKGMCKTVKYP 274
Query: 260 QAWKPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYH 319
+ WKPSLYMF+SLALN++THEGHFYWYTD GPAFSQEFVGIIYA+GA+AS+ GVLIYH
Sbjct: 275 EVWKPSLYMFISLALNISTHEGHFYWYTDPTAGPAFSQEFVGIIYAVGALASMFGVLIYH 334
Query: 320 KYLKNYTFRNLVFYAQLLYAISGLLDLM 347
K LK Y+FRN++F+AQLLY SG+LDL+
Sbjct: 335 KKLKGYSFRNILFFAQLLYVFSGMLDLV 362
>At1g79710 hypothetical protein
Length = 497
Score = 321 bits (823), Expect = 3e-88
Identities = 153/325 (47%), Positives = 217/325 (66%), Gaps = 3/325 (0%)
Query: 23 ILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQL 82
+L P+ WL ML L+ +F G+ +VYG+ QG L K+ T YY+KD QK+QPS Q+
Sbjct: 30 VLKRPLRWLRMLIEELHWSFFFGVIIVYGVSQGLGKGLSKVSTQYYFKDEQKIQPSQAQI 89
Query: 83 YVGIYFIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAALM 142
YVG+ IPWI+KP+WG+LTD P+LGYRRRPYF++AG + +S ++ + + + AL
Sbjct: 90 YVGLIQIPWIIKPLWGLLTDVVPVLGYRRRPYFILAGFLAMISMMVLWLHTNLHLALALS 149
Query: 143 CFVGVSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHL 202
C V SA +AIADVTIDAC+ + SI P LA D+QSLCG CS G+L+G+ SG VH +
Sbjct: 150 CLVAGSAGVAIADVTIDACVTQCSISHPTLAADMQSLCGLCSSIGSLVGFSLSGVLVHLV 209
Query: 203 GPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQAW 262
G +G GL+ V L +V+G V+ E + L + + G+ I +KT +Y + W
Sbjct: 210 GSKGVYGLLGVTAGLLVVVGMVLKESPSRSLGRKHVNDKFLDAGSAI---WKTFQYGEVW 266
Query: 263 KPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYL 322
+P L+M LS A+++ HEG FYWYTDSK GP+FS+E VG I + GA+ S++G+L+Y +L
Sbjct: 267 RPCLFMLLSAAVSLHIHEGMFYWYTDSKDGPSFSKEAVGSIMSFGAIGSLVGILLYQNFL 326
Query: 323 KNYTFRNLVFYAQLLYAISGLLDLM 347
KN+ FRN+VF+A L +SG LDL+
Sbjct: 327 KNFPFRNVVFWALSLSVLSGFLDLI 351
>At5g25050 putative membrane protein
Length = 499
Score = 308 bits (788), Expect = 4e-84
Identities = 152/326 (46%), Positives = 213/326 (64%), Gaps = 3/326 (0%)
Query: 22 SILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQ 81
+++ P+ WL ML++ L+ +F+ G+ +YGI QG GSL ++ T+YY KDVQK+QPS Q
Sbjct: 25 NVVCGPVRWLKMLSSELHWSFVFGVVSLYGINQGLGGSLGRVATEYYMKDVQKVQPSESQ 84
Query: 82 LYVGIYFIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAAL 141
I IPWI+KP+WGILTD PI G+ RRPYF++AGV+G VS +++ + + AL
Sbjct: 85 ALTAITKIPWIIKPLWGILTDVLPIFGFHRRPYFILAGVLGVVSLLFISLHSNLHLYLAL 144
Query: 142 MCFVGVSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHH 201
SA++AIADVTIDAC A NSI+ P LA D+QSLC S GALLG+ SG VH
Sbjct: 145 FWMTISSAAMAIADVTIDACTAYNSIKHPSLASDMQSLCSLSSSIGALLGFFMSGILVHL 204
