

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142498.7 + phase: 0
(1705 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
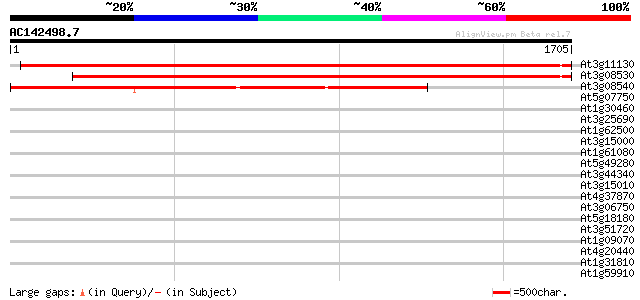
At3g11130 hypothetical protein 3097 0.0
At3g08530 hypothetical protein 2788 0.0
At3g08540 putative clathrin heavy chain, 3' partial 2268 0.0
At5g07750 putative protein 41 0.005
At1g30460 hypothetical protein 40 0.009
At3g25690 unknown protein 40 0.015
At1g62500 putative proline-rich cell wall protein (pir|IS52985);... 39 0.034
At3g15000 unknown protein 37 0.075
At1g61080 hypothetical protein 37 0.075
At5g49280 predicted GPI-anchored protein 37 0.13
At3g44340 putative protein 37 0.13
At3g15010 RNA-binding protein 37 0.13
At4g37870 phosphoenolpyruvate carboxykinase (ATP) -like protein 36 0.17
At3g06750 predicted GPI-anchored protein 36 0.17
At5g18180 putative protein 35 0.29
At3g51720 unknown protein 35 0.37
At1g09070 unknown protein 35 0.37
At4g20440 putative snRNP protein 34 0.64
At1g31810 hypothetical protein 34 0.83
At1g59910 hypothetical protein 33 1.1
>At3g11130 hypothetical protein
Length = 1673
Score = 3097 bits (8030), Expect = 0.0
Identities = 1540/1676 (91%), Positives = 1624/1676 (96%), Gaps = 7/1676 (0%)
Query: 34 MESDKYICVRETAPQNSVVIVDMNMPNQPLRRPITADSALMNPNSRILALKAQLQGTTQD 93
MESDKYICVRETAPQNSVVI+DMNMP QPLRRPITADSALMNPNSRILALKAQ+ GTTQD
Sbjct: 1 MESDKYICVRETAPQNSVVIIDMNMPMQPLRRPITADSALMNPNSRILALKAQVPGTTQD 60
Query: 94 HLQIFNIELKAKMKSYQMPEQVVFWKWISPKLLGLVTQTSVYHWSIEGDSEPVKMFERTA 153
HLQIFNIE KAK+KS+QMPEQV FWKWI+PK+LGLVTQTSVYHWSIEGDSEPVKMF+RTA
Sbjct: 61 HLQIFNIEAKAKLKSHQMPEQVAFWKWITPKMLGLVTQTSVYHWSIEGDSEPVKMFDRTA 120
Query: 154 NLANNQIINYRCDPTEKWLVLIGIAPGSPERPQLVKGNMQLFSVEQQRSQALEAHAASFA 213
NLANNQIINY+C P EKWLVLIGIAPGSPERPQLVKGNMQLFSV+QQRSQALEAHAASFA
Sbjct: 121 NLANNQIINYKCSPNEKWLVLIGIAPGSPERPQLVKGNMQLFSVDQQRSQALEAHAASFA 180
Query: 214 QFKVPGNENPSTLISFATKTVNAGQVISKLHVIELGAQPGKPSFTKKQADLFFPPDFADD 273
QFKVPGNENPS LISFA+K+ NAGQ+ SKLHVIELGAQPGKPSFTKKQADLFFPPDFADD
Sbjct: 181 QFKVPGNENPSILISFASKSFNAGQITSKLHVIELGAQPGKPSFTKKQADLFFPPDFADD 240
Query: 274 FPVSMQISHKYSLIYVITKLGLLFVYDLETATAVYRNRISPDPIFLTSEATSVGGFYAIN 333
FPV+MQ+SHK++LIYVITKLGLLFVYDLETA+A+YRNRISPDPIFLTSEA+SVGGFYAIN
Sbjct: 241 FPVAMQVSHKFNLIYVITKLGLLFVYDLETASAIYRNRISPDPIFLTSEASSVGGFYAIN 300
Query: 334 RRGQVLLATVNEQTIVNFVSGQLNNLELAVNLAKRGNLPGAEKLVVERFHELFAQTKYKE 393
RRGQVLLATVNE TI+ F+SGQLNNLELAVNLAKRGNLPGAE LVV+RF ELFAQTKYKE
Sbjct: 301 RRGQVLLATVNEATIIPFISGQLNNLELAVNLAKRGNLPGAENLVVQRFQELFAQTKYKE 360
Query: 394 AAELAAESPQGILRTPDTVAKFQSVPVQAGQTPPLLQYFGTLLTRGKLNAFESLELSRLV 453
AAELAAESPQGILRTPDTVAKFQSVPVQAGQTPPLLQYFGTLLTRGKLN++ESLELSRLV
Sbjct: 361 AAELAAESPQGILRTPDTVAKFQSVPVQAGQTPPLLQYFGTLLTRGKLNSYESLELSRLV 420
Query: 454 VNQNKKNLLENWLAEDKLECSEELGDLVKTVDNDLALKIYIKARATPKVVAAFAEKREFD 513
VNQNKKNLLENWLAEDKLECSEELGDLVKTVDNDLALKIYIKARATPKVVAAFAE+REFD
Sbjct: 421 VNQNKKNLLENWLAEDKLECSEELGDLVKTVDNDLALKIYIKARATPKVVAAFAERREFD 480
Query: 514 KILIYSKQVGYTPDYLFLLQSILRTDPQGAVNFALMMSQMEGGCPVDYNTITDLFLQRNL 573
KILIYSKQVGYTPDY+FLLQ+ILRTDPQGAVNFALMMSQMEGGCPVDYNTITDLFLQRNL
Sbjct: 481 KILIYSKQVGYTPDYMFLLQTILRTDPQGAVNFALMMSQMEGGCPVDYNTITDLFLQRNL 540
Query: 574 IREATAFLLDVLKPNLPEHGFLQTKVLEINLVTFPNVADAILANGMFSHYDRPRIAQLCE 633
IREATAFLLDVLKPNLPEH FLQTKVLEINLVTFPNVADAILANGMFSHYDRPR+AQLCE
Sbjct: 541 IREATAFLLDVLKPNLPEHAFLQTKVLEINLVTFPNVADAILANGMFSHYDRPRVAQLCE 600
Query: 634 KAGLYVRALQHYTELPDIKRVIVNTHAIEPQALVEFFGTLSKEWALECMKDLLLVNLRGN 693
KAGLY+++L+HY+ELPDIKRVIVNTHAIEPQALVEFFGTLS EWA+ECMKDLLLVNLRGN
Sbjct: 601 KAGLYIQSLKHYSELPDIKRVIVNTHAIEPQALVEFFGTLSSEWAMECMKDLLLVNLRGN 660
Query: 694 LQIIVQVAKEYCEQLGVDACIKIFEQFRSYEGLYFFLGSYLSSSEDPDIHFKYIEAAAKT 753
LQIIVQ KEYCEQLGVDACIK+FEQF+SYEGLYFFLGSYLS SEDP+IHFKYIEAAAKT
Sbjct: 661 LQIIVQACKEYCEQLGVDACIKLFEQFKSYEGLYFFLGSYLSMSEDPEIHFKYIEAAAKT 720
Query: 754 GQIKEVERVTRESSFYDPEKTKNFLMEAKLPDARPLINVCDRFGFVPDLTHYLYTSNMLR 813
GQIKEVERVTRES+FYD EKTKNFLMEAKLPDARPLINVCDRFGFVPDLTHYLYT+NMLR
Sbjct: 721 GQIKEVERVTRESNFYDAEKTKNFLMEAKLPDARPLINVCDRFGFVPDLTHYLYTNNMLR 780
