

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143337.8 - phase: 0
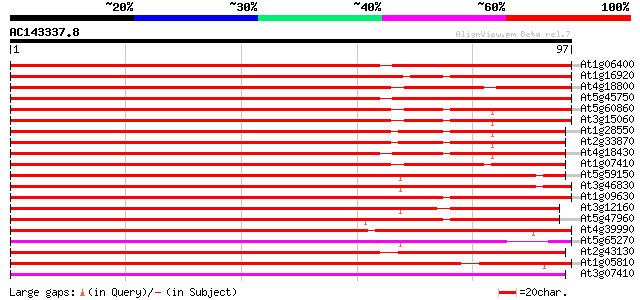
(97 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g06400 unknown protein 145 3e-36
At1g16920 unknown protein 139 2e-34
At4g18800 ras-like GTP-binding protein 135 5e-33
At5g45750 Rab-type small GTP-binding protein-like 134 8e-33
At5g60860 GTP-binding protein - like 123 2e-29
At3g15060 ras-related GTP-binding protein 121 7e-29
At1g28550 ras-related GTP-binding protein, putative 119 2e-28
At2g33870 putative GTP-binding protein 115 4e-27
At4g18430 membrane-bound small GTP-binding - like protein 114 1e-26
At1g07410 small G protein, putative 103 1e-23
At5g59150 GTP-binding protein rab11 - like 103 2e-23
At3g46830 GTP-binding protein Rab11 103 2e-23
At1g09630 putative RAS-related protein, RAB11C 99 3e-22
At3g12160 hypothetical protein 97 1e-21
At5g47960 RAS superfamily GTP-binding protein-like 96 2e-21
At4g39990 GTP-binding protein GB3 87 2e-18
At5g65270 GTP-binding protein 84 9e-18
At2g43130 Ras-related GTP-binding protein (ARA-4) 80 1e-16
At1g05810 RAS-related protein ARA-1 78 9e-16
At3g07410 GTP-binding protein like 77 1e-15
>At1g06400 unknown protein
Length = 216
Score = 145 bits (367), Expect = 3e-36
Identities = 73/97 (75%), Positives = 85/97 (87%), Gaps = 2/97 (2%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
MLIGNK DLRHLVAV TE+ K+FAERESLYFMETSAL+ATNVENAFTEVL+QI++IVSK+
Sbjct: 122 MLIGNKCDLRHLVAVKTEEAKAFAERESLYFMETSALDATNVENAFTEVLTQIHKIVSKR 181
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
+V+ GG S+ +P G+TINVKED SVLKR GCCSN
Sbjct: 182 SVDG--GGESADLPGKGETINVKEDGSVLKRMGCCSN 216
>At1g16920 unknown protein
Length = 216
Score = 139 bits (351), Expect = 2e-34
Identities = 73/97 (75%), Positives = 83/97 (85%), Gaps = 2/97 (2%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNKSDLRHL+AVPTEDGKS+AE+ESL FMETSALEATNVE+AF EVL+QIYRI SKK
Sbjct: 122 MLVGNKSDLRHLLAVPTEDGKSYAEQESLCFMETSALEATNVEDAFAEVLTQIYRITSKK 181
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
VEAGE G ++SVP G+ I VK D S LK+ GCCSN
Sbjct: 182 QVEAGEDG-NASVPK-GEKIEVKNDVSALKKLGCCSN 216
>At4g18800 ras-like GTP-binding protein
Length = 214
Score = 135 bits (339), Expect = 5e-33
Identities = 71/97 (73%), Positives = 82/97 (84%), Gaps = 4/97 (4%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNKSDLRHLVAV TED KSFAE ESLYFMETSALE+TNVENAF+EVL+QIY +VSKK
Sbjct: 122 MLVGNKSDLRHLVAVQTEDAKSFAENESLYFMETSALESTNVENAFSEVLTQIYHVVSKK 181
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
A+EAGE S +VPS G+ I+V D S +K+ GCCSN
Sbjct: 182 AMEAGE--DSGNVPSKGEKIDV--DVSAVKKTGCCSN 214
>At5g45750 Rab-type small GTP-binding protein-like
Length = 216
Score = 134 bits (337), Expect = 8e-33
Identities = 69/97 (71%), Positives = 81/97 (83%), Gaps = 2/97 (2%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNKSDLRHLVAV TED KSFAE+ESLYFMETSALEATNVENAF EVL+QI+ IVSKK