Query: 202 LGPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQA 261
+G +G GL+ AL V+G V E G ++ + G ++M++T+K
Sbjct: 205 VGSKGVFGLLTFPFALVSVVGIVFSEPHVPGFSYKQVNQKFTDAG---KAMWRTMKCSDV 261
Query: 262 WKPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKY 321
W+PSLYM++SL L + HEG FYW+TDSK GP F+QE VG I +IG++ SI+ +Y
Sbjct: 262 WRPSLYMYISLTLGLNIHEGLFYWFTDSKDGPLFAQETVGFILSIGSIGSILAATLYQLV 321
Query: 322 LKNYTFRNLVFYAQLLYAISGLLDLM 347
LK++ FR L + QLL+A+SG+LDL+
Sbjct: 322 LKDHPFRGLCLWTQLLFALSGMLDLI 347
>At5g25040 putative membrane protein
Length = 492
Score = 282 bits (722), Expect = 2e-76
Identities = 148/329 (44%), Positives = 206/329 (61%), Gaps = 7/329 (2%)
Query: 20 LLSILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPST 79
L S++ P+ WL ML + L+ +++ + GI QGF SL + TDYY KDVQK+QPS
Sbjct: 23 LRSVVCGPVRWLKMLASKLHWSYVFAVVSNCGINQGFGYSLGHVATDYYMKDVQKVQPSQ 82
Query: 80 VQLYVGIYFIPWIL-KPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVV 138
Q I I WI+ KP++GILTD PI G+ RRPYF++AGV+G VS +++ + +
Sbjct: 83 YQALSAITRISWIIFKPLFGILTDVLPIFGFHRRPYFILAGVIGVVSLLFISLQSNLHLY 142
Query: 139 AALMCFVGVSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFF 198
AL SA++AIADVTIDAC NS + P LA D+QSLC G LLG+ SG
Sbjct: 143 LALFWMTISSAAMAIADVTIDACTTYNSNKHPSLASDMQSLCSLSYSIGELLGFFMSGIL 202
Query: 199 VHHLGPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKY 258
VH +G +G GL+ AL V+G + E R G +N +++M++TIK
Sbjct: 203 VHLVGSKGVFGLLTFTFALVSVVGVLFSEPRVHGF------SFKQNFTNAMKAMWRTIKC 256
Query: 259 PQAWKPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIY 318
W+PSLYMF++ AL + EG FYW+TDSK GP F+QE VG I +IG++ SI+GVL+Y
Sbjct: 257 SDVWQPSLYMFITRALGLDIKEGLFYWFTDSKDGPLFAQETVGFILSIGSIGSILGVLLY 316
Query: 319 HKYLKNYTFRNLVFYAQLLYAISGLLDLM 347
+ LK++ FR L + QLL+A+SG+ DL+
Sbjct: 317 NLRLKDHPFRKLFLWTQLLFALSGMFDLI 345
>At2g32040 unknown protein
Length = 560
Score = 137 bits (346), Expect = 7e-33
Identities = 90/302 (29%), Positives = 165/302 (53%), Gaps = 18/302 (5%)
Query: 38 LNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIYFIPWILKPIW 97
L+P +A + +VY + QG G L +L +Y KD L P+ + G+ +PW++KP++
Sbjct: 123 LSPDNVA-VAMVYFV-QGVLG-LARLAVSFYLKDDLHLDPAETAVITGLSSLPWLVKPLY 179
Query: 98 GILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAALMCFVGVSASLAIADVT 157
G ++D+ P+ GYRRR Y V++G++G S +++A G S +A C + S S+A +DV
Sbjct: 180 GFISDSVPLFGYRRRSYLVLSGLLGAFSWSLMA-GFVDSKYSAAFCILLGSLSVAFSDVV 238
Query: 158 IDACIA-RNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGLMAVFPA 216