Query: 814 YIEGYVQKVNPGNAPLVVGQLLDDECPEDFIKGLILSVRSLLPVEPLVAECEKRNRLRLL 873
YIEGYVQKVNPGNAPLVVGQLLDDECPEDFIKGLILSVRSLLPVEPLVAECEKRNRLRLL
Sbjct: 781 YIEGYVQKVNPGNAPLVVGQLLDDECPEDFIKGLILSVRSLLPVEPLVAECEKRNRLRLL 840
Query: 874 TQFLEHLVSEGSQDVHVHNALGKIIIDSNNNPEHFLTTNPYYDSRVVGKYCEKRDPTLAV 933
TQFLEHLVSEGSQDVHVHNALGKIIIDSNNNPEHFLTTNPYYDS+VVGKYCEKRDPTLAV
Sbjct: 841 TQFLEHLVSEGSQDVHVHNALGKIIIDSNNNPEHFLTTNPYYDSKVVGKYCEKRDPTLAV 900
Query: 934 VAYRRGVCDDELINVTNKNSLFKLQARYVVERMDADLWEKVLNPDNTYRRQLIDQVVSTA 993
VAYRRG CD+ELINVTNKNSLFKLQARYVVERMD DLWEKVL +N YRRQLIDQVVSTA
Sbjct: 901 VAYRRGQCDEELINVTNKNSLFKLQARYVVERMDGDLWEKVLTEENEYRRQLIDQVVSTA 960
Query: 994 LPESKSPEQVSASVKAFMTADLPHELIELLEKIVLQNSAFSGNFNLQNLLILTAIKADSS 1053
LPESKSPEQVSA+VKAFMTADLPHELIELLEKIVLQNSAFSGNFNLQNLLILTAIKAD S
Sbjct: 961 LPESKSPEQVSAAVKAFMTADLPHELIELLEKIVLQNSAFSGNFNLQNLLILTAIKADPS 1020
Query: 1054 RVMDYVNRLDNFDGPQVGEVAVEAELYEEAFAIFKKFNLNVQAVNVLLDNIHSIDRAVEF 1113
RVMDY+NRLDNFDGP VGEVAV+A+LYEEAFAIFKKFNLNVQAVNVLLDN+ SI+RAVEF
Sbjct: 1021 RVMDYINRLDNFDGPAVGEVAVDAQLYEEAFAIFKKFNLNVQAVNVLLDNVRSIERAVEF 1080
Query: 1114 AFRVEEDAVWSQVAKAQLREGLVSDAIESFIRADDATQFLDVIRAAQDGNAYQDLVRYLL 1173
AFRVEEDAVWSQVAKAQLREGLVSDAIESFIRADD TQFL+VIRA++D N Y DLVRYLL
Sbjct: 1081 AFRVEEDAVWSQVAKAQLREGLVSDAIESFIRADDTTQFLEVIRASEDTNVYDDLVRYLL 1140
Query: 1174 MVRQKTKEPKVDSELIYAYAKIDRLSDIEEFILMPNVANLQNVGDRLYDEELYEAAKIIF 1233
MVRQK KEPKVDSELIYAYAKI+RL +IEEFILMPNVANLQ+VGDRLYDE LYEAAKII+
Sbjct: 1141 MVRQKVKEPKVDSELIYAYAKIERLGEIEEFILMPNVANLQHVGDRLYDEALYEAAKIIY 1200
Query: 1234 AFISNWAKLAVTLVKLKQFQGAVDAARKANSAKTWKEVCFACVDEEEFRLAQICGLNIII 1293
AFISNWAKLAVTLVKL+QFQGAVDAARKANSAKTWKEVCFACVD EEFRLAQICGLNIII
Sbjct: 1201 AFISNWAKLAVTLVKLQQFQGAVDAARKANSAKTWKEVCFACVDAEEFRLAQICGLNIII 1260
Query: 1294 QVDDLEEVSEYYQNRGCFNELISLMESGLGLERAHMGIFTELGVLYARYRPEKLMEHIKL 1353
QVDDLEEVSEYYQNRGCFNELISLMESGLGLERAHMGIFTELGVLYARYR EKLMEHIKL
Sbjct: 1261 QVDDLEEVSEYYQNRGCFNELISLMESGLGLERAHMGIFTELGVLYARYRYEKLMEHIKL 1320
Query: 1354 FATRLNIPKLIRACDEQQHWKELTYLYIQYDEFDNAATTIMNHSPEAWDHMQFKDVIAKV 1413
F+TRLNIPKLIRACDEQQHW+ELTYLYIQYDEFDNAATT+MNHSPEAW+HMQFKD++AKV
Sbjct: 1321 FSTRLNIPKLIRACDEQQHWQELTYLYIQYDEFDNAATTVMNHSPEAWEHMQFKDIVAKV 1380
Query: 1414 ANVELYYKAVHFYLQEHPDLINDVLNVLALRVDHARVVDIMRKAGHLRLVKPYMVAVQSN 1473
ANVELYYKAVHFYLQEHPD+IND+LNVLALR+DH RVVDIMRKAGHLRL+KPYMVAVQSN
Sbjct: 1381 ANVELYYKAVHFYLQEHPDIINDLLNVLALRLDHTRVVDIMRKAGHLRLIKPYMVAVQSN 1440
Query: 1474 NVSAVNEALNEIYVEEEDYDRLRESIDLHDNFDQIGLAQKIEKHELLEMRRVAAYIYKKA 1533
NVSAVNEALNEIY EEEDYDRLRESIDLHD+FDQIGLAQKIEKHEL+EMRRVAAYIYKKA
Sbjct: 1441 NVSAVNEALNEIYAEEEDYDRLRESIDLHDSFDQIGLAQKIEKHELVEMRRVAAYIYKKA 1500
Query: 1534 GRWKQSIALSKKDNLYKDAMETASQSGERELAEELLVYFIDQGKKECFASCLFVCYDLIR 1593
GRWKQSIALSKKDN+YKD METASQSG+ +LAE+LLVYFI+QGKKECFA+CLFVCYDLIR
Sbjct: 1501 GRWKQSIALSKKDNMYKDCMETASQSGDHDLAEQLLVYFIEQGKKECFATCLFVCYDLIR 1560
Query: 1594 VDVALELAWMHNMIDFAFPYLLQFIREYTGKVDELVKHKIESQNEVKAKEQEEKEVIAQQ 1653
DVALELAW++NMIDFAFPYLLQFIREY+GKVDEL+K K+E+Q EVKAKEQEEK+V++QQ
Sbjct: 1561 PDVALELAWINNMIDFAFPYLLQFIREYSGKVDELIKDKLEAQKEVKAKEQEEKDVMSQQ 1620
Query: 1654 NMYAQLLPLALPAPPMPGMGGGMGGGYAPPPPMGG--GM-GMPPMPPFGM-PMGGY 1705
NMYAQLLPLALPAPPMPGMG GGGY PPP MGG GM GMPPMPP+GM PMGGY
Sbjct: 1621 NMYAQLLPLALPAPPMPGMG---GGGYGPPPQMGGMPGMSGMPPMPPYGMPPMGGY 1673
>At3g08530 hypothetical protein
Length = 1516
Score = 2788 bits (7226), Expect = 0.0
Identities = 1388/1518 (91%), Positives = 1469/1518 (96%), Gaps = 6/1518 (0%)
Query: 192 MQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLISFATKTVNAGQVISKLHVIELGAQ 251
MQLFSV+QQRSQALEAHAASFAQFKVPGNENPS LISFA+K+ NAGQ+ SKLHVIELGAQ
Sbjct: 1 MQLFSVDQQRSQALEAHAASFAQFKVPGNENPSILISFASKSFNAGQITSKLHVIELGAQ 60
Query: 252 PGKPSFTKKQADLFFPPDFADDFPVSMQISHKYSLIYVITKLGLLFVYDLETATAVYRNR 311
PGKPSFTKKQADLFFPPDFADDFPV+MQ+SHK++LIYVITKLGLLFVYDLETA+A+YRNR
Sbjct: 61 PGKPSFTKKQADLFFPPDFADDFPVAMQVSHKFNLIYVITKLGLLFVYDLETASAIYRNR 120
Query: 312 ISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELAVNLAKRGNL 371
ISPDPIFLTSEA+SVGGFYAINRRGQVLLATVNE TI+ F+SGQLNNLELAVNLAKRGNL