Sbjct: 122 MLVGNKSDLRHLVAVQTEDAKSFAEKESLYFMETSALEATNVENAFAEVLTQIHHIVSKK 181
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
A+EA S++VPS G I++ +D S +K+ GCCSN
Sbjct: 182 AMEA--ASESANVPSKGDKIDIGKDVSAVKKGGCCSN 216
>At5g60860 GTP-binding protein - like
Length = 217
Score = 123 bits (308), Expect = 2e-29
Identities = 64/99 (64%), Positives = 81/99 (81%), Gaps = 5/99 (5%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
M +GNK+DLRHL AV TED K+FAERE+ +FMETSALE+ NVENAFTEVLSQIYR+VS+K
Sbjct: 122 MFVGNKADLRHLRAVSTEDAKAFAERENTFFMETSALESMNVENAFTEVLSQIYRVVSRK 181
Query: 61 AVEAGEGGSSSSVPSVGQTINV--KEDSSVLKRFGCCSN 97
A++ G+ +++P GQTINV K+D S +K+ GCCSN
Sbjct: 182 ALDIGD--DPAALPK-GQTINVGSKDDVSAVKKVGCCSN 217
>At3g15060 ras-related GTP-binding protein
Length = 217
Score = 121 bits (303), Expect = 7e-29
Identities = 64/99 (64%), Positives = 81/99 (81%), Gaps = 5/99 (5%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK+DLRHL AV TED K+FAERE+ +FMETSALEA NVENAFTEVLSQIYR+ SKK
Sbjct: 122 MLVGNKADLRHLRAVSTEDAKAFAERENTFFMETSALEALNVENAFTEVLSQIYRVASKK 181
Query: 61 AVEAGEGGSSSSVPSVGQTINV--KEDSSVLKRFGCCSN 97
A++ G+ +++P GQ+INV K+D S +K+ GCCS+
Sbjct: 182 ALDIGD--DHTTLPK-GQSINVGSKDDVSEVKKVGCCSS 217
>At1g28550 ras-related GTP-binding protein, putative
Length = 218
Score = 119 bits (299), Expect = 2e-28
Identities = 62/98 (63%), Positives = 82/98 (83%), Gaps = 4/98 (4%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK+DLRHL A+ TE+ K+FAERE+ +FMETSALEA NV+NAFTEVL+QIYR+VSKK
Sbjct: 122 MLVGNKADLRHLRAISTEEAKAFAERENTFFMETSALEAVNVDNAFTEVLTQIYRVVSKK 181
Query: 61 AVEAGEGGSSSSVPSVGQTINV--KEDSSVLKRFGCCS 96
A+EAG+ ++++P GQ INV ++D S +K+ GCCS
Sbjct: 182 ALEAGD-DPTTALPK-GQMINVGGRDDISAVKKPGCCS 217
>At2g33870 putative GTP-binding protein
Length = 219
Score = 115 bits (288), Expect = 4e-27
Identities = 60/98 (61%), Positives = 80/98 (81%), Gaps = 4/98 (4%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK+DL HL A+ TE+ K FAERE+ +FMETSALEA NVENAFTEVL+QIYR+VSKK
Sbjct: 123 MLVGNKADLNHLRAISTEEVKDFAERENTFFMETSALEAINVENAFTEVLTQIYRVVSKK 182
Query: 61 AVEAGEGGSSSSVPSVGQTINV--KEDSSVLKRFGCCS 96
A++AG+ ++++P GQ INV ++D S +K+ GCC+
Sbjct: 183 ALDAGD-DPTTALPK-GQMINVGSRDDVSAVKKSGCCA 218
>At4g18430 membrane-bound small GTP-binding - like protein
Length = 217
Score = 114 bits (284), Expect = 1e-26
Identities = 58/98 (59%), Positives = 79/98 (80%), Gaps = 5/98 (5%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK+DLRHL AVPTE+ +SF+ERE+++FMETSAL+ATNVE AFT VL+QIYR++S+K
Sbjct: 122 MLVGNKADLRHLRAVPTEEARSFSERENMFFMETSALDATNVEQAFTHVLTQIYRVMSRK 181
Query: 61 AVEAGEGGSSSSVPSVGQTINV--KEDSSVLKRFGCCS 96
A++ G S+P GQTI++ K+D + +K GCCS
Sbjct: 182 ALDG--TGDPMSLPK-GQTIDIGNKDDVTAVKSSGCCS 216
>At1g07410 small G protein, putative
Length = 214
Score = 103 bits (257), Expect = 1e-23
Identities = 54/96 (56%), Positives = 69/96 (71%), Gaps = 3/96 (3%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
M+ GNKSDL HL +V EDG+S AE+E L F+ETSALEATN+E AF +LS+IY I+SKK
