

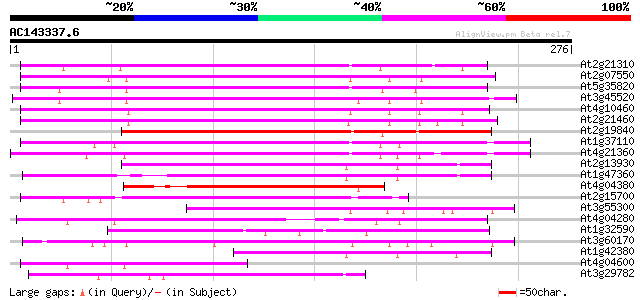
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143337.6 + phase: 0
(276 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g21310 putative retroelement pol polyprotein 159 2e-39
At2g07550 putative retroelement pol polyprotein 159 2e-39
At5g35820 copia-like retrotransposable element 157 6e-39
At3g45520 copia-like polyprotein 154 4e-38
At4g10460 putative retrotransposon 150 7e-37
At2g21460 putative retroelement pol polyprotein 146 1e-35
At2g19840 copia-like retroelement pol polyprotein 142 3e-34
At1g37110 132 2e-31
At4g21360 putative transposable element 130 6e-31
At2g13930 putative retroelement pol polyprotein 117 5e-27
At1g47360 polyprotein, putative 107 9e-24
At4g04380 putative polyprotein 96 2e-20
At2g15700 copia-like retroelement pol polyprotein 91 5e-19
At3g55300 putative protein 88 4e-18
At4g04280 putative transposon protein 86 3e-17
At1g32590 hypothetical protein, 5' partial 69 2e-12
At3g60170 putative protein 67 1e-11
At1g42380 hypothetical protein 64 7e-11
At4g04600 putative polyprotein 61 7e-10
At3g29782 hypothetical protein 51 6e-07
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 159 bits (402), Expect = 2e-39
Identities = 102/253 (40%), Positives = 147/253 (57%), Gaps = 25/253 (9%)
Query: 6 DIEKFTGDNDFGLWKVKMEA------VLIQQKCEKALKGEGSLPVTMSRAEKTK------ 53
++EK G+ D+ LWK K+ A +L + ++A++ E S T S KT+
Sbjct: 7 EVEKLDGEGDYVLWKEKLLAHIELLGLLEGLEEDEAIEEEESTAETDSLLTKTEDKVLKE 66
Query: 54 MVDKARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVE 113
KARS V+L LG+ VLR+V KE TAA M L+ L+M KSL +R +LKQ+LY ++M +
Sbjct: 67 KRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGYKMSD 126
Query: 114 SKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLE 173
S I E + +F K++ DLEN++V + DED+AI+LL +LPK F+ KDT+ YGK T+ L+
Sbjct: 127 SMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLKDTLKYGKT-TLALD 185
Query: 174 EVQAALRT---------KDLTKSKDLTHEHGEGLSVTRGNGGGRGNRRKSGNKSRFE--C 222
E+ A+R+ K L S D G S R R N+ +S +KSR + C
Sbjct: 186 EITGAIRSKVLELGASGKMLKNSSDALFVQDRGRSEKRDKSSER-NKSQSRSKSREKKVC 244
Query: 223 FNCHKMGHFKKDC 235
+ C K GHFKK C
Sbjct: 245 WVCGKEGHFKKQC 257
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 159 bits (401), Expect = 2e-39
Identities = 99/259 (38%), Positives = 149/259 (57%), Gaps = 25/259 (9%)
Query: 6 DIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMS-----------RAEKTKM 54
++EKF G D+ +WK K+ A + ALK S S + EK +
Sbjct: 7 EVEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEA 66
Query: 55 VD----KARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFR 110
++ KARSA+VL + D+VLR++ KE+TAA+M L+ LYM+K+L +R + KQ+LYSF+
Sbjct: 67 LEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLYSFK 126
Query: 111 MVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGK-EGT 169
M E+ ++ + EF +I+ DLEN+ V + DED+AILLL ALPK+F+ KDT+ Y +
Sbjct: 127 MSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLKYSSGKSI 186
