

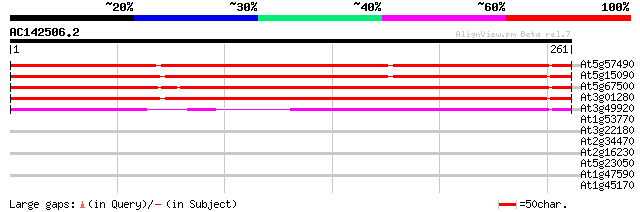
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142506.2 + phase: 1 /partial
(261 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g57490 porin-like protein 287 3e-78
At5g15090 voltage-dependent anion-selective channel protein hsr2 211 2e-55
At5g67500 porin-like protein 211 3e-55
At3g01280 putative porin 204 3e-53
At3g49920 porin - like protein 124 5e-29
At1g53770 unknown protein 36 0.024
At3g22180 hypothetical protein 30 1.7
At2g34470 putative urease accessory protein 29 2.2
At2g16230 putative beta-1,3-glucanase 29 2.9
At5g23050 acetyl-CoA synthetase-like protein 28 5.0
At1g47590 hypothetical protein 27 8.5
At1g45170 unknown protein 27 8.5
>At5g57490 porin-like protein
Length = 274
Score = 287 bits (735), Expect = 3e-78
Identities = 150/261 (57%), Positives = 189/261 (71%), Gaps = 5/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL KDY FDHKF+L++ S+TG ATGLK+D FF GD++TLYK N VD+K+++ S+V
Sbjct: 18 LLNKDYIFDHKFTLTMLSATGTEFVATGLKKDDFFFGDISTLYKGQNTIVDLKIDSHSSV 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
STKVTL + L S K +SF IPDHKSGKLDVQY+HPHA ++SSIGLNP P L+LSA I
Sbjct: 78 STKVTLKN--LLPSAKAVISFKIPDHKSGKLDVQYVHPHATLNSSIGLNPTPLLDLSATI 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
GS+++ +G EV FDT S+S + YNAGI FN SA L+L DKG+SL+A+Y+H V+
Sbjct: 136 GSQNVCLGGEVSFDTASSSLTKYNAGIGFNNQGVSAALILEDKGESLRATYVHTVN--PT 193
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
+ AEL + S+ N FT G+S S+D TV+KTRFS+ GKA QR WRP S IT SA
Sbjct: 194 TSFGAELIRRFSNYNNSFTVGSSHSVDQFTVVKTRFSNSGKAGMVVQREWRPKSHITFSA 253
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
EYDSK + SP K GLAL+LK
Sbjct: 254 EYDSKAVTSSP-KLGLALALK 273
>At5g15090 voltage-dependent anion-selective channel protein hsr2
Length = 274
Score = 211 bits (538), Expect = 2e-55
Identities = 111/261 (42%), Positives = 159/261 (60%), Gaps = 5/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLY+DY D KFS++ SSTG+ +T TG + F+GD+ T K+ N T DVKV+TDS++
Sbjct: 18 LLYRDYQGDQKFSVTTYSSTGVAITTTGTNKGSLFLGDVATQVKNNNFTADVKVSTDSSL 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T ++ + KV + +PDHKSGK +VQY H +A I +S+G P + S +
Sbjct: 78 LTTLTFDEPA--PGLKVIVQAKLPDHKSGKAEVQYFHDYAGISTSVGFTATPIVNFSGVV 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ +S+G +V ++T S +F +NAG F K D +A+L+L DKG+ L ASY V
Sbjct: 136 GTNGLSLGTDVAYNTESGNFKHFNAGFNFTKDDLTASLILNDKGEKLNASYYQIVS--PS 193
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AE++H ++ EN T GT ++DP T +K R ++ G A Q WRP S T+S
Sbjct: 194 TVVGAEISHNFTTKENAITVGTQHALDPLTTVKARVNNAGVANALIQHEWRPKSFFTVSG 253
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E DSK I S AK G+AL+LK
Sbjct: 254 EVDSKAIDKS-AKVGIALALK 273
>At5g67500 porin-like protein
Length = 276
Score = 211 bits (537), Expect = 3e-55
Identities = 113/261 (43%), Positives = 164/261 (62%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL +DYN D KFS+S S++G+ LT+T LK+ D+ T YK N DVK++TDS+V
Sbjct: 18 LLTRDYNSDQKFSISTYSASGVALTSTALKKGGVHAADVATQYKYKNALFDVKIDTDSSV 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T VTL ++ L +K +A SF +PD+ S KL+VQY H HA + ++ L P ++++A +
Sbjct: 78 LTTVTLTEI-LPSTKAIA-SFKVPDYNSAKLEVQYFHDHATVTAAAALKQNPLIDITATL 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
GS IS GAE G+DTTS +F+ YNAGI+ KPD +++L DKG SLKASY+H+ D