+D+ + R E ++ LQSLC S G ++ SG V G + G+ A+ P
Sbjct: 239 VDSMVVERARGESQSVSGSLQSLCWGSSAFGGIVSSYFSGSLVESYGVRFVFGVTALLPL 298
Query: 217 LTIVLGFVIYEKRTTGLHNEKKK-------GVVENVGTTIRSMYKTIKYPQAWKPSLYMF 269
+T + ++ E+R + +K+ G ++ + ++ IK P + P+L++F
Sbjct: 299 ITSAVAVLVNEQRVVRPASGQKENITLLSPGFLQTSKQNMIQLWGAIKQPNVFLPTLFIF 358
Query: 270 LSLALNVTTH-EGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNYTFR 328
L A T H + +++T +K+G F+ EF+G + + ++AS++GV +Y+ +LK R
Sbjct: 359 LWQA---TPHSDSAMFYFTTNKLG--FTPEFLGRVKLVTSIASLLGVGLYNGFLKTVPLR 413
Query: 329 NL 330
+
Sbjct: 414 KI 415
>At5g54860 unknown protein
Length = 491
Score = 103 bits (256), Expect = 2e-22
Identities = 93/377 (24%), Positives = 156/377 (40%), Gaps = 62/377 (16%)
Query: 28 IEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIY 87
I WL L + F+ + L+Y QGF ++ V+ Y KD +L PS Q +
Sbjct: 2 IHWLKQLRSAFGVAFLWLVCLIY-FTQGFRSFVWTAVS-YQLKDRLQLSPSASQFVFSVA 59
Query: 88 FIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGG-EVSVVAALMCFVG 146
F PW +KP++GI++D PI G +R PY V++ V+ V ++ + S LM F+
Sbjct: 60 FFPWSIKPLYGIISDCIPIGGKKRTPYLVISTVLSLVPWLVLGLDSTSRSSSLYLMIFLT 119
Query: 147 V-SASLAIADVTIDACIARN-SIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGP 204
V + A+ADV IDA IA +E A DLQS+ F G + G L G+ + +L
Sbjct: 120 VQNLGSAMADVVIDAMIAEAVRLEKASFAGDLQSVSWFAMAVGGVCGSLLGGYALSNLKI 179
Query: 205 QGSLGLMAVFPALTIVLGFVIYE--------------------------------KRTTG 232
+ L V PAL ++ ++ E K T
Sbjct: 180 ETIFLLFTVLPALQLLSCALVEESPANNEPLPELLDSNEFEEKSLTSNDNYPDTSKSNTR 239
Query: 233 LHNEKKKGV----------------------VENVGTTIRSMYKTIKYPQAWKPSLYMFL 270
+KKG +++ + + K P +P + F+
Sbjct: 240 RRKGQKKGKKGDSNGKSETQKKQSKSLASQWFQSLKAATFGLGRAFKQPIILRPMAWFFI 299
Query: 271 SLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNYTFRNL 330
+ FY+ T+ F+G +G + + G IY++YL++ T R
Sbjct: 300 AHITVPNLSTVMFYYQTEVL---QLDAAFLGTARVVGWLGLMFGTFIYNRYLQDMTLRKS 356
Query: 331 VFYAQLLYAISGLLDLM 347
+ +A + +++ LLD++
Sbjct: 357 LLFAHIGLSVTILLDMV 373
>At1g04570 unknown protein
Length = 542
Score = 85.5 bits (210), Expect = 4e-17
Identities = 74/305 (24%), Positives = 138/305 (44%), Gaps = 9/305 (2%)
Query: 45 GIFLVYGIGQGFTGSLFK--LVTDYYWKDVQKLQPSTVQLYVGIYFIPWILKPIWGILTD 102
GI ++ G+G GS L +++ LQPST+QL +P + KP++G+L+D
Sbjct: 86 GISVLCGLGYWVQGSRCFPWLALNFHMVHSLALQPSTLQLVQYSCSLPMVAKPLYGVLSD 145
Query: 103 AFPILGYRRRPYFVVAGVVGTVSATIVAV--GGEVSVVAALMCFVGVSASLAIADVTIDA 160