Query: 170 VTLEEVQAALRTKDLT--KSKDLTHEHGEGLSVT-------RGNGGGRGNRRKSGNKSRF 220
+TL+EV AA+ +K+L K EGL V +G G+G +K +K +
Sbjct: 187 LTLDEVAAAIYSKELELGSVKKSIKVQAEGLYVKDKNENKGKGEQKGKGKGKKGKSKKKP 246
Query: 221 ECFNCHKMGHFKKDCPKIN 239
C+ C + GHF+ CP N
Sbjct: 247 GCWTCGEEGHFRSSCPNQN 265
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 157 bits (397), Expect = 6e-39
Identities = 94/259 (36%), Positives = 145/259 (55%), Gaps = 30/259 (11%)
Query: 6 DIEKFTGDNDFGLWKVKM----------------EAVLIQQKCEKALKGEGSLPVTMSRA 49
++EKF GD D+ LWK K+ E +++ + G P T +
Sbjct: 7 EVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDPETATSK 66
Query: 50 EKTKMVD----KARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQ 105
+ K++ KARS ++L LG+ VLR+V K+ TAA M L+ L+M KSL +R +LKQ+
Sbjct: 67 LEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNRIYLKQR 126
Query: 106 LYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYG 165
LY ++M E+ + E + +F K++ DLEN++V + DED+AI+LL +LP+ F+ K+T+ Y
Sbjct: 127 LYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLKETLKYC 186
Query: 166 KEGTVTLEEVQAALRTK--DLTKSKDLTHEHGEGL-------SVTRGNGGGRGNRRKSGN 216
K T+ LEE+ +A+R+K +L S L + +GL S TRG G + R
Sbjct: 187 KT-TLHLEEITSAIRSKILELGASGKLLKNNSDGLFVQDRGRSETRGKGPNKNKSRSKSK 245
Query: 217 KSRFECFNCHKMGHFKKDC 235
+ C+ C K GHFKK C
Sbjct: 246 GAGKTCWICGKEGHFKKQC 264
>At3g45520 copia-like polyprotein
Length = 1363
Score = 154 bits (390), Expect = 4e-38
Identities = 100/273 (36%), Positives = 152/273 (55%), Gaps = 27/273 (9%)
Query: 2 GSKWDIEKFTGDNDFGLWKVKM---------EAVLIQQKCEKALKGEGSLPVTMSRAEKT 52
G++ ++EKF G D+ +WK K+ AVL + + + + + E+
Sbjct: 3 GARIEVEKFDGRGDYTMWKEKLLAHIDMLGLSAVLRESETPMGKERDSEKSDEDEKEERE 62
Query: 53 KMVD------KARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQL 106
KM KARS +VL + D+VLR++ KE +AA+M L+ LYM+K+L +R +LKQ+L
Sbjct: 63 KMEAFEEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQKL 122
Query: 107 YSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGK 166
YSF+M E+ +I + EF I+ DLEN+ V + DED+AILLL +LPK F+ KDT+ Y
Sbjct: 123 YSFKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYSS 182
Query: 167 EGTV-TLEEVQAALRTKDLT--KSKDLTHEHGEGLSVT-------RGNGGGRGNRRKSGN 216
TV +L+EV AA+ +++L K EGL V R +G ++S +
Sbjct: 183 GKTVLSLDEVAAAIYSRELEFGSVKKSIKGQAEGLYVKDKAENRGRSEQKDKGKGKRSKS 242
Query: 217 KSRFECFNCHKMGHFKKDCPKINGNSAQIVSEG 249
KS+ C+ C + GH K CP N N Q ++G
Sbjct: 243 KSKRGCWICGEDGHLKSTCP--NKNKPQFKNQG 273
>At4g10460 putative retrotransposon
Length = 1230
Score = 150 bits (379), Expect = 7e-37
Identities = 95/256 (37%), Positives = 143/256 (55%), Gaps = 25/256 (9%)
Query: 6 DIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDK-------- 57
++EKF G D+ LWK K+ A + AL+ S+ + E+ K +K
Sbjct: 7 EMEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGDKEALME 66
Query: 58 -----ARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMV 112
ARS +VL + D+VLR+ KE TA SM L+ LYM+K+L +R +LKQ+LYS++M