Sbjct: 121 ISPDPIFLTSEASSVGGFYAINRRGQVLLATVNEATIIPFISGQLNNLELAVNLAKRGNL 180
Query: 372 PGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQAGQTPPLLQY 431
PGAE LVV+RF ELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQAGQTPPLLQY
Sbjct: 181 PGAENLVVQRFQELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQAGQTPPLLQY 240
Query: 432 FGTLLTRGKLNAFESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVKTVDNDLALK 491
FGTLLTRGKLN++ESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVKTVDNDLALK
Sbjct: 241 FGTLLTRGKLNSYESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVKTVDNDLALK 300
Query: 492 IYIKARATPKVVAAFAEKREFDKILIYSKQVGYTPDYLFLLQSILRTDPQGAVNFALMMS 551
IYIKARATPKVVAAFAE+REFDKILIYSKQVGYTPDYLFLLQ+ILRTDPQGAVNFALMMS
Sbjct: 301 IYIKARATPKVVAAFAERREFDKILIYSKQVGYTPDYLFLLQTILRTDPQGAVNFALMMS 360
Query: 552 QMEGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPEHGFLQTKVLEINLVTFPNVA 611
QMEGG PVDYNTITDLFLQRNLIREAT+FLLDVLKPNLPEH FLQTKVLEINLVTFPNVA
Sbjct: 361 QMEGGSPVDYNTITDLFLQRNLIREATSFLLDVLKPNLPEHAFLQTKVLEINLVTFPNVA 420
Query: 612 DAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNTHAIEPQALVEFFG 671
DA+LANGMF+HYDRPRIAQLCEKAGLY+++L+HY+ELPDIKRVIVNTHAIEPQALVEFFG
Sbjct: 421 DAVLANGMFTHYDRPRIAQLCEKAGLYIQSLKHYSELPDIKRVIVNTHAIEPQALVEFFG 480
Query: 672 TLSKEWALECMKDLLLVNLRGNLQIIVQVA---KEYCEQLGVDACIKIFEQFRSYEGLYF 728
TLS EWA+ECMKDLLLVNLRGNLQIIVQ + KEYCEQLGVDACIK+FEQF+SYEGLYF
Sbjct: 481 TLSSEWAMECMKDLLLVNLRGNLQIIVQASGACKEYCEQLGVDACIKLFEQFKSYEGLYF 540
Query: 729 FLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFLMEAKLPDARP 788
FLGSYLS SEDP+IHFKYIEAAAKTGQIKEVERVTRES+FYD EKTKNFLMEAKLPDARP
Sbjct: 541 FLGSYLSMSEDPEIHFKYIEAAAKTGQIKEVERVTRESNFYDAEKTKNFLMEAKLPDARP 600
Query: 789 LINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQLLDDECPEDFIKGLI 848
LINVCDRF FVPDLTHYLYT+NMLRYIEGYVQKVNPGNAPLVVGQLLDDECPEDFIKGLI
Sbjct: 601 LINVCDRFSFVPDLTHYLYTNNMLRYIEGYVQKVNPGNAPLVVGQLLDDECPEDFIKGLI 660
Query: 849 LSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSNNNPEHF 908
LSVRSLLPVEPLV ECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSNNNPEHF
Sbjct: 661 LSVRSLLPVEPLVEECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSNNNPEHF 720
Query: 909 LTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNKNSLFKLQARYVVERMDA 968
LTTNPYYDS+VVGKYCEKRDPTLAVVAYRRG CD+ELINVTNKNSLFKLQARYVVERMD
Sbjct: 721 LTTNPYYDSKVVGKYCEKRDPTLAVVAYRRGQCDEELINVTNKNSLFKLQARYVVERMDG 780
Query: 969 DLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQVSASVKAFMTADLPHELIELLEKIVL 1028
DLW+KVL+ +N YRRQLIDQVVSTALPESKSPEQVSA+VKAFMTADLPHELIELLEKIVL
Sbjct: 781 DLWDKVLDENNDYRRQLIDQVVSTALPESKSPEQVSAAVKAFMTADLPHELIELLEKIVL 840
Query: 1029 QNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVGEVAVEAELYEEAFAIFK 1088
QNSAFSGNFNLQNLLILTAIKAD SRVMDY+NRLDNFDGP VGEVAVEA+LYEEAFAIFK
Sbjct: 841 QNSAFSGNFNLQNLLILTAIKADPSRVMDYINRLDNFDGPAVGEVAVEAQLYEEAFAIFK 900
Query: 1089 KFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSDAIESFIRADD 1148
KFNLNVQAVNVLLDN+ SI+RAVEFAFRVEED+VWSQVAKAQLREGLVSDAIESFIRADD
Sbjct: 901 KFNLNVQAVNVLLDNVRSIERAVEFAFRVEEDSVWSQVAKAQLREGLVSDAIESFIRADD 960
Query: 1149 ATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYAYAKIDRLSDIEEFILMP 1208
AT FL+VIR ++D + Y DLV+YLLMVRQK KEPKVDSELIYAYAKIDRL +IEEFILMP
Sbjct: 961 ATHFLEVIRVSEDTDVYDDLVKYLLMVRQKVKEPKVDSELIYAYAKIDRLGEIEEFILMP 1020
Query: 1209 NVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARKANSAKTW 1268
NVANLQ+VGDRLYDE LYEAAKII+AFISNW KLAVTLVKL+QFQGAVDAARKANSAKTW
Sbjct: 1021 NVANLQHVGDRLYDEALYEAAKIIYAFISNWGKLAVTLVKLQQFQGAVDAARKANSAKTW 1080
Query: 1269 KEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGLGLERAH 1328
KEVCFACVD EEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGLGLERAH
Sbjct: 1081 KEVCFACVDAEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGLGLERAH 1140
Query: 1329 MGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKELTYLYIQYDEFDN 1388
MGIFTELGVLYARYR EKLMEHIKLF+TRLNIPKLIRACDEQQHW+ELTYLYIQYDEFDN