I RR PY + + ++ + + G + + + C + + +I +V DA
Sbjct: 146 VLYIGSGRRVPYIAIGVFLQVLAWGSMGIFQGAREVLPSLVACVLLSNLGASITEVAKDA 205
Query: 161 CIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGLMAVFPALTIV 220
+A + LQS S +G +LG L G+ + P+ S + + +L +V
Sbjct: 206 LVAEYGLRY--RINGLQSYALMASAAGGVLGNLLGGYLLLTTPPKISFLVFSALLSLQLV 263
Query: 221 LGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQAWKPSLYMFLSLALNVTTHE 280
+ E+ + V+E+V I ++ + I+ + +P ++ +S+A+ V
Sbjct: 264 VSLSSKEESFGLPRIAETSSVLESVKKQISNLKEAIQADEISQPLIWAVVSIAM-VPLLS 322
Query: 281 GHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNYTFRNLVFYAQLLYAI 340
G + Y + +G+ IG + + ++Y +YLK R L+ QLLY +
Sbjct: 323 GSVFCYQTQVLN--LDPSVIGMSKVIGQLMLLCLTVVYDRYLKTLPMRPLIHIIQLLYGL 380
Query: 341 SGLLD 345
S LLD
Sbjct: 381 SILLD 385
>At1g64890 unknown protein
Length = 442
Score = 67.8 bits (164), Expect = 8e-12
Identities = 63/298 (21%), Positives = 126/298 (42%), Gaps = 11/298 (3%)
Query: 54 QGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIYFIPWILKPIWGILTDAFPILGYRRRP 113
QGF G + L +++ + ++ PS +QL +P + KPI+G+++D+ G R P
Sbjct: 30 QGFRGFPW-LGANFFLTEQLRVNPSVLQLLQNSANLPMVAKPIYGVVSDSVYFFGQHRIP 88
Query: 114 YFVVAGVVGTVSATIVAV--GGEVSVVAALMCFVGVSASLAIADVTIDACIARNSIEVPG 171
Y V ++ +S +A VS++A + + + ++ +V DA +A + +
Sbjct: 89 YIAVGALLQAISWLAIAFLSRSNVSILALSIYLLLSNLGASLVEVANDAIVAESGKQKTS 148
Query: 172 --LAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGLMAVFPALTIVLGFVIYEKR 229
+ +L S S G +LG L G + Q + + + L ++ I EK
Sbjct: 149 ETQSGELPSFVWMVSSLGGILGNLLGGIAIKTFSAQSTFLVFGILALLQFLVTINIREKS 208
Query: 230 TTGLHNEKKKGVVENVGTTIRSMYKTIKYPQAWKPSLYMFLSLALNVTTHEGHFYWYTDS 289
N G + + + ++ P+ ++ +S A+ V G ++Y
Sbjct: 209 LNLPENPSPAG---GIRKHLSDLSHVLRKPEISYSIAWIAVSTAV-VPVLTGTMFFYQTK 264
Query: 290 KVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNYTFRNLVFYAQLLYAISGLLDLM 347
+ +GI G +A ++ Y+++LK R L+ Q+ A + DL+
Sbjct: 265 FL--KIDASLLGISKVFGQIAMLLWGFAYNRWLKAMRPRKLLTAIQVTIAFFVISDLL 320
>At2g33280 hypothetical protein
Length = 408
Score = 64.3 bits (155), Expect = 9e-11
Identities = 64/263 (24%), Positives = 113/263 (42%), Gaps = 17/263 (6%)
Query: 92 ILKPIWGILTDAFPILGYRRRPYFVVAGVV-GTVSATIVAVGGEVSVVAALMCFVGVS-A 149
+ KP++G+L+D I G RR PY V ++ G ++ G V+ +LM F+ +S
Sbjct: 2 VAKPLYGVLSDVLYIGGARRVPYISVGVLLQGLAWGSLAIFPGAREVLPSLMAFILLSNL 61
Query: 150 SLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLG 209
+I +V+ DA +A + LQS S G +LG L G+ + P+