Sbjct: 67 EKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKLYSYKMQ 126
Query: 113 ESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGK-EGTVT 171
E+ ++ + EF +++ DLEN V + DED+AILLL +LPK F+ KDT+ YG T++
Sbjct: 127 ENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLKDTLKYGSGRTTLS 186
Query: 172 LEEVQAALRTKDLT--KSKDLTHEHGEGLSV-TRGNGGGRGNRRKSGNKSRFE------- 221
++EV AA+ +K+L +K EGL V + G +++ GNK R
Sbjct: 187 VDEVVAAIYSKELELGSNKKSIRGQAEGLYVKDKPETRGMSEQKEKGNKGRSRSRSKGWK 246
Query: 222 -CFNCHKMGHFKKDCP 236
C+ C + GHFK CP
Sbjct: 247 GCWICGEEGHFKTSCP 262
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 146 bits (369), Expect = 1e-35
Identities = 100/263 (38%), Positives = 150/263 (57%), Gaps = 28/263 (10%)
Query: 6 DIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDK-------- 57
++EKF G D+ +WK K+ A L ALK E L ++ + T+ +K
Sbjct: 7 EVEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEEVLRREL 66
Query: 58 -------ARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFR 110
ARSA+VL + D+VLR++ KE +AA+M L+ LYM+K+L +R + KQ+LYSF+
Sbjct: 67 LEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQKLYSFK 126
Query: 111 MVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYG-KEGT 169
M E+ +I + EF +I+ DLEN V + DED+AILLL +LPK F+ +DT+ YG T
Sbjct: 127 MSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLRDTLKYGLGRVT 186
Query: 170 VTLEEVQAALRTKDLT--KSKDLTHEHGEGLSV-----TRGNGGGRG---NRRKSGNKSR 219
++L+EV AA+ +K+L +K EGL V TRG RG N +KS +KSR
Sbjct: 187 LSLDEVVAAIYSKELELGSNKKSIKGQAEGLFVKEKTETRGRTEQRGNNNNNKKSRSKSR 246
Query: 220 FE--CFNCHKMGHFKKDCPKING 240
+ C+ C + + + + NG
Sbjct: 247 SKKGCWICGESSNGSSNYSEANG 269
>At2g19840 copia-like retroelement pol polyprotein
Length = 1137
Score = 142 bits (357), Expect = 3e-34
Identities = 74/184 (40%), Positives = 117/184 (63%), Gaps = 4/184 (2%)
Query: 56 DKARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESK 115
+KA + + +GDKVLR + TAA WA L+ LY+ KSL +R +L+ ++Y++RM +SK
Sbjct: 42 EKAMDMIFINVGDKVLRNIENSKTAAEAWATLDKLYLVKSLPNRVYLQLKVYNYRMQDSK 101
Query: 116 AIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEV 175
+ E + EF K++ DL N+++Q+ DE +AIL+L ALP S++ K+T+ YG+EG + L++V
Sbjct: 102 TLEENVDEFQKMISDLNNLQIQVPDEVQAILILSALPDSYDMLKETLKYGREG-IKLDDV 160
Query: 176 QAALRTK--DLTKSKDLTHEHGEGLSVTRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKK 233
+A ++K +L S + GEGL V RG RG+ + + C+ C K GHFK+
Sbjct: 161 ISAAKSKELELRDSSGGSRPVGEGLYV-RGKSQARGSDGPKSTEGKKVCWICGKEGHFKR 219
Query: 234 DCPK 237
C K
Sbjct: 220 QCYK 223
>At1g37110
Length = 1356
Score = 132 bits (333), Expect = 2e-31
Identities = 87/283 (30%), Positives = 151/283 (52%), Gaps = 36/283 (12%)
Query: 6 DIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEG---SLPVTMSRAE------------ 50
+I+ F GD DF LWK++++A L + L ++P+T S A+
Sbjct: 9 EIKVFNGDRDFSLWKIRIQAQLGVLGLKDTLTDFSLTKTVPLTKSEAKQESGDGESSGTK 68
Query: 51 ------KTKMVDKARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQ 104