Sbjct: 1141 MGIFTELGVLYARYRYEKLMEHIKLFSTRLNIPKLIRACDEQQHWQELTYLYIQYDEFDN 1200
Query: 1389 AATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVLNVLALRVDHA 1448
AATT+MNHSPEAW+HMQFKD++AKVANVELYYKAVHFYLQEHPD+IND+LNVLALR+DH
Sbjct: 1201 AATTVMNHSPEAWEHMQFKDIVAKVANVELYYKAVHFYLQEHPDIINDLLNVLALRLDHT 1260
Query: 1449 RVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHDNFDQI 1508
RVVDIMRKAGHLRL+KPYM+AVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHD+FDQI
Sbjct: 1261 RVVDIMRKAGHLRLIKPYMIAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHDSFDQI 1320
Query: 1509 GLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGERELAEEL 1568
GLAQKIEKHEL+EMRRVAAYIYKKAGRWKQSIALSKKDN+YKD METASQSGE ELAE+L
Sbjct: 1321 GLAQKIEKHELVEMRRVAAYIYKKAGRWKQSIALSKKDNMYKDCMETASQSGEHELAEQL 1380
Query: 1569 LVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYTGKVDEL 1628
LVYFI+QGKKECFA+CLFVCYDLIR DVALELAW++NM+DFAFPYLLQFIREY+GKVDEL
Sbjct: 1381 LVYFIEQGKKECFATCLFVCYDLIRPDVALELAWINNMMDFAFPYLLQFIREYSGKVDEL 1440
Query: 1629 VKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMGGGYAPPPPMGG 1688
+K K+E+Q EVKAKEQEEK+VI+QQNMYAQ+LPLALPAPPMPGMGG GGGY PPP MGG
Sbjct: 1441 IKDKLEAQKEVKAKEQEEKDVISQQNMYAQMLPLALPAPPMPGMGG--GGGYGPPPQMGG 1498
Query: 1689 GMGMPPMPPFGM-PMGGY 1705
GMPPMPP+GM PMGGY
Sbjct: 1499 MPGMPPMPPYGMPPMGGY 1516
>At3g08540 putative clathrin heavy chain, 3' partial
Length = 1263
Score = 2268 bits (5877), Expect = 0.0
Identities = 1144/1279 (89%), Positives = 1208/1279 (94%), Gaps = 28/1279 (2%)
Query: 3 AAANAPILMREALTLPSIGINPQHITFTHVTMESDKYICVRETAPQNSVVIVDMNMPNQP 62
AAANAPI M+E LTLPSIGIN Q ITFT+VTMESDKYICVRET+PQNSVVI+DMNMP QP
Sbjct: 2 AAANAPITMKEVLTLPSIGINQQFITFTNVTMESDKYICVRETSPQNSVVIIDMNMPMQP 61
Query: 63 LRRPITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKWIS 122
LRRPITADSALMNPNS+ILALKAQ+ GTTQDHLQIFNIE KAK+KS+QMPEQVVFWKWI+
Sbjct: 62 LRRPITADSALMNPNSKILALKAQVPGTTQDHLQIFNIEAKAKLKSHQMPEQVVFWKWIT 121
Query: 123 PKLLGLVTQTSVYHWSIEGDSEPVKMFERTANLANNQIINYRCDPTEKWLVLIGIAPGSP 182
PK+LGLVTQ SVYHWSIEGDSEPVKMF+RTANLANNQIINY+C P EKWLVLIGIAPGSP
Sbjct: 122 PKMLGLVTQNSVYHWSIEGDSEPVKMFDRTANLANNQIINYKCSPNEKWLVLIGIAPGSP 181
Query: 183 ERPQLVKGNMQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLISFATKTVNAGQVISK 242
ER QLVKGNMQLFSV+QQRSQALEAHAASFAQFKVPGNENPS LISFA+K+ NAGQ+ SK
Sbjct: 182 ERQQLVKGNMQLFSVDQQRSQALEAHAASFAQFKVPGNENPSILISFASKSFNAGQITSK 241
Query: 243 LHVIELGAQPGKPSFTKKQADLFFPPDFADDFPVSMQISHKYSLIYVITKLGLLFVYDLE 302
LHVIELGAQPGKPSFTKKQADLFFPPDFADDFPV+MQ+SHK++LIYVITKLGLLFVYDLE
Sbjct: 242 LHVIELGAQPGKPSFTKKQADLFFPPDFADDFPVAMQVSHKFNLIYVITKLGLLFVYDLE 301
Query: 303 TATAVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELA 362
TA+A+YRNRISPDPIFLTSEA+SVGGFYAINRRGQVLLATVNE TI+ F+SGQLNNLELA
Sbjct: 302 TASAIYRNRISPDPIFLTSEASSVGGFYAINRRGQVLLATVNEATIIPFISGQLNNLELA 361
Query: 363 VNLAKRGNLPGAEKL-----------VVERFHELFAQTKYKEAAELAAESPQGILRTPDT 411
VNLAKRGNLPGAE L VV+RF ELFAQTKYKEAAELAAESPQGILRTPDT
Sbjct: 362 VNLAKRGNLPGAENLVCDVYHGSVVSVVQRFQELFAQTKYKEAAELAAESPQGILRTPDT 421
Query: 412 VAKFQSVPVQAGQTPPLLQYFGTLLTRGKLNAFESLELSRLVVNQNKKNLLENWLAEDKL 471
VAKFQSVPVQAGQTPPLLQYFGTLLTRGKLN++ESLELSRLVVNQNKKNLLENWLAEDKL
Sbjct: 422 VAKFQSVPVQAGQTPPLLQYFGTLLTRGKLNSYESLELSRLVVNQNKKNLLENWLAEDKL 481
Query: 472 ECSEELGDLVKTVDNDLALKIYIKARATPKVVAAFAEKREFDKILIYSKQVGYTPDYLFL 531
ECSEELGDLVKTVDNDLALKIYIKARATPKVVAAFAE+REFDKILIYSKQVGYTPDYLFL
Sbjct: 482 ECSEELGDLVKTVDNDLALKIYIKARATPKVVAAFAERREFDKILIYSKQVGYTPDYLFL 541
Query: 532 LQSILRTDPQGAVNFALMMSQMEGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPE 591
LQ+ILRTDPQGAVNFALMMSQMEGG PVDYNTITDLFLQRNLIREAT+FLLDVLKPNLPE
Sbjct: 542 LQTILRTDPQGAVNFALMMSQMEGGSPVDYNTITDLFLQRNLIREATSFLLDVLKPNLPE 601
Query: 592 HGFLQTKVLEINLVTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDI 651
H FLQTKVLEINLVTFPNVADA+LANGMF+HYDRPRIAQLCEKAGLY+++L+HY+ELPDI
Sbjct: 602 HAFLQTKVLEINLVTFPNVADAVLANGMFTHYDRPRIAQLCEKAGLYIQSLKHYSELPDI 661
Query: 652 KRVIVNTHAIEPQALVEFFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVD 711