Sbjct: 62 GASITEVSQDALVAEYGLRYQ--INGLQSYALMASAVGGILGNLLGGYCLLKTPPRI--- 116
Query: 210 LMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKT-------IKYPQAW 262
L F AL + V + ++ + V + + + + K ++ +
Sbjct: 117 LFLAFTALLSLQLIVSLSSKEESVNLPRIGEVTPEISSVLGIVKKQFLDLKGIVQVDEIS 176
Query: 263 KPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYL 322
+P +++ S+AL V G + Y + +G+ IG + + ++Y +Y
Sbjct: 177 QPLIWIVSSIAL-VPLLSGSVFCYQTQVLN--LDPSVIGMSKVIGQLMLLCLTVVYDRYW 233
Query: 323 KNYTFRNLVFYAQLLYAISGLLD 345
K R L+ QLLYA S L D
Sbjct: 234 KKLPMRALIHIVQLLYAFSLLFD 256
>At5g65693 unknown protein
Length = 492
Score = 38.1 bits (87), Expect = 0.007
Identities = 36/135 (26%), Positives = 59/135 (43%), Gaps = 18/135 (13%)
Query: 99 ILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVS----VVAALMCFVGVSA----S 150
+ + F L R P+ ++ VG TI +G S ++A FVGV S
Sbjct: 95 VASPIFAGLSKRFNPFKLIG--VGLTVWTIAVIGCGFSYNFWMIAVFRMFVGVGEASFIS 152
Query: 151 LAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGL 210
LA + A +AR + + L C +G LGY+ G+ +HLG + + +
Sbjct: 153 LAAPYIDDSAPVARKNFWL--------GLFYMCIPAGVALGYVFGGYIGNHLGWRWAFYI 204
Query: 211 MAVFPALTIVLGFVI 225
A+ A+ ++L F I
Sbjct: 205 EAIAMAVFVILSFCI 219
>At1g75220 integral membrane protein, putative
Length = 487
Score = 31.6 bits (70), Expect = 0.67
Identities = 16/43 (37%), Positives = 25/43 (57%)
Query: 171 GLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGLMAV 213
GL S+ G S GA++G +ASG ++G +GSL + A+
Sbjct: 81 GLTVSEYSVFGSLSNVGAMVGAIASGQIAEYIGRKGSLMIAAI 123
>At1g70330 unknown protein (At1g70330)
Length = 450
Score = 31.2 bits (69), Expect = 0.88
Identities = 51/199 (25%), Positives = 85/199 (42%), Gaps = 17/199 (8%)
Query: 3 TQQDSTTTITKTNNKYPLLSILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFK 62
T D + I + P S+L P E T + + F I+ G+G + F
Sbjct: 24 TTTDKSAGIVTDSEAGPETSLLLNPHEGSTKKAPSDSYHFAYIIYFTLGVGFLLPWNAFI 83
Query: 63 LVTDYYWKDVQKLQPSTV--QLYVGIYFIPWILKPIWGILTDAFPILGYRR----RPYFV 116
DY+ L PST +++ IY + ++ ++ A L R FV
Sbjct: 84 TAVDYF----SYLYPSTAVDRIFAVIYMLVALVCLFVIVVFYAHKSLASFRINLGLLLFV 139
Query: 117 VAGVVGTVSATIVAVGGEVSVVAALMCFVGVSASLAIADVTIDACIARNSIEVPGLAPD- 175
+A +V V +V V G+V + A F SA++A++ + DA + I V G P+
Sbjct: 140 IALLVVPV-LDLVYVKGQVGLYAG---FDVTSAAVALSGLG-DALMQGGLIGVAGEMPER 194
Query: 176 -LQSLCGFCSGSGALLGYL 193
+Q++ +GSG L+ L
Sbjct: 195 YMQAVVAGTAGSGVLVSLL 213
>At1g19450 integral membrane protein, putative
Length = 488
Score = 31.2 bits (69), Expect = 0.88