K + ++A++ ++ + D VL +V AT A +WA L YM SL +R + +
Sbjct: 69 EVPDPVKIEQSEQAKNIIINHISDVVLLKVNHYATTADLWATLNKKYMETSLPNRIYTQL 128
Query: 105 QLYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLY 164
+LYSF+MV + I + + EF +I+ +L ++E+Q+++E +AIL+L +LP S K T+ Y
Sbjct: 129 KLYSFKMVSTMTIDQNVDEFLRIVAELGSLEIQVDEEVQAILILNSLPASHIQLKHTLKY 188
Query: 165 GKEGTVTLEEVQAALRT--KDLTKSKDL---------THEHGEGLSVTRGNGGGRGNRRK 213
G + T+T+++V ++ ++ ++L ++ DL T E G L GG R +
Sbjct: 189 GNK-TLTVQDVTSSAKSLERELAEAVDLDKGQAAVLYTTERGRPLVRNNQKGGQGKGRSR 247
Query: 214 SGNKSRFECFNCHKMGHFKKDCPKINGNSAQIVSEGYEDAGAL 256
S +K++ C+ C K GH KKDC ++ SEG +AG +
Sbjct: 248 SNSKTKVPCWYCKKEGHVKKDC---YSRKKKMESEGQGEAGVI 287
>At4g21360 putative transposable element
Length = 1308
Score = 130 bits (328), Expect = 6e-31
Identities = 92/293 (31%), Positives = 159/293 (53%), Gaps = 44/293 (15%)
Query: 1 MGSKWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKAL---------------KGEGSLPVT 45
M +K +I+ F GD DF LWK+++EA L + AL K E
Sbjct: 3 MSTKVEIKTFNGDRDFSLWKIRIEAQLGVLGLKPALSDFTLTKTILVVKSEKKESESEDD 62
Query: 46 MSRAEKTKMV---------DKARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSL 96
+ ++KT+ V D+A++ ++ + D VL +V TAA +WA L L+M SL
Sbjct: 63 ETDSKKTEEVPDPIKFEQSDQAKNFIINHITDTVLLKVQHCVTAAELWATLNKLFMETSL 122
Query: 97 AHRQFLKQQLYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFE 156
+R + + +LYSF+MV++ +I + EF +I+ +L ++++Q+ +E +AIL+L +LP S+
Sbjct: 123 PNRIYTQLRLYSFKMVDNLSIDQNTDEFLRIVAELGSLQIQVGEEVQAILILNSLPPSYI 182
Query: 157 SFKDTMLYGKEGTVTLEEVQAALRT--KDLTKSKD----------LTHEHGEGLSV-TRG 203
K T+ YG + T+++++V ++ ++ ++L++ K+ T E G + T+G
Sbjct: 183 QLKHTLKYGNK-TLSVQDVVSSAKSLERELSEQKETIRAPASTALYTAERGRPQTKNTQG 241
Query: 204 NGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKINGNSAQIVSEGYEDAGAL 256
G GRG +S +KSR C+ C K GH KKDC ++ +EG AG +
Sbjct: 242 QGKGRG---RSNSKSRLTCWFCKKEGHVKKDC---YAGKRKLENEGQGKAGVI 288
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 117 bits (294), Expect = 5e-27
Identities = 68/191 (35%), Positives = 107/191 (55%), Gaps = 10/191 (5%)
Query: 56 DKARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESK 115
DKA++ + L + DKVLR++ TAA W L+ L+M +SL HR + + Y+F+M E+K
Sbjct: 47 DKAKNVIFLNVADKVLRKIELCKTAAEAWETLDRLFMIRSLPHRVYTQLSFYTFKMQENK 106
Query: 116 AIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLY-GKEGTVTLEE 174
I E + +F KI+ DL ++++ + DE +AILLL +LP ++ +TM Y + L++
Sbjct: 107 KIDENIDDFLKIVADLNHLQIDVTDEVQAILLLSSLPARYDGLVETMKYSNSREKLRLDD 166
Query: 175 VQAALRTKDLTKSKD--------LTHEHGEGLSVTRGNGGGRGNRRKSGNKSRFECFNCH 226
V A R K+ S++ +G + +GN G +R KS + R C+ C
Sbjct: 167 VMVAARDKERELSQNNRPVVEGHFARGRPDGKNNNQGNKGKNRSRSKSADGKRV-CWICG 225
Query: 227 KMGHFKKDCPK 237
K GHFKK C K
Sbjct: 226 KEGHFKKQCYK 236
>At1g47360 polyprotein, putative
Length = 1182
Score = 107 bits (266), Expect = 9e-24
Identities = 73/240 (30%), Positives = 114/240 (47%), Gaps = 28/240 (11%)
Query: 7 IEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSAVVLCL 66