KRVIVNTHAIEPQALVEFFGTLS EWA+ECMKDLLL KEYCEQLGVD
Sbjct: 662 KRVIVNTHAIEPQALVEFFGTLSSEWAMECMKDLLL------------ACKEYCEQLGVD 709
Query: 712 ACIKIFEQFRSYEGLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDP 771
ACIK+FEQF+SYEGLYFFLGSYLS SEDP+IHFKYIEAAAKTGQIKEVERVTRES+FYD
Sbjct: 710 ACIKLFEQFKSYEGLYFFLGSYLSMSEDPEIHFKYIEAAAKTGQIKEVERVTRESNFYDA 769
Query: 772 EKTKNFLMEAKLPDARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVV 831
EKTKNFLMEAKLPDARPLINVCDRF FVPDLTHYLYT+NMLRYIEGYVQKVNPGNAPLVV
Sbjct: 770 EKTKNFLMEAKLPDARPLINVCDRFSFVPDLTHYLYTNNMLRYIEGYVQKVNPGNAPLVV 829
Query: 832 GQLLDDECPEDFIKGLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVH 891
GQLLDDECPEDFIKGLILSVRSLLPVEPLV ECEKRNRLRLLTQFLEHLVSEGSQDVHVH
Sbjct: 830 GQLLDDECPEDFIKGLILSVRSLLPVEPLVEECEKRNRLRLLTQFLEHLVSEGSQDVHVH 889
Query: 892 NALGKIIIDSNNNPEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNK 951
NALGKIIIDSNNNPEHFLTTNPYYDS+VVGKYCEKRDPTLAVVAYRRG CD+ELINVTNK
Sbjct: 890 NALGKIIIDSNNNPEHFLTTNPYYDSKVVGKYCEKRDPTLAVVAYRRGQCDEELINVTNK 949
Query: 952 NSLFKLQARYVVERMDADLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQVSASVKAFM 1011
NSLFKLQA RMD DLW+KVL+ +N YRRQLIDQVVSTALPESKSPEQVSA+VKAFM
Sbjct: 950 NSLFKLQA-----RMDGDLWDKVLDENNDYRRQLIDQVVSTALPESKSPEQVSAAVKAFM 1004
Query: 1012 TADLPHELIELLEKIVLQNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVG 1071
TADLPHELIELLEKIVLQNSAFSGNFNLQNLLILTAIKAD SRVMDY+NRLDNFDGP VG
Sbjct: 1005 TADLPHELIELLEKIVLQNSAFSGNFNLQNLLILTAIKADPSRVMDYINRLDNFDGPAVG 1064
Query: 1072 EVAVEAELYEEAFAIFKKFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVWSQVAKAQL 1131
EVAVEA+LYEEAFAIFKKFNLNVQAVNVLLDN+ SI+RAVEFAFRVEED+VWSQVAKAQL
Sbjct: 1065 EVAVEAQLYEEAFAIFKKFNLNVQAVNVLLDNVRSIERAVEFAFRVEEDSVWSQVAKAQL 1124
Query: 1132 REGLVSDAIESFIRADDATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYA 1191
REGLVSDAIESFIRADDAT FL+VIR ++D + Y DLV+YLLMVRQK KEPKVDSELIYA
Sbjct: 1125 REGLVSDAIESFIRADDATHFLEVIRVSEDTDVYDDLVKYLLMVRQKVKEPKVDSELIYA 1184
Query: 1192 YAKIDRLSDIEEFILMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQ 1251
YAKIDRL +IEEFILMPNVANLQ+VGDRLYDE LYEAAKII+AFISNW KLAVTLVKL+Q
Sbjct: 1185 YAKIDRLGEIEEFILMPNVANLQHVGDRLYDEALYEAAKIIYAFISNWGKLAVTLVKLQQ 1244
Query: 1252 FQGAVDAARKANSAKTWKE 1270
FQGAVDAARKANSAKTWKE
Sbjct: 1245 FQGAVDAARKANSAKTWKE 1263
>At5g07750 putative protein
Length = 1289
Score = 41.2 bits (95), Expect = 0.005
Identities = 22/45 (48%), Positives = 23/45 (50%), Gaps = 1/45 (2%)
Query: 1661 PLALPAPPMPGMGGGMGGGYAPPPPMGGGMGM-PPMPPFGMPMGG 1704
P+ APP P GG G G PPPP GG PP PP P GG
Sbjct: 1128 PMRGGAPPPPPPPGGRGPGAPPPPPPPGGRAPGPPPPPGPRPPGG 1172
Score = 38.9 bits (89), Expect = 0.026
Identities = 24/50 (48%), Positives = 24/50 (48%), Gaps = 13/50 (26%)
Query: 1665 PAPPMPGMGGGM---------GGGYAPPPPMGG-GMGMPPMPPFGMPMGG 1704
P PP P M GG GG PPPP GG G G PP PP P GG
Sbjct: 1110 PPPPPPPMHGGAPPPPPPPMRGGAPPPPPPPGGRGPGAPPPPP---PPGG 1156
Score = 37.7 bits (86), Expect = 0.058
Identities = 23/49 (46%), Positives = 23/49 (46%), Gaps = 10/49 (20%)
Query: 1665 PAPPMPGMGGG--------MGGGY--APPPPMGGGMGMPPMPPFGMPMG 1703
P PP P M GG M GG PPPPM GG PP PP G G
Sbjct: 1098 PPPPPPPMRGGAPPPPPPPMHGGAPPPPPPPMRGGAPPPPPPPGGRGPG 1146
Score = 37.4 bits (85), Expect = 0.075
Identities = 22/42 (52%), Positives = 22/42 (52%), Gaps = 4/42 (9%)
Query: 1663 ALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
A P PP P M GG PPPPM GG PP PP PM G
Sbjct: 1022 APPPPPPPPMHGG-APPPPPPPPMHGGAPPPPPPP---PMHG 1059
Score = 37.0 bits (84), Expect = 0.098
Identities = 21/40 (52%), Positives = 21/40 (52%), Gaps = 4/40 (10%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
P PP P M GG PPPPM GG PP PP PM G
Sbjct: 1011 PPPPPPPMHGGAPPP-PPPPPMHGGAPPPPPPP---PMHG 1046
Score = 36.6 bits (83), Expect = 0.13
Identities = 18/33 (54%), Positives = 18/33 (54%), Gaps = 2/33 (6%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP P M G G PPPPM GG PP PP
Sbjct: 1086 PPPPPPPMRG--GAPPPPPPPMRGGAPPPPPPP 1116
Score = 36.6 bits (83), Expect = 0.13