Sbjct: 121 MMAGNKSDLNHLRSVADEDGRSLAEKEGLSFLETSALEATNIEKAFQTILSEIYHIISKK 180
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
A+ A E ++ ++P G IN+ DSS R GCCS
Sbjct: 181 ALAAQE--AAGNLPGQGTAINI-SDSSATNRKGCCS 213
>At5g59150 GTP-binding protein rab11 - like
Length = 217
Score = 103 bits (256), Expect = 2e-23
Identities = 55/97 (56%), Positives = 71/97 (72%), Gaps = 2/97 (2%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
M+ GNK+DL HL +V EDG++ AE E L F+ETSALEATNVE AF VL++IY I+SKK
Sbjct: 121 MMAGNKADLNHLRSVAEEDGQTLAETEGLSFLETSALEATNVEKAFQTVLAEIYHIISKK 180
Query: 61 AVEAGE-GGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
A+ A E ++S++P G TINV++ S KR GCCS
Sbjct: 181 ALAAQEAAAANSAIPGQGTTINVEDTSGAGKR-GCCS 216
>At3g46830 GTP-binding protein Rab11
Length = 217
Score = 103 bits (256), Expect = 2e-23
Identities = 55/98 (56%), Positives = 70/98 (71%), Gaps = 2/98 (2%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
M+ GNKSDL HL +V EDG+S AE+E L F+ETSALEATNVE AF +L +IY I+SKK
Sbjct: 121 MMAGNKSDLNHLRSVAEEDGQSLAEKEGLSFLETSALEATNVEKAFQTILGEIYHIISKK 180
Query: 61 AVEAGE-GGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
A+ A E ++S++P G TINV + S KR CCS+
Sbjct: 181 ALAAQEAAAANSAIPGQGTTINVDDTSGGAKR-ACCSS 217
>At1g09630 putative RAS-related protein, RAB11C
Length = 217
Score = 99.4 bits (246), Expect = 3e-22
Identities = 50/97 (51%), Positives = 72/97 (73%), Gaps = 1/97 (1%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
MLIGNK+DL+HL AV TED +S+AE+E L F+ETSALEA NVE AF +LS++YRI+SKK
Sbjct: 121 MLIGNKTDLKHLRAVATEDAQSYAEKEGLSFIETSALEALNVEKAFQTILSEVYRIISKK 180
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
++ + + +++++ GQTI+V S + CCS+
Sbjct: 181 SISSDQTTANANIKE-GQTIDVAATSESNAKKPCCSS 216
>At3g12160 hypothetical protein
Length = 220
Score = 97.4 bits (241), Expect = 1e-21
Identities = 54/98 (55%), Positives = 69/98 (70%), Gaps = 5/98 (5%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
MLIGNK DL L AVPTED + FA+RE+L+FMETSALEATNVE AF +L++IYRI+SKK
Sbjct: 122 MLIGNKCDLGSLRAVPTEDAQEFAQRENLFFMETSALEATNVETAFLTILTEIYRIISKK 181
Query: 61 AVEAGE---GGSSSSVPSVGQTINVKEDSSVLKRFGCC 95
++ A + G+SS + G I + + KR GCC
Sbjct: 182 SLTADDDDADGNSSLLK--GTRIIIPSEQESGKRGGCC 217
>At5g47960 RAS superfamily GTP-binding protein-like
Length = 223
Score = 96.3 bits (238), Expect = 2e-21
Identities = 56/98 (57%), Positives = 68/98 (69%), Gaps = 4/98 (4%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
MLIGNK+DL L AVPTED K FA+RE+L+FMETSAL++ NVE +F VL++IYRIVSKK
Sbjct: 124 MLIGNKTDLGTLRAVPTEDAKEFAQRENLFFMETSALDSNNVEPSFLTVLTEIYRIVSKK 183
Query: 61 ---AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCC 95
A E GE G SS+ G I V + + K GCC
Sbjct: 184 NLVANEEGESGGDSSLLQ-GTKIVVAGEETESKGKGCC 220
>At4g39990 GTP-binding protein GB3
Length = 224
Score = 86.7 bits (213), Expect = 2e-18
Identities = 48/99 (48%), Positives = 65/99 (65%), Gaps = 3/99 (3%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+LIGNKSDL AVPTED K FAE+E L+F+ETSAL ATNVEN+F +++QIY V+KK
