

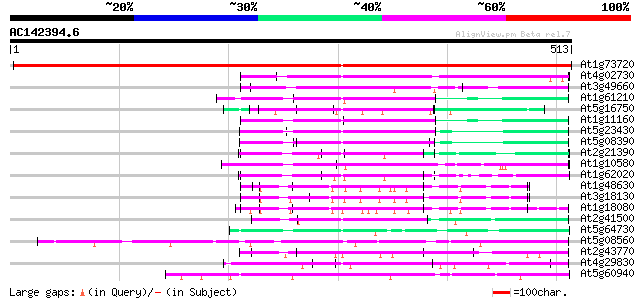
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142394.6 - phase: 0
(513 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g73720 unknown protein 908 0.0
At4g02730 putative WD-repeat protein 113 3e-25
At3g49660 putative WD-40 repeat - protein 112 4e-25
At1g61210 91 2e-18
At5g16750 WD40-repeat protein 89 4e-18
At1g11160 hypothetical protein 89 6e-18
At5g23430 unknown protein 86 4e-17
At5g08390 katanin p80 subunit - like protein 85 1e-16
At2g21390 coatomer alpha subunit 84 1e-16
At1g10580 putative pre-mRNA splicing factor 82 7e-16
At1g62020 hypothetical protein 81 2e-15
At1g48630 unknown protein 81 2e-15
At3g18130 protein kinase C-receptor/G-protein, putative 79 6e-15
At1g18080 putative guanine nucleotide-binding protein beta sub... 79 6e-15
At2g41500 putative U4/U6 small nuclear ribonucleoprotein 79 8e-15
At5g64730 unknown protein 78 1e-14
At5g08560 WD-repeat protein-like 77 2e-14
At2g43770 putative splicing factor 77 3e-14
At4g29830 unknown protein 76 5e-14
At5g60940 cleavage stimulation factor 50K chain 74 2e-13
>At1g73720 unknown protein
Length = 511
Score = 908 bits (2346), Expect = 0.0
Identities = 433/510 (84%), Positives = 481/510 (93%), Gaps = 1/510 (0%)
Query: 4 LEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILPQ 63
LEIEARDVIK +LQFCKENSL+QTFQTLQ+ECQVSLNTVDS+ETF++DIN GRWD++LPQ
Sbjct: 3 LEIEARDVIKIMLQFCKENSLNQTFQTLQSECQVSLNTVDSVETFISDINSGRWDSVLPQ 62
Query: 64 VSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLLV 123
VSQLKLPRN LEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQ ERYLR+EHLLV
Sbjct: 63 VSQLKLPRNKLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQAERYLRMEHLLV 122
Query: 124 RTYFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQ 183
R+YFDP+EAY +STKE++RAQIAQA+AAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQ
Sbjct: 123 RSYFDPHEAYGDSTKERKRAQIAQAVAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQ 182
Query: 184 FDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDY 243
FDLFRGTAAMKQDV+D +P L+HTIKFG KSHAECARFSPDGQ+L S S+DGFIEVWDY
Sbjct: 183 FDLFRGTAAMKQDVEDTHPNVLTHTIKFGKKSHAECARFSPDGQFLASSSVDGFIEVWDY 242
Query: 244 ISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLE 303
ISGKLKKDLQYQA+E+FMMHD+PVLC+DFSRDSEM+ASGS DGKIK+WRIRT C+RR +
Sbjct: 243 ISGKLKKDLQYQADESFMMHDDPVLCIDFSRDSEMLASGSQDGKIKIWRIRTGVCIRRFD 302
Query: 304 HAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTR 363
AHSQGVTS+SFSRDGSQLLSTSFD TARIHGLKSGK+LKEFRGHTSYVN A FT+DG+R
Sbjct: 303 -AHSQGVTSLSFSRDGSQLLSTSFDQTARIHGLKSGKLLKEFRGHTSYVNHAIFTSDGSR 361
Query: 364 VITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIY 423
+ITASSDCTVKVWD KTTDCLQTFKPPPPLRG DASVNS+ +FPKNTEHIVVCNKTSSIY
Sbjct: 362 IITASSDCTVKVWDSKTTDCLQTFKPPPPLRGTDASVNSIHLFPKNTEHIVVCNKTSSIY 421
Query: 424 IMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVH 483
IMTLQGQVVKSFSSG REGGDFVAAC+S KG+WIYC+GED+ +YCF+YQSG LEH M VH
Sbjct: 422 IMTLQGQVVKSFSSGNREGGDFVAACVSTKGDWIYCIGEDKKLYCFNYQSGGLEHFMMVH 481
Query: 484 DKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
+K++IG+THHPHRNL+AT+S+D TMKLWKP
Sbjct: 482 EKDVIGITHHPHRNLLATYSEDCTMKLWKP 511
>At4g02730 putative WD-repeat protein
Length = 333
Score = 113 bits (282), Expect = 3e-25
Identities = 78/305 (25%), Positives = 144/305 (46%), Gaps = 19/305 (6%)
Query: 212 GAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVD 271
G + C +FS DG L S S+D + +W + Y + H + +
Sbjct: 41 GHTAAISCVKFSNDGNLLASASVDKTMILWSATN--------YSLIHRYEGHSSGISDLA 92
Query: 272 FSRDSEMIASGSTDGKIKVWRIRTA-QCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDST 330
+S DS S S D +++W R+ +CL+ L H+ V V+F+ + ++S SFD T
Sbjct: 93 WSSDSHYTCSASDDCTLRIWDARSPYECLKVLR-GHTNFVFCVNFNPPSNLIVSGSFDET 151
Query: 331 ARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKPP 390
RI +K+GK ++ + H+ ++ F DG+ +++AS D + K+WD K CL+T
Sbjct: 152 IRIWEVKTGKCVRMIKAHSMPISSVHFNRDGSLIVSASHDGSCKIWDAKEGTCLKT---- 207
Query: 391 PPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMT-LQGQVVKSFSSGKREGGDFVAAC 449
L + S F N + I+V S++ + G+ +K ++ + +A
Sbjct: 208 --LIDDKSPAVSFAKFSPNGKFILVATLDSTLKLSNYATGKFLKVYTGHTNKVFCITSAF 265
Query: 450 ISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSD--DHT 507
G++I ED +Y + Q+ + + H +I V+ HP +N +++ + D T
Sbjct: 266 SVTNGKYIVSGSEDNCVYLWDLQARNILQRLEGHTDAVISVSCHPVQNEISSSGNHLDKT 325
Query: 508 MKLWK 512
+++WK
Sbjct: 326 IRIWK 330
Score = 94.4 bits (233), Expect = 1e-19
Identities = 72/272 (26%), Positives = 123/272 (44%), Gaps = 15/272 (5%)
Query: 245 SGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEH 304
SG + Y+ +T H + CV FS D ++AS S D + +W + R E
Sbjct: 24 SGNVPIYKPYRHLKTLEGHTAAISCVKFSNDGNLLASASVDKTMILWSATNYSLIHRYE- 82
Query: 305 AHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSG-KMLKEFRGHTSYVNDATFTNDGTR 363
HS G++ +++S D S S D T RI +S + LK RGHT++V F
Sbjct: 83 GHSSGISDLAWSSDSHYTCSASDDCTLRIWDARSPYECLKVLRGHTNFVFCVNFNPPSNL 142
Query: 364 VITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIY 423
+++ S D T+++W++KT C++ K ++SV F ++ IV + S
Sbjct: 143 IVSGSFDETIRIWEVKTGKCVRMIK------AHSMPISSVH-FNRDGSLIVSASHDGSCK 195
Query: 424 IM-TLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTV 482
I +G +K+ K F A SP G++I D + +Y +GK + T
Sbjct: 196 IWDAKEGTCLKTLIDDKSPAVSF--AKFSPNGKFILVATLDSTLKLSNYATGKFLKVYTG 253
Query: 483 HDKEIIGVTHH---PHRNLVATFSDDHTMKLW 511
H ++ +T + + + S+D+ + LW
Sbjct: 254 HTNKVFCITSAFSVTNGKYIVSGSEDNCVYLW 285
>At3g49660 putative WD-40 repeat - protein
Length = 317
Score = 112 bits (281), Expect = 4e-25
Identities = 81/294 (27%), Positives = 136/294 (45%), Gaps = 15/294 (5%)
Query: 221 RFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIA 280
+FS DG+ L S S D I Y + + +E F H+ + V FS D+ I
Sbjct: 31 KFSSDGRLLASASADKTIRT--YTINTINDPIAEPVQE-FTGHENGISDVAFSSDARFIV 87
Query: 281 SGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGK 340
S S D +K+W + T ++ L H+ V+F+ + ++S SFD T RI + +GK