Identities = 19/35 (54%), Positives = 19/35 (54%), Gaps = 2/35 (5%)
Query: 1663 ALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
A P PP P M G G PPPPM GG PP PP
Sbjct: 1048 APPPPPPPPMHG--GAPPPPPPPMFGGAQPPPPPP 1080
Score = 36.6 bits (83), Expect = 0.13
Identities = 19/35 (54%), Positives = 19/35 (54%), Gaps = 1/35 (2%)
Query: 1663 ALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
A P PP P M GG PPPPM GG PP PP
Sbjct: 1035 APPPPPPPPMHGG-APPPPPPPPMHGGAPPPPPPP 1068
Score = 36.2 bits (82), Expect = 0.17
Identities = 18/33 (54%), Positives = 18/33 (54%), Gaps = 2/33 (6%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP P M GG PPPPM GG PP PP
Sbjct: 1062 PPPPPPPMFGGAQP--PPPPPMRGGAPPPPPPP 1092
Score = 34.3 bits (77), Expect = 0.64
Identities = 17/31 (54%), Positives = 17/31 (54%), Gaps = 2/31 (6%)
Query: 1667 PPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
PP P M G G PPPPM GG PP PP
Sbjct: 1076 PPPPPMRG--GAPPPPPPPMRGGAPPPPPPP 1104
Score = 33.5 bits (75), Expect = 1.1
Identities = 16/34 (47%), Positives = 16/34 (47%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPF 1698
P PP P G G PPPP G PP PPF
Sbjct: 970 PPPPPPPPGYGSPPPPPPPPPSYGSPPPPPPPPF 1003
Score = 32.0 bits (71), Expect = 3.2
Identities = 17/33 (51%), Positives = 17/33 (51%), Gaps = 3/33 (9%)
Query: 1666 APPMPGMGGGMGGGYAPPP---PMGGGMGMPPM 1695
APP P GG G PPP P GGG PPM
Sbjct: 1147 APPPPPPPGGRAPGPPPPPGPRPPGGGPPPPPM 1179
Score = 31.6 bits (70), Expect = 4.1
Identities = 20/47 (42%), Positives = 20/47 (42%), Gaps = 10/47 (21%)
Query: 1663 ALPAPPMPGMGGG-------MGGGYAPPPP---MGGGMGMPPMPPFG 1699
A P PP P GG M GG PPPP GG PP P G
Sbjct: 1073 AQPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMHG 1119
Score = 31.6 bits (70), Expect = 4.1
Identities = 17/40 (42%), Positives = 17/40 (42%), Gaps = 5/40 (12%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
P PP P G PPPP G G PP PP P G
Sbjct: 959 PPPPPPSYGSP-----PPPPPPPPGYGSPPPPPPPPPSYG 993
>At1g30460 hypothetical protein
Length = 678
Score = 40.4 bits (93), Expect = 0.009
Identities = 33/86 (38%), Positives = 40/86 (46%), Gaps = 16/86 (18%)
Query: 1634 ESQNEVKAKEQEEKEVIAQQN-------MYAQLLPLALPAPPMPGMG----GGMGGGYAP 1682
E + E +++E+EE Q M+ +PL PMPGMG G MG G A
Sbjct: 466 EEEEEDESEEEEESMAGGPQGRGRGRGIMWPPQMPLGRGIRPMPGMGGFPLGVMGPGDAF 525
Query: 1683 PPPMGGGMGMPPMPPFGM---PMGGY 1705
P GG GMP PFGM P G Y
Sbjct: 526 PYGPGGYNGMP--DPFGMGPRPFGPY 549
>At3g25690 unknown protein
Length = 939
Score = 39.7 bits (91), Expect = 0.015
Identities = 21/44 (47%), Positives = 22/44 (49%), Gaps = 6/44 (13%)
Query: 1660 LPLALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMG 1703
LP P PP P GGG PPPP GGG PP PP + G
Sbjct: 609 LPGGGPPPPPPPPGGG------PPPPPGGGPPPPPPPPGALGRG 646
Score = 35.8 bits (81), Expect = 0.22
Identities = 24/80 (30%), Positives = 31/80 (38%), Gaps = 12/80 (15%)
Query: 1634 ESQNEVKAKEQEEKEVIAQQNMY-AQLLPLALPAPPMPGMGGG-----------MGGGYA 1681
ES + K E + + + + P +P PP GGG + GG
Sbjct: 555 ESNESNEGKASENAATVTKMKLVDIEKRPPRVPRPPPRSAGGGKSTNLPSARPPLPGGGP 614
Query: 1682 PPPPMGGGMGMPPMPPFGMP 1701
PPPP G G PP P G P
Sbjct: 615 PPPPPPPGGGPPPPPGGGPP 634
>At1g62500 putative proline-rich cell wall protein (pir|IS52985);
similar to ESTs gb|AI239404, gb|R89984, and emb|Z17709
Length = 297
Score = 38.5 bits (88), Expect = 0.034
Identities = 21/46 (45%), Positives = 23/46 (49%), Gaps = 2/46 (4%)
Query: 1661 PLALPAPPMPG--MGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
P P PP+P GG GGG + PPP GG G PP G GG
Sbjct: 29 PCTRPHPPVPKPPQHGGGGGGGSKPPPHHGGKGGGKPPPHGGKGGG 74
Score = 32.7 bits (73), Expect = 1.9
Identities = 21/51 (41%), Positives = 21/51 (41%), Gaps = 11/51 (21%)
Query: 1665 PAPPMPGMGGGMGGGYAPPP-----------PMGGGMGMPPMPPFGMPMGG 1704
P P P GGG GGG PPP P GG G PP G GG
Sbjct: 36 PVPKPPQHGGGGGGGSKPPPHHGGKGGGKPPPHGGKGGGPPHHGGGGGGGG 86
>At3g15000 unknown protein
Length = 395
Score = 37.4 bits (85), Expect = 0.075
Identities = 33/117 (28%), Positives = 47/117 (39%), Gaps = 23/117 (19%)