Identities = 16/43 (37%), Positives = 25/43 (57%)
Query: 171 GLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGLMAV 213
GL S+ G S GA++G +ASG ++G +GSL + A+
Sbjct: 82 GLTVSEYSVFGSLSNVGAMVGAIASGQIAEYVGRKGSLMIAAI 124
>At3g22140 hypothetical protein
Length = 1237
Score = 30.8 bits (68), Expect = 1.2
Identities = 29/101 (28%), Positives = 43/101 (41%), Gaps = 5/101 (4%)
Query: 98 GILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAALMCFVGVSAS-LAIADV 156
G T F I G + V +G ++ VAVGG VV GV+ +A+ V
Sbjct: 374 GFSTGGFAIGGVATGGFAVGGVAIGGIATGGVAVGG---VVVGGFAIGGVATGEVAVGGV 430
Query: 157 TIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGF 197
T +A + + G+A + GF +G A+ G GF
Sbjct: 431 TTGG-VAIGEVAIGGVAVGGVATGGFATGGVAIGGIATGGF 470
Score = 29.3 bits (64), Expect = 3.3
Identities = 26/85 (30%), Positives = 40/85 (46%), Gaps = 6/85 (7%)
Query: 115 FVVAGV-VGTVSATIVAVGGEVSVVAALMCFVGVSAS-LAIADVTIDACIARNSIEVPGL 172
F V GV +G ++ VAVGG +V GV+ +A+ VT +A + + G+
Sbjct: 640 FAVGGVAIGGIATGGVAVGG---IVVGGFAIGGVATGGVAVGGVTTGG-VAIGGVAIGGV 695
Query: 173 APDLQSLCGFCSGSGALLGYLASGF 197
A + GF +G A+ G GF
Sbjct: 696 AVGGVATGGFATGGVAIGGIATGGF 720
>At1g31300 unknown protein
Length = 278
Score = 30.4 bits (67), Expect = 1.5
Identities = 22/90 (24%), Positives = 40/90 (44%), Gaps = 7/90 (7%)
Query: 266 LYMFLSLALNVTTHEGHFYWYTDS-----KVGPAFSQEFVGIIYAIGAVASIIGVLIYHK 320
LY ++ L +TT E + WY D+ + + F+ + + + + I + YH
Sbjct: 170 LYTYMVLISEITTPEINLRWYLDTAGMKKSLAYVVNGVFIFLAWLVARILLFI-YMFYHV 228
Query: 321 YLK-NYTFRNLVFYAQLLYAISGLLDLMCL 349
YL N R +F L++ + L +M L
Sbjct: 229 YLHYNQVMRMHIFGYVLVFGVPAALGVMNL 258
>At1g22710 putative sucrose transport protein, SUC2
Length = 512
Score = 30.4 bits (67), Expect = 1.5
Identities = 23/78 (29%), Positives = 35/78 (44%), Gaps = 19/78 (24%)
Query: 75 LQPSTVQLYVGIYFIP-------WILKPIWGILTDAFPILGY----------RRRPYFVV 117
LQ S + YV + IP W+ PI G+L PI+GY RRRP+ V
Sbjct: 48 LQLSLLTPYVQLLGIPHKWASLIWLCGPISGMLVQ--PIVGYHSDRCTSRFGRRRPFIVA 105
Query: 118 AGVVGTVSATIVAVGGEV 135
+ TV+ ++ ++
Sbjct: 106 GAGLVTVAVFLIGYAADI 123
>At5g38030 putative transmembrane protein
Length = 498
Score = 28.9 bits (63), Expect = 4.4
Identities = 20/58 (34%), Positives = 34/58 (58%), Gaps = 2/58 (3%)
Query: 134 EVSVVAALMCFVGVSASLAIADVTID-ACIARNSIEVP-GLAPDLQSLCGFCSGSGAL 189
+ S+ AA F G +++A+A V+++ + IA S V G+ L++LCG G+G L
Sbjct: 63 QYSLGAATQVFAGHISTIALAAVSVENSVIAGFSFGVMLGMGSALETLCGQAFGAGKL 120
>At1g23020 putative superoxide-generating NADPH oxidase