Sbjct: 88 SASDDKTLKLWDVETGSLIKTLI-GHTNYAFCVNFNPQSNMIVSGSFDETVRIWDVTTGK 146
Query: 341 MLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTF--KPPPPLRGGDA 398
LK H+ V F DG+ ++++S D ++WD T C++T PP+
Sbjct: 147 CLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRIWDSGTGHCVKTLIDDENPPVSFVRF 206
Query: 399 SVNSVFIFPKNTEHIV-VCNKTSSIYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWI 457
S N FI ++ + + N +S+ ++ T G V + +A G+ I
Sbjct: 207 SPNGKFILVGTLDNTLRLWNISSAKFLKTYTGHVNAQYCIS--------SAFSVTNGKRI 258
Query: 458 YCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLW 511
ED ++ + S KL + H + ++ V HP NL+A+ S D T+++W
Sbjct: 259 VSGSEDNCVHMWELNSKKLLQKLEGHTETVMNVACHPTENLIASGSLDKTVRIW 312
Score = 105 bits (263), Expect = 5e-23
Identities = 64/206 (31%), Positives = 102/206 (49%), Gaps = 19/206 (9%)
Query: 212 GAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVD 271
G ++A C F+P +VS S D + +WD +GK K L H +PV VD
Sbjct: 111 GHTNYAFCVNFNPQSNMIVSGSFDETVRIWDVTTGKCLKVLP--------AHSDPVTAVD 162
Query: 272 FSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTA 331
F+RD +I S S DG ++W T C++ L + V+ V FS +G +L + D+T
Sbjct: 163 FNRDGSLIVSSSYDGLCRIWDSGTGHCVKTLIDDENPPVSFVRFSPNGKFILVGTLDNTL 222
Query: 332 RIHGLKSGKMLKEFRGHTS---YVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFK 388
R+ + S K LK + GH + ++ A +G R+++ S D V +W+L + LQ
Sbjct: 223 RLWNISSAKFLKTYTGHVNAQYCISSAFSVTNGKRIVSGSEDNCVHMWELNSKKLLQ--- 279
Query: 389 PPPPLRGGDASVNSVFIFPKNTEHIV 414
L G +V +V P TE+++
Sbjct: 280 ---KLEGHTETVMNVACHP--TENLI 300
>At1g61210
Length = 282
Score = 90.9 bits (224), Expect = 2e-18
Identities = 56/200 (28%), Positives = 97/200 (48%), Gaps = 14/200 (7%)
Query: 190 TAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLK 249
T+ MK D Y +L G S + F +++ + G I++WD K+
Sbjct: 69 TSLMKNDAIAFYWQSLC-----GHTSAVDSVAFDSAEVLVLAGASSGVIKLWDVEEAKMV 123
Query: 250 KDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQG 309
+ F H V+F E +ASGS+D +K+W IR C++ + HS+G
Sbjct: 124 R--------AFTGHRSNCSAVEFHPFGEFLASGSSDANLKIWDIRKKGCIQTYK-GHSRG 174
Query: 310 VTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASS 369
++++ F+ DG ++S D+ ++ L +GK+L EF+ H + F + T S+
Sbjct: 175 ISTIRFTPDGRWVVSGGLDNVVKVWDLTAGKLLHEFKFHEGPIRSLDFHPLEFLLATGSA 234
Query: 370 DCTVKVWDLKTTDCLQTFKP 389
D TVK WDL+T + + + +P
Sbjct: 235 DRTVKFWDLETFELIGSTRP 254
Score = 73.9 bits (180), Expect = 2e-13
Identities = 59/262 (22%), Positives = 99/262 (37%), Gaps = 61/262 (23%)
Query: 260 FMMHDEPVLCVDFSRD-SEMIASGSTDGKIKVWRIRTAQCLRRLEH---------AHSQG 309
F+ H V C+ + S + +G D K+ +W I L + + H+
Sbjct: 31 FLAHSANVNCLSIGKKTSRLFITGGDDYKVNLWAIGKPTSLMKNDAIAFYWQSLCGHTSA 90
Query: 310 VTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASS 369
V SV+F +L+ + ++ ++ KM++ F GH S + F G + + SS
Sbjct: 91 VDSVAFDSAEVLVLAGASSGVIKLWDVEEAKMVRAFTGHRSNCSAVEFHPFGEFLASGSS 150
Query: 370 DCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQG 429
D +K+WD++ C+QT+ K S I T++
Sbjct: 151 DANLKIWDIRKKGCIQTY------------------------------KGHSRGISTIR- 179
Query: 430 QVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIG 489
+P G W+ G D + + +GKL H H+ I
Sbjct: 180 --------------------FTPDGRWVVSGGLDNVVKVWDLTAGKLLHEFKFHEGPIRS 219
Query: 490 VTHHPHRNLVATFSDDHTMKLW 511
+ HP L+AT S D T+K W
Sbjct: 220 LDFHPLEFLLATGSADRTVKFW 241
>At5g16750 WD40-repeat protein
Length = 876
Score = 89.4 bits (220), Expect = 4e-18
Identities = 48/162 (29%), Positives = 86/162 (52%), Gaps = 3/162 (1%)
Query: 228 YLVSCSIDGFIEVW--DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTD 285
+ VS S D ++VW D IS ++ + + HD+ + V +R+ ++ +GS D
Sbjct: 461 FFVSGSGDRTLKVWSLDGISEDSEEPINLKTRSVVAAHDKDINSVAVARNDSLVCTGSED 520
Query: 286 GKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEF 345
+WR+ + L+ H + + SV FS +++ S D T +I + G LK F
Sbjct: 521 RTASIWRLPDLVHVVTLK-GHKRRIFSVEFSTVDQCVMTASGDKTVKIWAISDGSCLKTF 579
Query: 346 RGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTF 387
GHTS V A+F DGT+ ++ +D +K+W++ T++C+ T+
Sbjct: 580 EGHTSSVLRASFITDGTQFVSCGADGLLKLWNVNTSECIATY 621
Score = 60.5 bits (145), Expect = 2e-09
Identities = 70/325 (21%), Positives = 119/325 (36%), Gaps = 42/325 (12%)
Query: 196 DVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQ 255
++ D +++ TI+ G SPD + L S I VWD + K +
Sbjct: 43 NIVDSTDSSVKSTIE-GESDTLTALALSPDDKLLFSAGHSRQIRVWDLETLKCIR----- 96
Query: 256 AEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSF 315
++ H+ PV+ + ++A+ D K+ VW + C H V+S+ F
Sbjct: 97 ---SWKGHEGPVMGMACHASGGLLATAGADRKVLVWDVDGGFCTHYFR-GHKGVVSSILF 152
Query: 316 SRDGSQ--LLSTSFDSTARIHGLKSG----KMLKEFRGHTSYVNDATFTNDGTRVITASS 369
D ++ L+S S D+T R+ L + K L H S V + DG + +A
Sbjct: 153 HPDSNKNILISGSDDATVRVWDLNAKNTEKKCLAIMEKHFSAVTSIALSEDGLTLFSAGR 212
Query: 370 DCTVKVWDLKTTDC------------LQTFKPPPPLRGGDASVN-------------SVF 404
D V +WDL C + T P AS++ + F
Sbjct: 213 DKVVNLWDLHDYSCKATVATYEVLEAVTTVSSGTPFASFVASLDQKKSKKKESDSQATYF 272
Query: 405 IFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDR 464
I + + SI + + + S + F AA + P + CV D+
Sbjct: 273 ITVGERGVVRIWKSEGSICLYEQKSSDITVSSDDEESKRGFTAAAMLPSDHGLLCVTADQ 332
Query: 465 NMYCFSYQSGKLEHLMTVHDKEIIG 489
+ +S +E V K ++G
Sbjct: 333 QFFFYSVVE-NVEETELVLSKRLVG 356
Score = 60.1 bits (144), Expect = 3e-09
Identities = 41/136 (30%), Positives = 64/136 (46%), Gaps = 10/136 (7%)
Query: 263 HDEPVLCVDFSRDS-EMIASGSTDGKIKVWRIRTAQ---------CLRRLEHAHSQGVTS 312
H+ +L V F++ S SGS D +KVW + R + AH + + S
Sbjct: 445 HNGDILAVAFAKKSFSFFVSGSGDRTLKVWSLDGISEDSEEPINLKTRSVVAAHDKDINS 504
Query: 313 VSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCT 372
V+ +R+ S + + S D TA I L + +GH + F+ V+TAS D T
Sbjct: 505 VAVARNDSLVCTGSEDRTASIWRLPDLVHVVTLKGHKRRIFSVEFSTVDQCVMTASGDKT 564
Query: 373 VKVWDLKTTDCLQTFK 388
VK+W + CL+TF+
Sbjct: 565 VKIWAISDGSCLKTFE 580
Score = 43.9 bits (102), Expect = 2e-04
Identities = 23/77 (29%), Positives = 37/77 (47%), Gaps = 8/77 (10%)
Query: 220 ARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMI 279