Query: 1610 AFPYLLQFIREY---TGKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPA 1666
A PY ++ E+ + +E + +N +++ E + Q P+ P
Sbjct: 193 AVPYDPKYHEEWIRNNARANERNRRNDRPRNNDRSRNFERRRENMAGGPPPQRPPMGGPP 252
Query: 1667 PPMPGMGGG------MGGGYAPPPPMGGGMGMPP---------MPPFGMP----MGG 1704
PP P +GG MGG PPP MG G PP PP+G P MGG
Sbjct: 253 PP-PHIGGSAPPPPHMGGSAPPPPHMGQNYGPPPPNNMGGPRHPPPYGAPPQNNMGG 308
>At1g61080 hypothetical protein
Length = 907
Score = 37.4 bits (85), Expect = 0.075
Identities = 19/40 (47%), Positives = 19/40 (47%), Gaps = 3/40 (7%)
Query: 1667 PPMPGMGGGMGGGYAPPPPM---GGGMGMPPMPPFGMPMG 1703
PPMP G GG PPPPM G PP PP M G
Sbjct: 578 PPMPMGNSGSGGPPPPPPPMPLANGATPPPPPPPMAMANG 617
Score = 33.5 bits (75), Expect = 1.1
Identities = 16/37 (43%), Positives = 16/37 (43%)
Query: 1661 PLALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P A APP P G PPPP G PP PP
Sbjct: 530 PRAAVAPPPPPPPPGTAAAPPPPPPPPGTQAAPPPPP 566
Score = 33.1 bits (74), Expect = 1.4
Identities = 17/39 (43%), Positives = 17/39 (43%), Gaps = 4/39 (10%)
Query: 1663 ALPAPPMPGMGGGMGGGYAPPPP----MGGGMGMPPMPP 1697
A P PP P M G PPPP M G PP PP
Sbjct: 603 ATPPPPPPPMAMANGAAGPPPPPPRMGMANGAAGPPPPP 641
Score = 31.6 bits (70), Expect = 4.1
Identities = 20/43 (46%), Positives = 20/43 (46%), Gaps = 6/43 (13%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPM---GGGMGMPPMPP-FGMPMG 1703
P PPMP G PPPPM G G PP PP GM G
Sbjct: 593 PPPPMPLANGATPP--PPPPPMAMANGAAGPPPPPPRMGMANG 633
Score = 31.2 bits (69), Expect = 5.4
Identities = 18/41 (43%), Positives = 18/41 (43%), Gaps = 3/41 (7%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPP---MGGGMGMPPMPPFGMPM 1702
P PP P M G PPPP M G PP PP M M
Sbjct: 590 PPPPPPPMPLANGATPPPPPPPMAMANGAAGPPPPPPRMGM 630
Score = 30.8 bits (68), Expect = 7.1
Identities = 19/53 (35%), Positives = 20/53 (36%), Gaps = 13/53 (24%)
Query: 1663 ALPAPPMPGMGGGMGGGYAPPPPM-------------GGGMGMPPMPPFGMPM 1702
A P PP P G PPPPM G G PP PP MP+
Sbjct: 547 AAPPPPPPPPGTQAAPPPPPPPPMQNRAPSPPPMPMGNSGSGGPPPPPPPMPL 599
>At5g49280 predicted GPI-anchored protein
Length = 162
Score = 36.6 bits (83), Expect = 0.13
Identities = 19/43 (44%), Positives = 20/43 (46%), Gaps = 1/43 (2%)
Query: 1663 ALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGGY 1705
A P PP P GG Y PPP GG P PP+G GY
Sbjct: 64 ACPPPPSPPSSGGGSSYYYPPPSQSGGGSKYP-PPYGGGGQGY 105
>At3g44340 putative protein
Length = 1122
Score = 36.6 bits (83), Expect = 0.13
Identities = 22/49 (44%), Positives = 23/49 (46%), Gaps = 8/49 (16%)
Query: 1661 PLALPAPPMPG------MGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMG 1703
P PA PG G G+ PPPMG GM MP PP GMP G
Sbjct: 144 PGGFPASGPPGGVPSGPPSGARPIGFGSPPPMGPGMSMP--PPSGMPGG 190
Score = 31.6 bits (70), Expect = 4.1
Identities = 18/46 (39%), Positives = 19/46 (41%), Gaps = 11/46 (23%)
Query: 1667 PPMPGMGGGMGGGYAPPPPMG-----------GGMGMPPMPPFGMP 1701
PP GM GG PPP MG G M P PP+G P
Sbjct: 210 PPPSGMPGGPLSNGPPPPMMGPGAFPRGSQFTSGPMMAPPPPYGQP 255
Score = 30.8 bits (68), Expect = 7.1
Identities = 17/37 (45%), Positives = 17/37 (45%)
Query: 1667 PPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMG 1703
PP GM GG PP M GG PP GMP G
Sbjct: 182 PPPSGMPGGPLSNGPPPSGMHGGHLSNGPPPSGMPGG 218
>At3g15010 RNA-binding protein
Length = 404
Score = 36.6 bits (83), Expect = 0.13
Identities = 19/38 (50%), Positives = 21/38 (55%), Gaps = 3/38 (7%)
Query: 1667 PPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
P G+ GG+GGGY P G G G MPP MP GG
Sbjct: 325 PGGTGVYGGLGGGYGGP---GTGSGQYRMPPSSMPGGG 359
>At4g37870 phosphoenolpyruvate carboxykinase (ATP) -like protein
Length = 671
Score = 36.2 bits (82), Expect = 0.17
Identities = 30/105 (28%), Positives = 44/105 (41%), Gaps = 21/105 (20%)
Query: 734 LSSSEDPDIHFKYIEAAAKTGQIKE---VERVTRESSFYDPEKTKN--------FLMEAK 782
LS ++PDI A K G + E + TRE + D T+N F+ AK
Sbjct: 413 LSREKEPDIW-----NAIKFGTVLENVVFDEHTREVDYSDKSVTENTRAAYPIEFIPNAK 467
Query: 783 LPDARP-----LINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKV 822
+P P ++ CD FG +P ++ M +I GY V