flavocytochrome
Length = 693
Score = 28.9 bits (63), Expect = 4.4
Identities = 37/142 (26%), Positives = 58/142 (40%), Gaps = 22/142 (15%)
Query: 205 QGSLGLMAVFPALT--IVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQAW 262
Q L +AV LT I LGF+ Y + ++ VG T S + KY W
Sbjct: 163 QARLDSIAVRLGLTGNICLGFLFYPVA-------RGSSLLAAVGLTSES---STKY-HIW 211
Query: 263 KPSLYMFLSLALNVTTHEGHF--YWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHK 320
+L M L T+H + YW + ++V + GI + G +A + G+L++
Sbjct: 212 LGNLVMTL-----FTSHGLCYCIYWISTNQVSQMLEWDRTGISHLAGEIALVAGLLMWAT 266
Query: 321 YLK--NYTFRNLVFYAQLLYAI 340
F + FY LY +
Sbjct: 267 TFPAIRRRFFEVFFYTHYLYMV 288
>At4g05120 putative protein
Length = 418
Score = 28.1 bits (61), Expect = 7.5
Identities = 45/190 (23%), Positives = 80/190 (41%), Gaps = 27/190 (14%)
Query: 49 VYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIYFIPWILKPIWGILTDAFPILG 108
+ GIG + + + DYY+K PS V + + + P+ L IL A+
Sbjct: 25 ILGIGSLVSWNSMLTIADYYYKVFPDYHPSRV---LTLVYQPFALGT---ILILAYHESK 78
Query: 109 YRRRPYFVVAGVVGTVSATIVAV--------GGEVSVVAALMCFVGVSASLAIADVTID- 159
R ++ ++ T+S ++ V GG + + V AS +AD T+
Sbjct: 79 INTRKRNLIGYILFTISTFLLIVLDLATKGRGG----IGPYIGLCAVVASFGLADATVQG 134
Query: 160 ACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYL----ASGFFVHHLGP-QGSLGLMAVF 214
I S+ P L +QS G + SGAL L + F + GP +G++ +A+
Sbjct: 135 GMIGDLSLMCPEL---VQSFMGGLAVSGALTSALRLITKAAFEKTNDGPRKGAMMFLAIS 191
Query: 215 PALTIVLGFV 224
+ ++ F+
Sbjct: 192 TCIELLCVFL 201
>At4g28700 ammonium transporter - like protein
Length = 504
Score = 27.7 bits (60), Expect = 9.7
Identities = 12/47 (25%), Positives = 23/47 (48%), Gaps = 11/47 (23%)
Query: 282 HFYWYTDSKVGPAFSQEFV-----------GIIYAIGAVASIIGVLI 317
H++W +D PA S+ + G+++ +G +A + G LI
Sbjct: 181 HWFWSSDGWASPARSENLLFQSGVIDFAGSGVVHMVGGIAGLWGALI 227
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.141 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,601,071
Number of Sequences: 26719
Number of extensions: 320581
Number of successful extensions: 1017
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 987
Number of HSP's gapped (non-prelim): 26
length of query: 349
length of database: 11,318,596
effective HSP length: 100
effective length of query: 249
effective length of database: 8,646,696
effective search space: 2153027304
effective search space used: 2153027304
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC143339.10