Sbjct: 126 ILIGNKSDLEDQRAVPTEDAKEFAEKEGLFFLETSALNATNVENSFNTLMTQIYNTVNKK 185
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVL--KRFGCCSN 97
+ A EG S++ G+ I + + K CC++
Sbjct: 186 NL-ASEGDSNNPGSLAGKKILIPGSGQEIPAKTSTCCTS 223
>At5g65270 GTP-binding protein
Length = 226
Score = 84.3 bits (207), Expect = 9e-18
Identities = 49/108 (45%), Positives = 65/108 (59%), Gaps = 18/108 (16%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
+LIGNKSDL A+PTED K FAE+E L+F+ETSA ATNVE+AF+ VL++I+ IV+KK
Sbjct: 126 ILIGNKSDLVDQRAIPTEDAKEFAEKEGLFFLETSAFNATNVESAFSTVLTEIFNIVNKK 185
Query: 61 AVEAGE-----------GGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 97
++ A E G VP GQ I K + CC++
Sbjct: 186 SLAASEDQENGNPGSLAGKKIDIVPGPGQVIPNKSNM-------CCNS 226
>At2g43130 Ras-related GTP-binding protein (ARA-4)
Length = 214
Score = 80.5 bits (197), Expect = 1e-16
Identities = 44/96 (45%), Positives = 60/96 (61%), Gaps = 3/96 (3%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
MLIGNK DL + AV E+GKS AE E L+FMETSAL++TNV+ AF V+ +IY +S+K
Sbjct: 121 MLIGNKCDLESIRAVSVEEGKSLAESEGLFFMETSALDSTNVKTAFEMVIREIYSNISRK 180
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
+ + S +V + VK ++ K F CCS
Sbjct: 181 QLNS---DSYKEELTVNRVSLVKNENEGTKTFSCCS 213
>At1g05810 RAS-related protein ARA-1
Length = 261
Score = 77.8 bits (190), Expect = 9e-16
Identities = 44/100 (44%), Positives = 63/100 (63%), Gaps = 6/100 (6%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK DL ++ AV E+GK+ AE E L+F+ETSAL++TNV+ AF V+ IY VS+K
Sbjct: 164 MLVGNKCDLENIRAVSVEEGKALAEEEGLFFVETSALDSTNVKTAFEMVILDIYNNVSRK 223
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKR---FGCCSN 97
+ + +V V VK+D+S K+ F CCS+
Sbjct: 224 QLNSDTYKDELTVNRVSL---VKDDNSASKQSSGFSCCSS 260
>At3g07410 GTP-binding protein like
Length = 217
Score = 77.4 bits (189), Expect = 1e-15
Identities = 42/96 (43%), Positives = 57/96 (58%)
Query: 1 MLIGNKSDLRHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKK 60
ML+GNK DL + AV E+GK+ AE E L+FMETSAL+ATNV+ AF V+ +I+ VS+K
Sbjct: 121 MLVGNKCDLEDIRAVSVEEGKALAEEEGLFFMETSALDATNVDKAFEIVIREIFNNVSRK 180
Query: 61 AVEAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCS 96
+ + + SV V N S + CCS
Sbjct: 181 LLNSDAYKAELSVNRVSLVNNQDGSESSWRNPSCCS 216
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.127 0.336
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,979,072
Number of Sequences: 26719
Number of extensions: 62992
Number of successful extensions: 234
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 158
Number of HSP's gapped (non-prelim): 79
length of query: 97
length of database: 11,318,596
effective HSP length: 73
effective length of query: 24
effective length of database: 9,368,109
effective search space: 224834616
effective search space used: 224834616
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC143337.8