Sbjct: 136 GSPVISFGAEAGYDTTSKTFTKYNAGISVTKPDACLSIILGDKGDSLKASYLHHFDEFKR 195
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
E+ K S++EN T G +ID T +K + ++ G Q P SL+T+S+
Sbjct: 196 TAAVGEVYRKFSTNENTITVGGLYAIDHSTAVKAKLNNHGTLGALLQHEVLPRSLVTVSS 255
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K + P +FGL+L+LK
Sbjct: 256 EIDTKALEKHP-RFGLSLALK 275
>At3g01280 putative porin
Length = 276
Score = 204 bits (520), Expect = 3e-53
Identities = 110/261 (42%), Positives = 158/261 (60%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLYKD+N D KFS++ S G+ +T+TG K+ +GD+ + N+T D+KV TDS
Sbjct: 18 LLYKDHNSDQKFSITTFSPAGVAITSTGTKKGDLLLGDVAFQSRRKNITTDLKVCTDSTF 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T+++ + + SF +PD SGK+++QYLH +A I +S+GL P + S I
Sbjct: 78 LITATVDEAA--PGLRSIFSFKVPDQNSGKVELQYLHEYAGISTSMGLTQNPTVNFSGVI 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
GS +++G +V FDT S +F+ NAG++F K D A+L + DKG L ASY H V+
Sbjct: 136 GSNVLAVGTDVSFDTKSGNFTKINAGLSFTKEDLIASLTVNDKGDLLNASYYHIVNPLFN 195
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AE++HKLSS ++ T GT S+DP T +K R + G A+ Q W+P S T+S
Sbjct: 196 TAVGAEVSHKLSSKDSTITVGTQHSLDPLTSVKARVNSAGIASALIQHEWKPKSFFTISG 255
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S AK GLAL+LK
Sbjct: 256 EVDTKSIDKS-AKVGLALALK 275
>At3g49920 porin - like protein
Length = 226
Score = 124 bits (311), Expect = 5e-29
Identities = 81/261 (31%), Positives = 123/261 (47%), Gaps = 53/261 (20%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL +DY+ D KFS+S S +G+ LT+T LK ++ T YK N DVK++TD NV
Sbjct: 18 LLTRDYSTDQKFSISTNSVSGVALTSTALKNGVLHAANVATQYKYRNTFFDVKIDTDFNV 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
+ V P +K +D L
Sbjct: 78 KSLV------------------YPMNKFVSIDHNTL------------------------ 95
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+DTTS +F+ YN G++ KPD +++L DKG S+KASY++Y+D
Sbjct: 96 ----------TGYDTTSRTFTKYNVGVSVTKPDQCVSIILGDKGDSIKASYVYYLDESTR 145
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
E+ K+S++E T G ++D T +K + + +GK Q P S++T+S
Sbjct: 146 SATVGEVIRKISTNETTVTVGGLYAVDHLTNVKAKLNSNGKFGALLQHEGLPKSIVTISG 205
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K + P + GL+LSLK
Sbjct: 206 EIDTKTLDKYP-RLGLSLSLK 225
>At1g53770 unknown protein
Length = 563
Score = 35.8 bits (81), Expect = 0.024
Identities = 26/98 (26%), Positives = 47/98 (47%), Gaps = 9/98 (9%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
L+Y +F + FS+ +P+S T TG + DL LY D+ + V
Sbjct: 83 LIYLSTDFSNYFSVKVPNSAFRSNTLTG----RVHESDLQALYLLRKQESDLFSIWNHTV 138
Query: 61 STKVTLNDVSLSHSKKVALSFNIPD-----HKSGKLDV 93
S T++DV + ++++L+ I + HK+G +D+
Sbjct: 139 SNLSTIDDVKSAVFRQISLNRQIQNALLSPHKTGNVDI 176
>At3g22180 hypothetical protein
Length = 706
Score = 29.6 bits (65), Expect = 1.7
Identities = 42/200 (21%), Positives = 84/200 (42%), Gaps = 30/200 (15%)
Query: 40 NTLYKSGNVTVDVKVNTDSNV--STKVTLNDV--SLSHSKKVALSFNIPDHKSGKLDVQY 95
N L G V++ V+TD+NV S ++ ND+ SLS + + ++ +G +
Sbjct: 437 NDLSSIGTVSIISSVSTDANVAASKEIRNNDLRSSLSRNSFAPSQGSRDEYDTGSHGMSN 496
Query: 96 LHPHAAIDSSIGLNPAPKLELSAAIGSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFS 155
L + + S+ L P P + +G++ + T+ ST++ + D
Sbjct: 497 LSSPSHVHESVTLAPLP--QNPTIVGNRFTA--------TSHHMHSTFDDKVLHRGNDAD 546
Query: 156 ATLMLADKGQSL----KASYI------HYVDRPDGLTVAAELAHKLSSSENRFTFGTSQS 205