I F DF LWK ++ A L + + G+ P+T E + + LC
Sbjct: 6 IAVFDESGDFSLWKTRIMAHLSVIGLKDVVIGKTITPLTAEEEEDPE------KKIELC- 58
Query: 67 GDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTEFNK 126
TA W L+ L+M +SL HR + + Y+F+M E+K I E + +F K
Sbjct: 59 -----------QTAQEAWETLDRLFMIRSLPHRVYTQLSFYTFKMQENKKIDENIDDFLK 107
Query: 127 ILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLY-GKEGTVTLEEVQAALRTKDLT 185
I+ DL ++++++ DE +AILLL +LP ++ +TM Y + L++V R K+
Sbjct: 108 IVADLNHLQIEVTDEVQAILLLSSLPARYDGLVETMKYSNSREKLRLDDVMVGARDKERE 167
Query: 186 KSKD--------LTHEHGEGLSVTRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPK 237
S++ EG + +GN G +R KS + R C+ C K GHFKK C K
Sbjct: 168 LSQNNRPVAEGHYARGRPEGKNNNQGNKGKNRSRSKSADGKRV-CWICGKEGHFKKQCYK 226
>At4g04380 putative polyprotein
Length = 778
Score = 95.9 bits (237), Expect = 2e-20
Identities = 57/129 (44%), Positives = 84/129 (64%), Gaps = 14/129 (10%)
Query: 57 KARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKA 116
K RS +VL + D+VL EA+ + LYM+K+L ++ +LKQ+LY F+M E+ +
Sbjct: 55 KTRSTIVLSVSDRVL-----EAS--------DRLYMSKALPNQIYLKQKLYRFKMSENLS 101
Query: 117 IMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTV-TLEEV 175
+ + EF I+ DLEN+ V + DED+ ILLL +LPKSF+ KDT+ Y TV TL+EV
Sbjct: 102 MEGNIDEFLHIVADLENLNVLVSDEDQTILLLMSLPKSFDQLKDTLQYSSGKTVLTLDEV 161
Query: 176 QAALRTKDL 184
AA+ +K+L
Sbjct: 162 TAAIYSKEL 170
>At2g15700 copia-like retroelement pol polyprotein
Length = 1166
Score = 91.3 bits (225), Expect = 5e-19
Identities = 69/217 (31%), Positives = 116/217 (52%), Gaps = 35/217 (16%)
Query: 6 DIEKFTGDNDFGLWKVKMEA--------------------VLIQQKCEKALK----GEGS 41
++++F G DF LWKV+M A +I+++ A + G+
Sbjct: 8 EVDRFDGTGDFSLWKVRMLAHYGVLGLKGILNDEQLLRDPPVIEEEAAVAGRDYHVGDFE 67
Query: 42 LP--VTMSRAEKTKMVDKARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHR 99
LP V + + EK+ +KA+ +VL +G++VLR++ TAA+MW+ L+ LYM SL +R
Sbjct: 68 LPSNVDLIKFEKS---EKAKDLIVLNVGNQVLRKIKNCETAAAMWSTLKRLYMETSLPNR 124
Query: 100 QFLKQQLYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFK 159
+L+ + Y+++M +S++I + +F K++ DL NI V + +E +AILLL +L E+ +
Sbjct: 125 IYLQLKFYTYKMTDSRSIDGNVDDFLKLVTDLNNIGVDVTEEVQAILLLSSLYDRKENSQ 184
Query: 160 DTMLYGKEGTVTLEEVQAALRTKDLTKSKDLTHEHGE 196
EG L Q A + + L K K HGE
Sbjct: 185 KVESSQSEG---LYVEQEADQRRGLKKEK---ANHGE 215
>At3g55300 putative protein
Length = 262
Score = 88.2 bits (217), Expect = 4e-18
Identities = 59/181 (32%), Positives = 96/181 (52%), Gaps = 20/181 (11%)
Query: 88 ESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILL 147
E Y TK+LA+ +LK + SF+MVE+K+I E L K++ DL ++++ DED+A+
Sbjct: 37 EGNYQTKTLANMIYLKHKFTSFKMVENKSIEENLDTSLKLVADLASLDINTSDEDQALQF 96
Query: 148 LCALPKSFESFKDTMLYGK-EGTVTLEEVQAALRTKDL-TKSKDLTH----EHGEGLSVT 201
+ LP ++S DT+ YG + T+TL V ++ +K++ K K L + + L
Sbjct: 97 MAGLPPQYDSLVDTLKYGSGKDTLTLNAVISSAYSKEIELKEKGLLNVSKPDSEALLGDF 156
Query: 202 RGNGGGRGNRR-----KSGN-------KSRFECFNCHKMGHFKKDCP--KINGNSAQIVS 247