A F DG VSC DG +++W+ + + T+ H++ V + + +EMI
Sbjct: 589 ASFITDGTQFVSCGADGLLKLWNVNTS--------ECIATYDQHEDKVWALAVGKKTEMI 640
Query: 280 ASGSTDGKIKVWRIRTA 296
A+G D I +W TA
Sbjct: 641 ATGGGDAVINLWHDSTA 657
Score = 38.9 bits (89), Expect = 0.007
Identities = 61/333 (18%), Positives = 115/333 (34%), Gaps = 49/333 (14%)
Query: 223 SPDGQYLVSCSIDGFIEVWDYISGKLKKDLQ-YQAEE--TFMMHDEPVLCVDFSRDSEMI 279
S DG L S D + +WD K + Y+ E T + P S D +
Sbjct: 201 SEDGLTLFSAGRDKVVNLWDLHDYSCKATVATYEVLEAVTTVSSGTPFASFVASLDQKKS 260
Query: 280 ASGSTD------------GKIKVWRIRTAQCLRRLEHAH----------SQGVTSVSFSR 317
+D G +++W+ + CL + + +G T+ +
Sbjct: 261 KKKESDSQATYFITVGERGVVRIWKSEGSICLYEQKSSDITVSSDDEESKRGFTAAAMLP 320
Query: 318 DGSQLLSTSFDSTARIHGLKSGK------MLKEFRGHTSYVNDATFTNDGTRVITASSDC 371
LL + D + + + K G+ + D F D + + +++
Sbjct: 321 SDHGLLCVTADQQFFFYSVVENVEETELVLSKRLVGYNEEIADMKFLGDEEQFLAVATNL 380
Query: 372 T-VKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQ 430
V+V+D+ T C L G V S+ ++ ++++ + +
Sbjct: 381 EEVRVYDVATMSCSYV------LAGHKEVVLSLDTCVSSSGNVLIVTGSKDKTVRLWNAT 434
Query: 431 VVKSFSSGKREGGDFVAACISPKGEWIYCVGE-DRNMYCFSYQ----------SGKLEHL 479
G GD +A + K + G DR + +S + K +
Sbjct: 435 SKSCIGVGTGHNGDILAVAFAKKSFSFFVSGSGDRTLKVWSLDGISEDSEEPINLKTRSV 494
Query: 480 MTVHDKEIIGVTHHPHRNLVATFSDDHTMKLWK 512
+ HDK+I V + +LV T S+D T +W+
Sbjct: 495 VAAHDKDINSVAVARNDSLVCTGSEDRTASIWR 527
Score = 37.0 bits (84), Expect = 0.026
Identities = 35/184 (19%), Positives = 71/184 (38%), Gaps = 20/184 (10%)
Query: 334 HGLKSG----KMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKP 389
H LK + LK+F G ++ ++DG+ + A D + + D + T +
Sbjct: 4 HSLKKNYRCSRSLKQFYGGGPFI----VSSDGSFIACACGD-VINIVDSTDSSVKSTIE- 57
Query: 390 PPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDFVAAC 449
G++ + + + + + I + L+ +K S K G +
Sbjct: 58 ------GESDTLTALALSPDDKLLFSAGHSRQIRVWDLE--TLKCIRSWKGHEGPVMGMA 109
Query: 450 ISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHP--HRNLVATFSDDHT 507
G + G DR + + G H H + + HP ++N++ + SDD T
Sbjct: 110 CHASGGLLATAGADRKVLVWDVDGGFCTHYFRGHKGVVSSILFHPDSNKNILISGSDDAT 169
Query: 508 MKLW 511
+++W
Sbjct: 170 VRVW 173
Score = 34.3 bits (77), Expect = 0.17
Identities = 43/220 (19%), Positives = 85/220 (38%), Gaps = 22/220 (10%)
Query: 306 HSQGVTSVSFSRDGSQLLSTSFD-STARIHGLKSGKMLKEFRGHTSYV---NDATFTNDG 361
+++ + + F D Q L+ + + R++ + + GH V + ++
Sbjct: 357 YNEEIADMKFLGDEEQFLAVATNLEEVRVYDVATMSCSYVLAGHKEVVLSLDTCVSSSGN 416
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
++T S D TV++W+ + C+ G + + +V K+ V + +
Sbjct: 417 VLIVTGSKDKTVRLWNATSKSCIGVGT------GHNGDILAVAFAKKSFSFFVSGSGDRT 470
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAA-------CISPKGEWIYCVG-EDRNMYCFSYQS 473
+ + +L G S + VAA + + + C G EDR +
Sbjct: 471 LKVWSLDGISEDSEEPINLKTRSVVAAHDKDINSVAVARNDSLVCTGSEDRTASIWRLPD 530
Query: 474 GKLEHLMTV--HDKEIIGVTHHPHRNLVATFSDDHTMKLW 511
L H++T+ H + I V V T S D T+K+W
Sbjct: 531 --LVHVVTLKGHKRRIFSVEFSTVDQCVMTASGDKTVKIW 568
>At1g11160 hypothetical protein
Length = 974
Score = 89.0 bits (219), Expect = 6e-18
Identities = 50/178 (28%), Positives = 90/178 (50%), Gaps = 9/178 (5%)
Query: 212 GAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVD 271
G S + F+ + +++ + G I++WD K+ + F H V+
Sbjct: 5 GHTSPVDSVAFNSEEVLVLAGASSGVIKLWDLEESKMVR--------AFTGHRSNCSAVE 56
Query: 272 FSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTA 331
F E +ASGS+D ++VW R C++ + H++G++++ FS DG ++S D+
Sbjct: 57 FHPFGEFLASGSSDTNLRVWDTRKKGCIQTYK-GHTRGISTIEFSPDGRWVVSGGLDNVV 115
Query: 332 RIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKP 389
++ L +GK+L EF+ H + F + T S+D TVK WDL+T + + T +P
Sbjct: 116 KVWDLTAGKLLHEFKCHEGPIRSLDFHPLEFLLATGSADRTVKFWDLETFELIGTTRP 173
Score = 64.7 bits (156), Expect = 1e-10
Identities = 49/206 (23%), Positives = 80/206 (38%), Gaps = 51/206 (24%)
Query: 306 HSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVI 365
H+ V SV+F+ + +L+ + ++ L+ KM++ F GH S + F G +
Sbjct: 6 HTSPVDSVAFNSEEVLVLAGASSGVIKLWDLEESKMVRAFTGHRSNCSAVEFHPFGEFLA 65
Query: 366 TASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIM 425
+ SSD ++VWD + C+QT+ K + I
Sbjct: 66 SGSSDTNLRVWDTRKKGCIQTY------------------------------KGHTRGIS 95
Query: 426 TLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDK 485
T++ SP G W+ G D + + +GKL H H+
Sbjct: 96 TIE---------------------FSPDGRWVVSGGLDNVVKVWDLTAGKLLHEFKCHEG 134
Query: 486 EIIGVTHHPHRNLVATFSDDHTMKLW 511
I + HP L+AT S D T+K W
Sbjct: 135 PIRSLDFHPLEFLLATGSADRTVKFW 160
>At5g23430 unknown protein
Length = 837
Score = 86.3 bits (212), Expect = 4e-17
Identities = 51/179 (28%), Positives = 84/179 (46%), Gaps = 9/179 (5%)
Query: 211 FGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCV 270
+G S + F + + + G I++WD K+ + T H + V
Sbjct: 56 YGHSSGIDSVTFDASEVLVAAGAASGTIKLWDLEEAKIVR--------TLTGHRSNCISV 107
Query: 271 DFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDST 330
DF E ASGS D +K+W IR C+ + H++GV + F+ DG ++S D+
Sbjct: 108 DFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYK-GHTRGVNVLRFTPDGRWVVSGGEDNI 166
Query: 331 ARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKP 389
++ L +GK+L EF+ H + F + T S+D TVK WDL+T + + + P
Sbjct: 167 VKVWDLTAGKLLTEFKSHEGQIQSLDFHPHEFLLATGSADRTVKFWDLETFELIGSGGP 225
Score = 80.9 bits (198), Expect = 2e-15
Identities = 60/259 (23%), Positives = 101/259 (38%), Gaps = 54/259 (20%)
Query: 254 YQAEETFMMHDEPVLCVDFSR-DSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTS 312
Y+ +E F+ H V C+ R S ++ +G D K+ +W I + L + HS G+ S
Sbjct: 7 YKLQE-FVAHSAAVNCLKIGRKSSRVLVTGGEDHKVNLWAIGKPNAILSL-YGHSSGIDS 64
Query: 313 VSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCT 372
V+F + + + T ++ L+ K+++ GH S F G + S D
Sbjct: 65 VTFDASEVLVAAGAASGTIKLWDLEEAKIVRTLTGHRSNCISVDFHPFGEFFASGSLDTN 124
Query: 373 VKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVV 432
+K+WD++ C+ T+ +G VN +
Sbjct: 125 LKIWDIRKKGCIHTY------KGHTRGVNVLRF--------------------------- 151