+ A L K S + YV P +T +E+ ++L + ++ T+ +
Sbjct: 547 PLFLFAPATSHLRDVRKTSVVWDPEAGRYVSAP--VTTTSEVRNRLLNPSSQ----TAST 600
Query: 206 IDPKTVLKTRFSDDGKAAFQ 225
+P+ +L S G +A +
Sbjct: 601 QNPRPILPAHDSSSGSSALR 620
>At2g34470 putative urease accessory protein
Length = 275
Score = 29.3 bits (64), Expect = 2.2
Identities = 43/161 (26%), Positives = 59/161 (35%), Gaps = 24/161 (14%)
Query: 5 DYNFDHK-FSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNVSTK 63
D NF + F++ I G G TA L +F L Y VT D+ D K
Sbjct: 66 DRNFSERAFTVGIGGPVGTGKTALMLALCRF----LRDKYSLAAVTNDIFTKEDGEFLVK 121
Query: 64 VTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAIGSK 123
+ +P+ + ++ PHAAI I +N P ELS K
Sbjct: 122 ----------------NGALPEERIRAVETGGC-PHAAIREDISINLGPLEELSNLF--K 162
Query: 124 DISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKG 164
+ E G D +A+FS A D SA + KG
Sbjct: 163 ADLLLCESGGDNLAANFSRELADYIIYIIDVSAGDKIPRKG 203
>At2g16230 putative beta-1,3-glucanase
Length = 456
Score = 28.9 bits (63), Expect = 2.9
Identities = 32/123 (26%), Positives = 52/123 (42%), Gaps = 12/123 (9%)
Query: 27 TGLKRDKFFVGDLNTLYKS----GNVTVDVKVNTDSNVSTKVTLNDVSLSHSKKVALSFN 82
TG+K F ++ ++ + G V+V V ST + N+V S A + N
Sbjct: 230 TGIKYMNMFDAQVDAVHSALKSIGFEKVEVLVAETGWPSTGDS-NEVGPSVENAKAYNGN 288
Query: 83 IPDHKSGKLDVQYLHPHAAIDSSI------GLNPAPKLELSAAIGSKDISMGAEVGFDTT 136
+ H + L P +ID+ I L P P E S + D+SM ++G T
Sbjct: 289 LIAHLRSMVGTP-LMPGKSIDTYIFALFDENLKPGPSFEQSFGLFKPDLSMAYDIGLTKT 347
Query: 137 SAS 139
++S
Sbjct: 348 TSS 350
>At5g23050 acetyl-CoA synthetase-like protein
Length = 668
Score = 28.1 bits (61), Expect = 5.0
Identities = 19/52 (36%), Positives = 26/52 (49%)
Query: 26 ATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNVSTKVTLNDVSLSHSKKV 77
A K +F DLN L KS N + K+N VS+ VTL + + + KV
Sbjct: 599 AVVFKSPEFSNPDLNLLKKSFNSEIQKKLNPLFKVSSVVTLPSLPRTATNKV 650
>At1g47590 hypothetical protein
Length = 341
Score = 27.3 bits (59), Expect = 8.5
Identities = 11/28 (39%), Positives = 19/28 (67%)
Query: 51 DVKVNTDSNVSTKVTLNDVSLSHSKKVA 78
+++ +TDSN+ +T N +L+H K VA
Sbjct: 63 ELRDHTDSNIVIMITRNKANLNHLKSVA 90
>At1g45170 unknown protein
Length = 212
Score = 27.3 bits (59), Expect = 8.5
Identities = 32/149 (21%), Positives = 64/149 (42%), Gaps = 26/149 (17%)
Query: 43 YKSGNVTVDVKVNTDSNVSTKVTLNDVSLSHSKK--VALSFNIPDHK------------S 88
+ +GN + + TD++ + N +SL+ K + +N+P
Sbjct: 22 FNAGNAKLRATM-TDASFVAGPSFNGLSLAVEKPGFFIIDYNVPKKDVRFQFMNTIRIAE 80
Query: 89 GKLDVQYLHPHA----AIDSSIGLNPAPKLELSAAIGSKDISMGAEVGFDTTSASFSTYN 144
L++ Y+H +D S ++PA KL + +G+K+ ++ + +T+
Sbjct: 81 KPLNLTYIHMRGDNRTIVDGSFVIDPANKLSANYMVGTKN----CKLKYTYVHGGIATFE 136
Query: 145 A--GIAFNKPDFSATLMLADKGQSLKASY 171
+A N DF+ + L G +LKA+Y
Sbjct: 137 PCYDVAKNMWDFAISHKLYG-GDNLKATY 164
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.131 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,552,647
Number of Sequences: 26719
Number of extensions: 220199
Number of successful extensions: 412
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 397
Number of HSP's gapped (non-prelim): 14
length of query: 261
length of database: 11,318,596
effective HSP length: 97
effective length of query: 164
effective length of database: 8,726,853
effective search space: 1431203892
effective search space used: 1431203892
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC142506.2