+G +GN+R SGN + CF C K H+K++CP K + A I +
Sbjct: 157 KGRSNNKGNKRPCTGSSSGNGKFQSRPSNNTSCFICGKEDHWKRECPQRKKHAELANIAT 216
Query: 248 E 248
E
Sbjct: 217 E 217
>At4g04280 putative transposon protein
Length = 1104
Score = 85.5 bits (210), Expect = 3e-17
Identities = 69/260 (26%), Positives = 119/260 (45%), Gaps = 44/260 (16%)
Query: 4 KWDIEKFTGDNDFGLWKVKMEAVL-------------------IQQKCEKALKGEGSLPV 44
K +I+ F GD DF WK+++EA L + +K EK + E
Sbjct: 2 KVEIKTFNGDRDFSFWKIRIEAQLGVLGLKNSLTDFKLTKTVPVAKKEEKESEYEDDASD 61
Query: 45 TMSRAE-----KTKMVDKARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHR 99
E K + ++A++ ++ + D VL +V TAA +WA L L+M SL +R
Sbjct: 62 IKQATEEPDPIKFEQSEQAKNFIINHITDTVLLKVQHCKTAAEIWATLNKLFMETSLLNR 121
Query: 100 QFLKQQLYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFK 159
+ + +LYSF+MV++ +I + + EF +IL D + + LP S+ K
Sbjct: 122 IYTQLKLYSFKMVDTLSIDQNVDEFLRILADCLDQNL--------------LPSSYIQLK 167
Query: 160 DTMLYGKEGTVTLEEVQAAL-RTKDLTKSKDL---THEHGEGLSVTRGNGGGRGNRRKSG 215
T+ + + + ++ L T++ K+ + T + G + G NR +S
Sbjct: 168 HTL--NTDVVSSAKSLERKLSETQESNKNVSMALYTTDRGRPQVRNQDKQGQGKNRGRSN 225
Query: 216 NKSRFECFNCHKMGHFKKDC 235
+K+R C+ C K H K+DC
Sbjct: 226 SKTRVTCWFCKKEAHVKRDC 245
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 69.3 bits (168), Expect = 2e-12
Identities = 51/210 (24%), Positives = 94/210 (44%), Gaps = 24/210 (11%)
Query: 49 AEKTKMVDKARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYS 108
AEKT K ++ + + +L+ + ++ T+ +W ++ Y L++ S
Sbjct: 19 AEKTVKDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQGNDRVQSAQLQRLRRS 78
Query: 109 FRMVESKAIMEQLTEF----NKILDDLENIEVQLEDE-----------DKAILLLCALPK 153
F ++E K I E +T + +I +D+ N+ + D +K ++CA+ +
Sbjct: 79 FEVLEMK-IGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTLVEKFTYVVCAIEE 137
Query: 154 SFESFKDTMLYGKEGTVTLEE-------VQAALRTKDLTKSKDLTHEHGEGLSVTRGNGG 206
S + K+ + G + ++ + E V+ + + D G S RG GG
Sbjct: 138 S-NNIKELTVDGLQSSLMVHEQNLSRHDVEERVLKAETQWRPDGGRGRGGSPSRGRGRGG 196
Query: 207 GRGNRRKSGNKSRFECFNCHKMGHFKKDCP 236
+G R N+ ECF CHKMGH+K +CP
Sbjct: 197 YQGRGRGYVNRDTVECFKCHKMGHYKAECP 226
>At3g60170 putative protein
Length = 1339
Score = 66.6 bits (161), Expect = 1e-11
Identities = 62/271 (22%), Positives = 123/271 (44%), Gaps = 31/271 (11%)
Query: 7 IEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGE------GSLPVT---MSRAEKTKMVD- 56
I +F G DF W + ME L ++ + ++ G+ PV+ S E+ K+ D
Sbjct: 12 IPRFDGYYDF--WSMTMENFLRSRELWRLVEEGIPAIVVGTTPVSEAQRSAVEEAKLKDL 69
Query: 57 KARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHR---QFLKQQLYSFRMVE 113
K ++ + + ++L + ++T+ ++W ++ Y + R Q L+++ M E
Sbjct: 70 KVKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEFELLAMKE 129
Query: 114 SKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKE-GTVTL 172
+ I L +++ ++ +E +L +L F ++ + T+++
Sbjct: 130 GEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEESNDLSTLSI 189