Query: 433 KSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTH 492
+P G W+ GED + + +GKL H+ +I +
Sbjct: 152 ------------------TPDGRWVVSGGEDNIVKVWDLTAGKLLTEFKSHEGQIQSLDF 193
Query: 493 HPHRNLVATFSDDHTMKLW 511
HPH L+AT S D T+K W
Sbjct: 194 HPHEFLLATGSADRTVKFW 212
>At5g08390 katanin p80 subunit - like protein
Length = 823
Score = 84.7 bits (208), Expect = 1e-16
Identities = 60/253 (23%), Positives = 98/253 (38%), Gaps = 53/253 (20%)
Query: 260 FMMHDEPVLCVDFSR-DSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRD 318
F+ H V C+ R S ++ +G D K+ +W I + L + HS G+ SV+F
Sbjct: 105 FVAHSAAVNCLKIGRKSSRVLVTGGEDHKVNLWAIGKPNAILSL-YGHSSGIDSVTFDAS 163
Query: 319 GSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDL 378
+ + + T ++ L+ K+++ GH S F G + S D +K+WD+
Sbjct: 164 EGLVAAGAASGTIKLWDLEEAKVVRTLTGHRSNCVSVNFHPFGEFFASGSLDTNLKIWDI 223
Query: 379 KTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSG 438
+ C+ T+ +G VN +
Sbjct: 224 RKKGCIHTY------KGHTRGVNVLRF--------------------------------- 244
Query: 439 KREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNL 498
+P G WI GED + + +GKL H H+ +I + HPH L
Sbjct: 245 ------------TPDGRWIVSGGEDNVVKVWDLTAGKLLHEFKSHEGKIQSLDFHPHEFL 292
Query: 499 VATFSDDHTMKLW 511
+AT S D T+K W
Sbjct: 293 LATGSADKTVKFW 305
Score = 83.6 bits (205), Expect = 2e-16
Identities = 49/174 (28%), Positives = 82/174 (46%), Gaps = 9/174 (5%)
Query: 211 FGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCV 270
+G S + F + + + G I++WD K+ + T H + V
Sbjct: 149 YGHSSGIDSVTFDASEGLVAAGAASGTIKLWDLEEAKVVR--------TLTGHRSNCVSV 200
Query: 271 DFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDST 330
+F E ASGS D +K+W IR C+ + H++GV + F+ DG ++S D+
Sbjct: 201 NFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYK-GHTRGVNVLRFTPDGRWIVSGGEDNV 259
Query: 331 ARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCL 384
++ L +GK+L EF+ H + F + T S+D TVK WDL+T + +
Sbjct: 260 VKVWDLTAGKLLHEFKSHEGKIQSLDFHPHEFLLATGSADKTVKFWDLETFELI 313
Score = 73.2 bits (178), Expect = 3e-13
Identities = 40/126 (31%), Positives = 60/126 (46%), Gaps = 1/126 (0%)
Query: 263 HDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQL 322
H + V F ++A+G+ G IK+W + A+ +R L H SV+F G
Sbjct: 151 HSSGIDSVTFDASEGLVAAGAASGTIKLWDLEEAKVVRTLT-GHRSNCVSVNFHPFGEFF 209
Query: 323 LSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTD 382
S S D+ +I ++ + ++GHT VN FT DG +++ D VKVWDL
Sbjct: 210 ASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTPDGRWIVSGGEDNVVKVWDLTAGK 269
Query: 383 CLQTFK 388
L FK
Sbjct: 270 LLHEFK 275
>At2g21390 coatomer alpha subunit
Length = 1218
Score = 84.3 bits (207), Expect = 1e-16
Identities = 56/170 (32%), Positives = 81/170 (46%), Gaps = 10/170 (5%)
Query: 210 KFGAKSH-AECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVL 268
KF KS+ + F P ++++ G I++WDY G L + F H+ PV
Sbjct: 4 KFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLI--------DRFDEHEGPVR 55
Query: 269 CVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFD 328
V F + SG D KIKVW +T +CL L H + +V F + ++S S D
Sbjct: 56 GVHFHNSQPLFVSGGDDYKIKVWNYKTHRCLFTLL-GHLDYIRTVQFHHENPWIVSASDD 114
Query: 329 STARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDL 378
T RI +S + GH YV A+F V++AS D TV+VWD+
Sbjct: 115 QTIRIWNWQSRTCISVLTGHNHYVMCASFHPKEDLVVSASLDQTVRVWDI 164
Score = 54.7 bits (130), Expect = 1e-07
Identities = 47/227 (20%), Positives = 88/227 (38%), Gaps = 25/227 (11%)
Query: 286 GKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEF 345
G I++W R + R + H V V F +S D ++ K+ + L
Sbjct: 31 GVIQLWDYRMGTLIDRFDE-HEGPVRGVHFHNSQPLFVSGGDDYKIKVWNYKTHRCLFTL 89
Query: 346 RGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFI 405
GH Y+ F ++ +++AS D T+++W+ ++ C+ L G + V
Sbjct: 90 LGHLDYIRTVQFHHENPWIVSASDDQTIRIWNWQSRTCISV------LTGHNHYVMCASF 143
Query: 406 FPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRN 465
PK E +VV + G + K +S + F N
Sbjct: 144 HPK--EDLVVSASLDQTVRVWDIGALKKKSASPADDLMRF----------------SQMN 185
Query: 466 MYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLWK 512
F +++++ HD+ + + HP L+ + +DD +KLW+
Sbjct: 186 SDLFGGVDAIVKYVLEGHDRGVNWASFHPTLPLIVSGADDRQVKLWR 232
Score = 50.1 bits (118), Expect = 3e-06
Identities = 45/181 (24%), Positives = 72/181 (38%), Gaps = 30/181 (16%)
Query: 212 GAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVD 271
G + CA F P +VS S+D + VWD G LKK A+ D
Sbjct: 133 GHNHYVMCASFHPKEDLVVSASLDQTVRVWDI--GALKKKSASPAD-------------D 177
Query: 272 FSRDSEMIAS--GSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDS 329
R S+M + G D +K + H +GV SF ++S + D
Sbjct: 178 LMRFSQMNSDLFGGVDAIVKY-----------VLEGHDRGVNWASFHPTLPLIVSGADDR 226
Query: 330 TARIHGLKSGKM--LKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTF 387
++ + K + RGH + V+ F +++ S D +++VWD +QTF
Sbjct: 227 QVKLWRMNETKAWEVDTLRGHMNNVSSVMFHAKQDIIVSNSEDKSIRVWDATKRTGIQTF 286
Query: 388 K 388
+
Sbjct: 287 R 287
Score = 44.7 bits (104), Expect = 1e-04
Identities = 40/205 (19%), Positives = 73/205 (35%), Gaps = 51/205 (24%)
Query: 307 SQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVIT 366
S V +SF +L++ ++ + G ++ F H V F N ++
Sbjct: 9 SNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDRFDEHEGPVRGVHFHNSQPLFVS 68
Query: 367 ASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMT 426
D +KVW+ KT CL T H+ YI T
Sbjct: 69 GGDDYKIKVWNYKTHRCLFTL----------------------LGHLD--------YIRT 98
Query: 427 LQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKE 486
+Q + WI +D+ + +++QS ++T H+
Sbjct: 99 VQ---------------------FHHENPWIVSASDDQTIRIWNWQSRTCISVLTGHNHY 137
Query: 487 IIGVTHHPHRNLVATFSDDHTMKLW 511
++ + HP +LV + S D T+++W
Sbjct: 138 VMCASFHPKEDLVVSASLDQTVRVW 162
>At1g10580 putative pre-mRNA splicing factor
Length = 616
Score = 82.0 bits (201), Expect = 7e-16
Identities = 78/327 (23%), Positives = 134/327 (40%), Gaps = 25/327 (7%)
Query: 194 KQDVDDMY-PTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWD-YISGKLKKD 251