Sbjct: 468 IPCVGPHPTNVILLACDAFGVLPPVSKLNLAQTMYHFISGYTALV 512
>At3g06750 predicted GPI-anchored protein
Length = 147
Score = 36.2 bits (82), Expect = 0.17
Identities = 18/42 (42%), Positives = 20/42 (46%), Gaps = 1/42 (2%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMP-MGGY 1705
P PP P GG G Y PPP G+ PP+G GGY
Sbjct: 51 PPPPPPSSSGGGGSYYYSPPPPSSSGGVKYPPPYGGDGYGGY 92
>At5g18180 putative protein
Length = 189
Score = 35.4 bits (80), Expect = 0.29
Identities = 32/100 (32%), Positives = 42/100 (42%), Gaps = 17/100 (17%)
Query: 1623 GKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMY----AQLLPLALPAPPMPGMGGGMGG 1678
G+VDE+ ES +K +E +Q + + +LLPL+ P G GG G
Sbjct: 84 GRVDEIFGPINESLFSIKMREGIVATSYSQGDKFFISPEKLLPLSRFLPQPKGQSGGRGE 143
Query: 1679 GYAPP----PPMG----GGMGMP-----PMPPFGMPMGGY 1705
G PP PP G G G P P G P GG+
Sbjct: 144 GRVPPRGRGPPRGRGNFRGRGAPRGASRGFQPRGGPRGGF 183
>At3g51720 unknown protein
Length = 407
Score = 35.0 bits (79), Expect = 0.37
Identities = 28/115 (24%), Positives = 55/115 (47%), Gaps = 2/115 (1%)
Query: 1470 VQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHDNFDQIGLAQKIEKHELLEMRRVAAYI 1529
+ S N + ++ + + E +++ R E++ N + Q K E EMR +AA
Sbjct: 150 LNSENAAEMSMEIQRLSYEAKEFSRTGENVRYAVNKAVAEIEQTRNKIEAAEMRLIAARK 209
Query: 1530 YKKAGRWKQSIALSK-KDNLYKDAMETASQSGERELAEELLVYFIDQGKKECFAS 1583
K+A R +++A+++ K + +GE + EE+L ID+ +E +S
Sbjct: 210 MKEAARAAEAVAIAEIKAVTRRGRRRRRGGNGEETMQEEIL-ETIDETAREIRSS 263
>At1g09070 unknown protein
Length = 324
Score = 35.0 bits (79), Expect = 0.37
Identities = 19/40 (47%), Positives = 20/40 (49%), Gaps = 3/40 (7%)
Query: 1668 PMPGMGGGMGG--GYAPPPPMGGGMGMPPMPPFGMPMGGY 1705
P PG GG GY PP GG G PP P+G P GY
Sbjct: 216 PPPGAYPQQGGYPGY-PPQQQGGYPGYPPQGPYGYPQQGY 254
>At4g20440 putative snRNP protein
Length = 257
Score = 34.3 bits (77), Expect = 0.64
Identities = 22/61 (36%), Positives = 22/61 (36%), Gaps = 24/61 (39%)
Query: 1664 LPAPPMPGMGGGMGGGYAPP------------------------PPMGGGMGMPPMPPFG 1699
LP PP G G MG G PP PP GG M PP PP G
Sbjct: 166 LPPPPFGGQGPPMGRGPPPPYGMRPPPQQFSGPPPPQYGQRPMIPPPGGMMRGPPPPPHG 225
Query: 1700 M 1700
M
Sbjct: 226 M 226
Score = 33.1 bits (74), Expect = 1.4
Identities = 20/43 (46%), Positives = 22/43 (50%), Gaps = 7/43 (16%)
Query: 1661 PLALPAPPMPGMGGGMGGGYAPPPPMGG---GMGMPPMPPFGM 1700
P L APP+ G M PPPP GG MG P PP+GM
Sbjct: 150 PPQLSAPPIIRPPGQM----LPPPPFGGQGPPMGRGPPPPYGM 188
Score = 30.4 bits (67), Expect = 9.2
Identities = 18/35 (51%), Positives = 19/35 (53%), Gaps = 4/35 (11%)
Query: 1667 PPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMP 1701
P +P GG M G PPPP G G PP P GMP
Sbjct: 207 PMIPPPGGMMRG---PPPPPHGMQGPPPPRP-GMP 237
>At1g31810 hypothetical protein
Length = 1201
Score = 33.9 bits (76), Expect = 0.83
Identities = 17/35 (48%), Positives = 19/35 (53%), Gaps = 5/35 (14%)
Query: 1667 PPMPGMGGGMGGGYAPPPPMG--GGMGMPPMPPFG 1699
PP PG+G G G PPP+G G PP PP G
Sbjct: 740 PPPPGLGRGTSSG---PPPLGAKGSNAPPPPPPAG 771
>At1g59910 hypothetical protein
Length = 929
Score = 33.5 bits (75), Expect = 1.1
Identities = 21/43 (48%), Positives = 21/43 (48%), Gaps = 6/43 (13%)
Query: 1661 PLALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMG 1703
P A P PP PG G G PPPPM PP PP G P G
Sbjct: 405 PAAPPPPPPPGK---KGAGPPPPPPM--SKKGPPKPP-GNPKG 441
Score = 32.0 bits (71), Expect = 3.2
Identities = 16/37 (43%), Positives = 16/37 (43%)
Query: 1661 PLALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P A PP P G PPPP G G PP PP
Sbjct: 390 PSAAAPPPPPPPKKGPAAPPPPPPPGKKGAGPPPPPP 426
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,111,264
Number of Sequences: 26719
Number of extensions: 1626067
Number of successful extensions: 6377
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 5912
Number of HSP's gapped (non-prelim): 280
length of query: 1705
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1592
effective length of database: 8,299,349
effective search space: 13212563608
effective search space used: 13212563608
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC142498.7