Query: 173 EEVQAALRTKDLT--------KSKDLTHEH----GEGLSVTRGNGG-GRGNRRKSGNKSR 219
+E+ +L + ++ +THE G G V RG+ G GRG R N++
Sbjct: 190 DELHGSLLVHEQRLNGHVQEEQALKVTHEERPSQGRGRGVFRGSRGRGRGRGRSGTNRAI 249
Query: 220 FECFNCHKMGHFKKDCP--KINGNSAQIVSE 248
EC+ CH +GHF+ +CP + N N A++ E
Sbjct: 250 VECYKCHNLGHFQYECPEWEKNANYAELEEE 280
>At1g42380 hypothetical protein
Length = 143
Score = 64.3 bits (155), Expect = 7e-11
Identities = 42/135 (31%), Positives = 69/135 (51%), Gaps = 8/135 (5%)
Query: 111 MVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLY-GKEGT 169
M E+K I E + +F KI+ DL +I++++ DE +AILLL +LP ++ +TM Y
Sbjct: 1 MQENKKIDENIDDFLKIVVDLNHIQIEVRDEVQAILLLSSLPSRYDGLVETMKYSNSREK 60
Query: 170 VTLEEVQAALRTKDLTKSKD----LTHEHGEGLSVTRGNGGGRGNRRKSGNKS---RFEC 222
+ L +V A R K+ S++ + V + N G + +S +KS + C
Sbjct: 61 LRLNDVMVAARDKEREMSQNNRLIAEGHYARRRPVGKNNNQGNKGKNRSWSKSADGKRVC 120
Query: 223 FNCHKMGHFKKDCPK 237
+ C K HF + C K
Sbjct: 121 WICGKEKHFNEQCYK 135
>At4g04600 putative polyprotein
Length = 922
Score = 60.8 bits (146), Expect = 7e-10
Identities = 42/138 (30%), Positives = 64/138 (45%), Gaps = 26/138 (18%)
Query: 6 DIEKFTGDNDFGLWKVKMEAVL------------IQQKCEKALKGEGSLPVTMSRAEKTK 53
+I+ F GDNDF LWK+++ A L K E K E + + +
Sbjct: 8 EIKAFDGDNDFSLWKMRIMAQLGVLGLKGTLTDFALTKTEPLTKSEEKQVASGDESSDSS 67
Query: 54 MV--------------DKARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHR 99
V ++A + ++ + D VLR+V TAA++W L LYM SL +R
Sbjct: 68 AVLTKEVPDPIKIEKSEQAINIIINHISDTVLRKVNHYKTAATLWELLNELYMETSLPNR 127
Query: 100 QFLKQQLYSFRMVESKAI 117
+ + + YSFRM+ SK I
Sbjct: 128 IYAQLKFYSFRMMTSKTI 145
>At3g29782 hypothetical protein
Length = 1130
Score = 51.2 bits (121), Expect = 6e-07
Identities = 40/174 (22%), Positives = 81/174 (45%), Gaps = 9/174 (5%)
Query: 10 FTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSL-PVTMSRAEKTKMVDKARSAVVLCLG- 67
F G + F W+ KM L+ E+ L + + P + +D C G
Sbjct: 71 FDGKSGFKTWQEKMRYYLVSINMERYLPEDPPIVPQGTTYVYTVGGMDTWAQGDYCCKGL 130
Query: 68 --DKVLREV----AKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQL 121
+++L ++ +K ++ ++W LE+ Y T ++F + +F+MV+SK IMEQ+
Sbjct: 131 ILNRLLNDLFDLYSKAKSSKTLWLTLENKYKTDESRMQRFSTAKFLNFKMVDSKPIMEQV 190
Query: 122 TEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEV 175
+I ++E + + + K L+ LP + FK+ + + K +T +++
Sbjct: 191 EALQRICQEIELEGMSICNVFKTNCLIEKLPPGWSDFKNYLNF-KRKAMTFDDL 243
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,200,248
Number of Sequences: 26719
Number of extensions: 259126
Number of successful extensions: 1341
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 79
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 1139
Number of HSP's gapped (non-prelim): 200
length of query: 276
length of database: 11,318,596
effective HSP length: 98
effective length of query: 178
effective length of database: 8,700,134
effective search space: 1548623852
effective search space used: 1548623852
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC143337.6