K + D Y P L HT K + F G L+S +D +++WD Y SGK +
Sbjct: 262 KANNDHCYIPKRLVHTWSGHTKGVSAIRFFPKQGHLLLSAGMDCKVKIWDVYNSGKCMR- 320
Query: 252 LQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVT 311
T+M H + V + FS D + D IK W T Q + V
Sbjct: 321 -------TYMGHAKAVRDICFSNDGSKFLTAGYDKNIKYWDTETGQVISTFSTGKIPYVV 373
Query: 312 SVSFSRDGSQLLSTSFDSTARIH-GLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSD 370
++ D +L + + +G++ +E+ H VN TF ++ R +T+S D
Sbjct: 374 KLNPDDDKQNILLAGMSDKKIVQWDINTGEVTQEYDQHLGAVNTITFVDNNRRFVTSSDD 433
Query: 371 CTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQ 430
+++VW+ ++ P S+ S+ + P N + + + I I + + +
Sbjct: 434 KSLRVWEFGIPVVIKYISEPHM-----HSMPSISVHP-NGNWLAAQSLDNQILIYSTRER 487
Query: 431 VVKSFSSGKREGGDFVA--AC---ISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDK 485
+ KR G VA AC SP G ++ + + + ++S K+ + H+
Sbjct: 488 F--QLNKKKRFAGHIVAGYACQVNFSPDGRFVMSGDGEGKCWFWDWKSCKVFRTLKCHNG 545
Query: 486 EIIGVTHHP-HRNLVATFSDDHTMKLW 511
IG HP ++ VAT D +K W
Sbjct: 546 VCIGAEWHPLEQSKVATCGWDGLIKYW 572
Score = 55.5 bits (132), Expect = 7e-08
Identities = 49/221 (22%), Positives = 87/221 (39%), Gaps = 60/221 (27%)
Query: 300 RRLEHA---HSQGVTSVSF-SRDGSQLLSTSFDSTARIHGL-KSGKMLKEFRGHTSYVND 354
+RL H H++GV+++ F + G LLS D +I + SGK ++ + GH V D
Sbjct: 272 KRLVHTWSGHTKGVSAIRFFPKQGHLLLSAGMDCKVKIWDVYNSGKCMRTYMGHAKAVRD 331
Query: 355 ATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIV 414
F+NDG++ +TA D +K WD +T
Sbjct: 332 ICFSNDGSKFLTAGYDKNIKYWDTET---------------------------------- 357
Query: 415 VCNKTSSIYIMTLQGQVVKSFSSGKREGGDFVAACISP---KGEWIYCVGEDRNMYCFSY 471
GQV+ +FS+GK ++P K + D+ + +
Sbjct: 358 --------------GQVISTFSTGKIP----YVVKLNPDDDKQNILLAGMSDKKIVQWDI 399
Query: 472 QSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLWK 512
+G++ H + +T + T SDD ++++W+
Sbjct: 400 NTGEVTQEYDQHLGAVNTITFVDNNRRFVTSSDDKSLRVWE 440
>At1g62020 hypothetical protein
Length = 1216
Score = 80.9 bits (198), Expect = 2e-15
Identities = 55/170 (32%), Positives = 80/170 (46%), Gaps = 10/170 (5%)
Query: 210 KFGAKSH-AECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVL 268
KF KS+ + F P ++++ G I++WDY G L + F H+ PV
Sbjct: 4 KFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLI--------DRFDEHEGPVR 55
Query: 269 CVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFD 328
V F + SG D KIKVW + +CL L H + +V F + ++S S D
Sbjct: 56 GVHFHNSQPLFVSGGDDYKIKVWNYKNHRCLFTLL-GHLDYIRTVQFHHEYPWIVSASDD 114
Query: 329 STARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDL 378
T RI +S + GH YV A+F V++AS D TV+VWD+
Sbjct: 115 QTIRIWNWQSRTCVSVLTGHNHYVMCASFHPKEDLVVSASLDQTVRVWDI 164
Score = 53.5 bits (127), Expect = 3e-07
Identities = 49/227 (21%), Positives = 93/227 (40%), Gaps = 25/227 (11%)
Query: 286 GKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEF 345
G I++W R + R + H V V F +S D ++ K+ + L
Sbjct: 31 GVIQLWDYRMGTLIDRFDE-HEGPVRGVHFHNSQPLFVSGGDDYKIKVWNYKNHRCLFTL 89
Query: 346 RGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFI 405
GH Y+ F ++ +++AS D T+++W+ ++ C+ L G + V
Sbjct: 90 LGHLDYIRTVQFHHEYPWIVSASDDQTIRIWNWQSRTCVSV------LTGHNHYVMCASF 143
Query: 406 FPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRN 465
PK E +VV ++S+ Q V+ + G +SP + + N
Sbjct: 144 HPK--EDLVV---SASL------DQTVRVWDIGALR-----KKTVSPADDIMRLT--QMN 185
Query: 466 MYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLWK 512
F +++++ HD+ + HP L+ + +DD +KLW+
Sbjct: 186 SDLFGGVDAIVKYVLEGHDRGVNWAAFHPTLPLIVSGADDRQVKLWR 232
Score = 48.1 bits (113), Expect = 1e-05
Identities = 44/205 (21%), Positives = 78/205 (37%), Gaps = 36/205 (17%)
Query: 212 GAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVD 271
G + +F + ++VS S D I +W++ S L H+ V+C
Sbjct: 91 GHLDYIRTVQFHHEYPWIVSASDDQTIRIWNWQSRTCVSVLTG--------HNHYVMCAS 142
Query: 272 FSRDSEMIASGSTDGKIKVWRIRT--------AQCLRRLEH------------------A 305
F +++ S S D ++VW I A + RL
Sbjct: 143 FHPKEDLVVSASLDQTVRVWDIGALRKKTVSPADDIMRLTQMNSDLFGGVDAIVKYVLEG 202
Query: 306 HSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKM--LKEFRGHTSYVNDATFTNDGTR 363
H +GV +F ++S + D ++ + K + RGH + V+ F
Sbjct: 203 HDRGVNWAAFHPTLPLIVSGADDRQVKLWRMNETKAWEVDTLRGHMNNVSSVMFHAKQDI 262
Query: 364 VITASSDCTVKVWDLKTTDCLQTFK 388
+++ S D +++VWD LQTF+
Sbjct: 263 IVSNSEDKSIRVWDATKRTGLQTFR 287
Score = 40.0 bits (92), Expect = 0.003
Identities = 34/174 (19%), Positives = 75/174 (42%), Gaps = 15/174 (8%)
Query: 341 MLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTF-KPPPPLRGGDA- 398
ML +F ++ V +F ++ + +++WD + + F + P+RG
Sbjct: 1 MLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDRFDEHEGPVRGVHFH 60
Query: 399 SVNSVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDFVAAC-ISPKGEWI 457
+ +F+ + I V N + + TL G + D++ + WI
Sbjct: 61 NSQPLFVSGGDDYKIKVWNYKNHRCLFTLLGHL------------DYIRTVQFHHEYPWI 108
Query: 458 YCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLW 511
+D+ + +++QS ++T H+ ++ + HP +LV + S D T+++W
Sbjct: 109 VSASDDQTIRIWNWQSRTCVSVLTGHNHYVMCASFHPKEDLVVSASLDQTVRVW 162
>At1g48630 unknown protein
Length = 326
Score = 80.9 bits (198), Expect = 2e-15
Identities = 49/160 (30%), Positives = 80/160 (49%), Gaps = 13/160 (8%)
Query: 223 SPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASG 282
S DGQ+ +S S DG + +WD +G ++ F+ H + VL V FS D+ I S
Sbjct: 72 SSDGQFALSGSWDGELRLWDLATG--------ESTRRFVGHTKDVLSVAFSTDNRQIVSA 123
Query: 283 STDGKIKVWRIRTAQCLRRLEHA--HSQGVTSVSFSRDG--SQLLSTSFDSTARIHGLKS 338
S D IK+W +C + A H + V+ V FS + ++S S+D T ++ L++
Sbjct: 124 SRDRTIKLWN-TLGECKYTISEADGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQN 182
Query: 339 GKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDL 378
K+ GH+ Y+N + DG+ + D + +WDL
Sbjct: 183 CKLRNTLAGHSGYLNTVAVSPDGSLCASGGKDGVILLWDL 222
Score = 71.2 bits (173), Expect = 1e-12
Identities = 65/251 (25%), Positives = 113/251 (44%), Gaps = 23/251 (9%)
Query: 229 LVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMM-HDEPVLCVDFSRDSEMIASGSTDGK 287
+V+ S D I +W K+D Y + M H V V S D + SGS DG+
Sbjct: 31 IVTSSRDKSIILWKLT----KEDKSYGVAQRRMTGHSHFVQDVVLSSDGQFALSGSWDGE 86
Query: 288 IKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGL--KSGKMLKEF 345
+++W + T + RR H++ V SV+FS D Q++S S D T ++ + + E
Sbjct: 87 LRLWDLATGESTRRFV-GHTKDVLSVAFSTDNRQIVSASRDRTIKLWNTLGECKYTISEA 145
Query: 346 RGHTSYVNDATFTNDG--TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSV 403
GH +V+ F+ + +++AS D TVKVW+L+ T L G +N+V
Sbjct: 146 DGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQNCKLRNT------LAGHSGYLNTV 199
Query: 404 FIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDFV-AACISPKGEWIYCVGE 462
+ P + + + ++ + K +S E G + + C SP W+ C
Sbjct: 200 AVSPDGS--LCASGGKDGVILLWDLAEGKKLYS---LEAGSIIHSLCFSPNRYWL-CAAT 253
Query: 463 DRNMYCFSYQS 473
+ ++ + +S
Sbjct: 254 ENSIRIWDLES 264
Score = 65.5 bits (158), Expect = 7e-11
Identities = 58/224 (25%), Positives = 95/224 (41%), Gaps = 14/224 (6%)
Query: 259 TFMMHDEPVLCVDFSRD-SEMIASGSTDGKIKVWRI----RTAQCLRRLEHAHSQGVTSV 313
T H + V + D S++I + S D I +W++ ++ +R HS V V
Sbjct: 10 TMCAHTDMVTAIATPVDNSDVIVTSSRDKSIILWKLTKEDKSYGVAQRRMTGHSHFVQDV 69
Query: 314 SFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTV 373
S DG LS S+D R+ L +G+ + F GHT V F+ D ++++AS D T+
Sbjct: 70 VLSSDGQFALSGSWDGELRLWDLATGESTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTI 129
Query: 374 KVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNT--EHIVVCNKTSSIYIMTLQGQV 431
K+W+ +C T G S F NT IV + ++ + LQ
Sbjct: 130 KLWN-TLGECKYTISE----ADGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQNCK 184
Query: 432 VKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGK 475
+++ +G G +SP G G+D + + GK
Sbjct: 185 LRNTLAG--HSGYLNTVAVSPDGSLCASGGKDGVILLWDLAEGK 226
Score = 47.0 bits (110), Expect = 3e-05
Identities = 46/188 (24%), Positives = 80/188 (42%), Gaps = 32/188 (17%)
Query: 212 GAKSHAECARFSPDGQY--LVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLC 269
G K C RFSP+ +VS S D ++VW+ + KL+ T H +
Sbjct: 147 GHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQNCKLRN--------TLAGHSGYLNT 198
Query: 270 VDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDS 329
V S D + ASG DG I +W + + L LE + S+ FS + L + + ++
Sbjct: 199 VAVSPDGSLCASGGKDGVILLWDLAEGKKLYSLEAGSI--IHSLCFSPNRYWLCAAT-EN 255
Query: 330 TARIHGLKSGKMLKEFR-----------GHTS--------YVNDATFTNDGTRVITASSD 370
+ RI L+S ++++ + G T Y ++ DG + + +D
Sbjct: 256 SIRIWDLESKSVVEDLKVDLKAEAEKTDGSTGIGNKTKVIYCTSLNWSADGNTLFSGYTD 315
Query: 371 CTVKVWDL 378
++VW +
Sbjct: 316 GVIRVWGI 323
Score = 29.3 bits (64), Expect = 5.4
Identities = 24/107 (22%), Positives = 45/107 (41%), Gaps = 5/107 (4%)
Query: 409 NTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKRE---GGDFVA-ACISPKGEWIYCVGEDR 464
N++ IV ++ SI + L + KS+ +R FV +S G++ D
Sbjct: 27 NSDVIVTSSRDKSIILWKLTKED-KSYGVAQRRMTGHSHFVQDVVLSSDGQFALSGSWDG 85
Query: 465 NMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLW 511
+ + +G+ H K+++ V + + S D T+KLW
Sbjct: 86 ELRLWDLATGESTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTIKLW 132
>At3g18130 protein kinase C-receptor/G-protein, putative
Length = 326
Score = 79.0 bits (193), Expect = 6e-15
Identities = 50/171 (29%), Positives = 80/171 (46%), Gaps = 13/171 (7%)
Query: 212 GAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVD 271
G E S DGQ+ +S S DG + +WD +G + F+ H + VL V
Sbjct: 61 GHSHFVEDVVLSSDGQFALSGSWDGELRLWDLATG--------ETTRRFVGHTKDVLSVA 112
Query: 272 FSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEH--AHSQGVTSVSFSRDG--SQLLSTSF 327
FS D+ I S S D IK+W +C + H + V+ V FS + ++S S+
Sbjct: 113 FSTDNRQIVSASRDRTIKLWN-TLGECKYTISEGDGHKEWVSCVRFSPNTLVPTIVSASW 171
Query: 328 DSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDL 378
D T ++ L++ K+ GH+ Y+N + DG+ + D + +WDL
Sbjct: 172 DKTVKVWNLQNCKLRNSLVGHSGYLNTVAVSPDGSLCASGGKDGVILLWDL 222
Score = 69.3 bits (168), Expect = 5e-12
Identities = 67/252 (26%), Positives = 114/252 (44%), Gaps = 25/252 (9%)
Query: 229 LVSCSIDGFIEVWDYISGKLKKDLQYQ--AEETFMMHDEPVLCVDFSRDSEMIASGSTDG 286
+V+ S D I +W KL KD + A+ H V V S D + SGS DG
Sbjct: 31 IVTASRDKSIILW-----KLTKDDKSYGVAQRRLTGHSHFVEDVVLSSDGQFALSGSWDG 85
Query: 287 KIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGL--KSGKMLKE 344
++++W + T + RR H++ V SV+FS D Q++S S D T ++ + + E
Sbjct: 86 ELRLWDLATGETTRRFV-GHTKDVLSVAFSTDNRQIVSASRDRTIKLWNTLGECKYTISE 144
Query: 345 FRGHTSYVNDATFTNDG--TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNS 402
GH +V+ F+ + +++AS D TVKVW+ LQ K L G +N+
Sbjct: 145 GDGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWN------LQNCKLRNSLVGHSGYLNT 198
Query: 403 VFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDFV-AACISPKGEWIYCVG 461
V + P + + + ++ + K +S E G + + C SP W+ C
Sbjct: 199 VAVSPDGS--LCASGGKDGVILLWDLAEGKKLYS---LEAGSIIHSLCFSPNRYWL-CAA 252
Query: 462 EDRNMYCFSYQS 473
+ ++ + +S
Sbjct: 253 TENSIRIWDLES 264
Score = 63.2 bits (152), Expect = 3e-10
Identities = 54/207 (26%), Positives = 89/207 (42%), Gaps = 13/207 (6%)
Query: 275 DSEMIASGSTDGKIKVWRI----RTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDST 330
+S++I + S D I +W++ ++ +R HS V V S DG LS S+D
Sbjct: 27 NSDIIVTASRDKSIILWKLTKDDKSYGVAQRRLTGHSHFVEDVVLSSDGQFALSGSWDGE 86
Query: 331 ARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKPP 390
R+ L +G+ + F GHT V F+ D ++++AS D T+K+W+ +C T
Sbjct: 87 LRLWDLATGETTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTIKLWN-TLGECKYTISE- 144
Query: 391 PPLRGGDASVNSVFIFPKNT--EHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDFVAA 448
G S F NT IV + ++ + LQ +++ G G
Sbjct: 145 ---GDGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQNCKLRNSLVG--HSGYLNTV 199
Query: 449 CISPKGEWIYCVGEDRNMYCFSYQSGK 475
+SP G G+D + + GK
Sbjct: 200 AVSPDGSLCASGGKDGVILLWDLAEGK 226
Score = 46.6 bits (109), Expect = 3e-05
Identities = 45/188 (23%), Positives = 80/188 (41%), Gaps = 32/188 (17%)
Query: 212 GAKSHAECARFSPDGQY--LVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLC 269
G K C RFSP+ +VS S D ++VW+ + KL+ L + H +
Sbjct: 147 GHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQNCKLRNSL--------VGHSGYLNT 198
Query: 270 VDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDS 329
V S D + ASG DG I +W + + L LE + S+ FS + L + + ++
Sbjct: 199 VAVSPDGSLCASGGKDGVILLWDLAEGKKLYSLEAGSI--IHSLCFSPNRYWLCAAT-EN 255
Query: 330 TARIHGLKSGKMLKEFRGHTS-------------------YVNDATFTNDGTRVITASSD 370
+ RI L+S ++++ + Y ++ DG+ + + +D
Sbjct: 256 SIRIWDLESKSVVEDLKVDLKSEAEKNEGGVGTGNQKKVIYCTSLNWSADGSTLFSGYTD 315
Query: 371 CTVKVWDL 378
V+VW +
Sbjct: 316 GVVRVWGI 323
Score = 29.3 bits (64), Expect = 5.4
Identities = 24/107 (22%), Positives = 44/107 (40%), Gaps = 5/107 (4%)
Query: 409 NTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKRE---GGDFVA-ACISPKGEWIYCVGEDR 464
N++ IV ++ SI + L KS+ +R FV +S G++ D
Sbjct: 27 NSDIIVTASRDKSIILWKLTKDD-KSYGVAQRRLTGHSHFVEDVVLSSDGQFALSGSWDG 85
Query: 465 NMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLW 511
+ + +G+ H K+++ V + + S D T+KLW
Sbjct: 86 ELRLWDLATGETTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTIKLW 132
>At1g18080 putative guanine nucleotide-binding protein beta
subunit
Length = 327
Score = 79.0 bits (193), Expect = 6e-15
Identities = 53/172 (30%), Positives = 79/172 (45%), Gaps = 14/172 (8%)
Query: 212 GAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVD 271
G E S DGQ+ +S S DG + +WD +G + F+ H + VL V
Sbjct: 61 GHSHFVEDVVLSSDGQFALSGSWDGELRLWDLAAG--------VSTRRFVGHTKDVLSVA 112
Query: 272 FSRDSEMIASGSTDGKIKVWRIRTAQCLRRLE---HAHSQGVTSVSFSRDGSQ--LLSTS 326
FS D+ I S S D IK+W +C + H V+ V FS + Q ++S S
Sbjct: 113 FSLDNRQIVSASRDRTIKLWN-TLGECKYTISEGGEGHRDWVSCVRFSPNTLQPTIVSAS 171
Query: 327 FDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDL 378
+D T ++ L + K+ GHT YV+ + DG+ + D V +WDL
Sbjct: 172 WDKTVKVWNLSNCKLRSTLAGHTGYVSTVAVSPDGSLCASGGKDGVVLLWDL 223
Score = 64.3 bits (155), Expect = 2e-10
Identities = 65/262 (24%), Positives = 111/262 (41%), Gaps = 19/262 (7%)
Query: 259 TFMMHDEPVLCVDFSRDS-EMIASGSTDGKIKVWRI----RTAQCLRRLEHAHSQGVTSV 313
T H + V + D+ ++I S S D I +W++ + +R HS V V
Sbjct: 10 TMRAHTDMVTAIATPIDNADIIVSASRDKSIILWKLTKDDKAYGVAQRRLTGHSHFVEDV 69
Query: 314 SFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTV 373
S DG LS S+D R+ L +G + F GHT V F+ D ++++AS D T+
Sbjct: 70 VLSSDGQFALSGSWDGELRLWDLAAGVSTRRFVGHTKDVLSVAFSLDNRQIVSASRDRTI 129
Query: 374 KVWDLKTTDCLQTFKPPPPLRGGDASVN--SVFIFPKNT--EHIVVCNKTSSIYIMTLQG 429
K+W+ +C T GG+ + S F NT IV + ++ + L
Sbjct: 130 KLWN-TLGECKYTIS-----EGGEGHRDWVSCVRFSPNTLQPTIVSASWDKTVKVWNLSN 183
Query: 430 QVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIG 489
++S +G G +SP G G+D + + GK + + + I
Sbjct: 184 CKLRSTLAG--HTGYVSTVAVSPDGSLCASGGKDGVVLLWDLAEGKKLYSLEA-NSVIHA 240
Query: 490 VTHHPHRNLVATFSDDHTMKLW 511
+ P+R + + +H +K+W
Sbjct: 241 LCFSPNRYWLCA-ATEHGIKIW 261
Score = 62.8 bits (151), Expect = 4e-10
Identities = 66/253 (26%), Positives = 110/253 (43%), Gaps = 26/253 (10%)
Query: 229 LVSCSIDGFIEVWDYISGKLKKDLQYQ--AEETFMMHDEPVLCVDFSRDSEMIASGSTDG 286
+VS S D I +W KL KD + A+ H V V S D + SGS DG
Sbjct: 31 IVSASRDKSIILW-----KLTKDDKAYGVAQRRLTGHSHFVEDVVLSSDGQFALSGSWDG 85
Query: 287 KIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIH---GLKSGKMLK 343
++++W + RR H++ V SV+FS D Q++S S D T ++ G + +
Sbjct: 86 ELRLWDLAAGVSTRRFV-GHTKDVLSVAFSLDNRQIVSASRDRTIKLWNTLGECKYTISE 144
Query: 344 EFRGHTSYVNDATFTNDGTR--VITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVN 401
GH +V+ F+ + + +++AS D TVKVW+L T L G V+
Sbjct: 145 GGEGHRDWVSCVRFSPNTLQPTIVSASWDKTVKVWNLSNCKLRST------LAGHTGYVS 198
Query: 402 SVFIFPKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDFV-AACISPKGEWIYCV 460
+V + P + + + ++ + K +S E + A C SP W+ C
Sbjct: 199 TVAVSPDGS--LCASGGKDGVVLLWDLAEGKKLYS---LEANSVIHALCFSPNRYWL-CA 252
Query: 461 GEDRNMYCFSYQS 473
+ + + +S
Sbjct: 253 ATEHGIKIWDLES 265
Score = 44.7 bits (104), Expect = 1e-04
Identities = 45/195 (23%), Positives = 81/195 (41%), Gaps = 33/195 (16%)
Query: 207 HTIKFGAKSHAE---CARFSPDGQY--LVSCSIDGFIEVWDYISGKLKKDLQYQAEETFM 261
+TI G + H + C RFSP+ +VS S D ++VW+ + KL+ T
Sbjct: 140 YTISEGGEGHRDWVSCVRFSPNTLQPTIVSASWDKTVKVWNLSNCKLRS--------TLA 191
Query: 262 MHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQ 321
H V V S D + ASG DG + +W + + L LE + + ++ FS +
Sbjct: 192 GHTGYVSTVAVSPDGSLCASGGKDGVVLLWDLAEGKKLYSLEA--NSVIHALCFSPNRYW 249
Query: 322 LLSTS--------FDSTARIHGLKSGKMLKEFRGHTS----------YVNDATFTNDGTR 363
L + + +S + + LK + + S Y ++ DG+
Sbjct: 250 LCAATEHGIKIWDLESKSIVEDLKVDLKAEAEKADNSGPAATKRKVIYCTSLNWSADGST 309
Query: 364 VITASSDCTVKVWDL 378
+ + +D ++VW +
Sbjct: 310 LFSGYTDGVIRVWGI 324
>At2g41500 putative U4/U6 small nuclear ribonucleoprotein
Length = 554
Score = 78.6 bits (192), Expect = 8e-15
Identities = 71/297 (23%), Positives = 117/297 (38%), Gaps = 31/297 (10%)
Query: 222 FSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMM---HDEPVLCVDFSRDSEM 278
FS DG+ L +CS+ G ++W+ Q T + H E V FS +
Sbjct: 263 FSRDGKILATCSLSGVTKLWE----------MPQVTNTIAVLKDHKERATDVVFSPVDDC 312
Query: 279 IASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKS 338
+A+ S D K+W+ T L + H + V+F G L +TS+D T R+ + +
Sbjct: 313 LATASADRTAKLWK--TDGTLLQTFEGHLDRLARVAFHPSGKYLGTTSYDKTWRLWDINT 370
Query: 339 GKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKPP-PPLRGGD 397
G L GH+ V F DG + D +VWDL+T + F+ P+ +
Sbjct: 371 GAELLLQEGHSRSVYGIAFQQDGALAASCGLDSLARVWDLRTGRSILVFQGHIKPVFSVN 430
Query: 398 ASVNSVFIF---PKNTEHIVVCNKTSSIYIMTLQGQVVKSFSSGKREGGDFVAACISPKG 454
S N + N I S+YI+ +V +E G
Sbjct: 431 FSPNGYHLASGGEDNQCRIWDLRMRKSLYIIPAHANLVSQVKYEPQE------------G 478
Query: 455 EWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLW 511
++ D + +S + L + H+ ++ + + +AT S D T+KLW
Sbjct: 479 YFLATASYDMKVNIWSGRDFSLVKSLAGHESKVASLDITADSSCIATVSHDRTIKLW 535
Score = 76.3 bits (186), Expect = 4e-14
Identities = 47/162 (29%), Positives = 75/162 (46%), Gaps = 10/162 (6%)
Query: 222 FSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIAS 281
F DG SC +D VWD +G+ + F H +PV V+FS + +AS
Sbjct: 389 FQQDGALAASCGLDSLARVWDLRTGR--------SILVFQGHIKPVFSVNFSPNGYHLAS 440
Query: 282 GSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFS-RDGSQLLSTSFDSTARIHGLKSGK 340
G D + ++W +R + L + AH+ V+ V + ++G L + S+D I +
Sbjct: 441 GGEDNQCRIWDLRMRKSLYIIP-AHANLVSQVKYEPQEGYFLATASYDMKVNIWSGRDFS 499
Query: 341 MLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTD 382
++K GH S V T D + + T S D T+K+W D
Sbjct: 500 LVKSLAGHESKVASLDITADSSCIATVSHDRTIKLWTSSGND 541
Score = 59.3 bits (142), Expect = 5e-09
Identities = 59/250 (23%), Positives = 99/250 (39%), Gaps = 13/250 (5%)
Query: 264 DEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLL 323
D P+ FSRD +++A+ S G K+W + + H + T V FS L
Sbjct: 255 DRPLTGCSFSRDGKILATCSLSGVTKLWEMPQVTNTIAVLKDHKERATDVVFSPVDDCLA 314
Query: 324 STSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDC 383
+ S D TA++ G +L+ F GH + F G + T S D T ++WD+ T
Sbjct: 315 TASADRTAKL-WKTDGTLLQTFEGHLDRLARVAFHPSGKYLGTTSYDKTWRLWDINTGAE 373
Query: 384 LQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQ-GQVVKSFSSGKREG 442
L L+ G + F ++ C S + L+ G+ + F +
Sbjct: 374 LL-------LQEGHSRSVYGIAFQQDGALAASCGLDSLARVWDLRTGRSILVFQGHIK-- 424
Query: 443 GDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRN-LVAT 501
+ SP G + GED + + K +++ H + V + P +AT
Sbjct: 425 -PVFSVNFSPNGYHLASGGEDNQCRIWDLRMRKSLYIIPAHANLVSQVKYEPQEGYFLAT 483
Query: 502 FSDDHTMKLW 511
S D + +W
Sbjct: 484 ASYDMKVNIW 493
>At5g64730 unknown protein
Length = 299
Score = 78.2 bits (191), Expect = 1e-14
Identities = 73/324 (22%), Positives = 131/324 (39%), Gaps = 45/324 (13%)
Query: 202 PTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFM 261
PT +H +K G + ARF+ DG Y ++C D I +W+ G L K T+
Sbjct: 7 PTKEAHILK-GHEGAVLAARFNGDGNYALTCGKDRTIRLWNPHRGILIK--------TYK 57
Query: 262 MHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQ 321
H V V + D+ S D ++ W + T + +R+ H V +V F+ S
Sbjct: 58 SHGREVRDVHVTSDNAKFCSCGGDRQVYYWDVSTGRVIRKFR-GHDGEVNAVKFNDSSSV 116
Query: 322 LLSTSFDSTARI-----HGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVW 376
++S FD + R+ H ++ +++ F V T T +I S D TV+ +
Sbjct: 117 VVSAGFDRSLRVWDCRSHSVEPVQIIDTF---LDTVMSVVLTK--TEIIGGSVDGTVRTF 171
Query: 377 DLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVC--------NKTSSIYIMTLQ 428
D++ + P VN + I + C ++T+ + +
Sbjct: 172 DMRIGREMSDNLGQP--------VNCISISNDGNCVLAGCLDSTLRLLDRTTGELLQVYK 223
Query: 429 GQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEII 488
G + KSF + C++ + ED ++ + K+ HD +
Sbjct: 224 GHISKSFKTD---------CCLTNSDAHVIGGSEDGLVFFWDLVDAKVLSKFRAHDLVVT 274
Query: 489 GVTHHPHRNLVATFSDDHTMKLWK 512
V++HP + + T S D T+++WK
Sbjct: 275 SVSYHPKEDCMLTSSVDGTIRVWK 298
>At5g08560 WD-repeat protein-like
Length = 589
Score = 77.0 bits (188), Expect = 2e-14
Identities = 102/510 (20%), Positives = 208/510 (40%), Gaps = 57/510 (11%)
Query: 26 QTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILPQVSQLKLPRNTLED-----LYEQ 80
+T L+ E +SL+ +I+ F+ + G+WD + + ++ P L EQ
Sbjct: 83 KTGAMLEEESGISLHN-STIKLFLQQVKDGKWDQSVKTLHRIGFPDEKAVKAASFLLLEQ 141
Query: 81 IVLEMIELRELDTARAILRQTQVMGVMKQEQPERY-LRLEHLLVRTYFDPNEAYQESTKE 139
LE +++ ++ A LR P R + H L + P+ +T
Sbjct: 142 KFLEFLKVEKIADALRTLRNEMA--------PLRINTKRVHELASSLISPSSFISHTTST 193
Query: 140 KRRAQI---------AQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFDLFRGT 190
+ + Q + ++P RL L+ +L Q+ + DL
Sbjct: 194 PGKESVNSRSKVLEELQTLLPASVIIPEKRLECLVENSLHIQRDSCVFHNTLDSDL--SL 251
Query: 191 AAMKQDVDDMYPTNLSHTIKFGAKSHAE---CARFSPDGQYLVSCSIDGFIEVWDYISGK 247
+ Q P+ + ++ SH + +FS +G+YL S S D +W+
Sbjct: 252 YSDHQCGKHQIPSQTAQILE----SHTDEVWFLQFSHNGKYLASSSKDQTAIIWE----- 302
Query: 248 LKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHS 307
+ D + T + H +PV+ + +S D + + + I+ W + + C+ E
Sbjct: 303 ISADGHISLKHTLVGHHKPVIAILWSPDDRQVLTCGAEEVIRRWDVDSGDCVHMYEKG-- 360
Query: 308 QGVTSVS--FSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRG-HTSYVNDATFTNDGTRV 364
G++ +S + DG +++ D + + L G+ + ++G T V+D T+DG +
Sbjct: 361 -GISPISCGWYPDGQGIIAGMTDRSICMWDL-DGREKECWKGQRTQKVSDIAMTDDGKWL 418
Query: 365 ITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYI 424
++ D + ++D + T L + + S F + ++I+V I +
Sbjct: 419 VSVCKDSVISLFDREAT--------VERLIEEEDMITS-FSLSNDNKYILVNLLNQEIRL 469
Query: 425 MTLQG--QVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTV 482
++G ++V + KR + K +I ED +Y + +GKL +
Sbjct: 470 WNIEGDPKIVSRYKGHKRSRFIIRSCFGGYKQAFIASGSEDSQVYIWHRSTGKLIVELPG 529
Query: 483 HDKEIIGVTHHP-HRNLVATFSDDHTMKLW 511
H + V+ P + +++A+ SDD T+++W
Sbjct: 530 HAGAVNCVSWSPTNLHMLASASDDGTIRIW 559
>At2g43770 putative splicing factor
Length = 343
Score = 76.6 bits (187), Expect = 3e-14
Identities = 61/259 (23%), Positives = 108/259 (41%), Gaps = 21/259 (8%)
Query: 263 HDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQL 322
H V + F+ +IASGS D +I +WR+ + H + + ++ DGSQ+
Sbjct: 52 HPSAVYTMKFNPAGTLIASGSHDREIFLWRVHGDCKNFMVLKGHKNAILDLHWTSDGSQI 111
Query: 323 LSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG-TRVITASSDCTVKVWDLKTT 381
+S S D T R +++GK +K+ H+S+VN T G +I+ S D T K+WD++
Sbjct: 112 VSASPDKTVRAWDVETGKQIKKMAEHSSFVNSCCPTRRGPPLIISGSDDGTAKLWDMRQR 171
Query: 382 DCLQTFKPPPPLRGGDASVNSVFIFPKNTEH-IVVCNKTSSIYIMTLQGQVVKSFSSGKR 440
+QTF + S + IF ++ + V + MTL+G
Sbjct: 172 GAIQTFPDKYQITAVSFSDAADKIFTGGVDNDVKVWDLRKGEATMTLEG----------- 220
Query: 441 EGGDFVAACISPKGEWIYCVGEDRNMYCFSYQ----SGKLEHLMTVH----DKEIIGVTH 492
+SP G ++ G D + + + + + H +K ++ +
Sbjct: 221 HQDTITGMSLSPDGSYLLTNGMDNKLCVWDMRPYAPQNRCVKIFEGHQHNFEKNLLKCSW 280
Query: 493 HPHRNLVATFSDDHTMKLW 511
P V S D + +W
Sbjct: 281 SPDGTKVTAGSSDRMVHIW 299
Score = 69.7 bits (169), Expect = 4e-12
Identities = 64/246 (26%), Positives = 100/246 (40%), Gaps = 52/246 (21%)
Query: 222 FSPDGQYLVSCSIDGFIEVWDYISGK-LKK------------------------------ 250
++ DG +VS S D + WD +GK +KK
Sbjct: 104 WTSDGSQIVSASPDKTVRAWDVETGKQIKKMAEHSSFVNSCCPTRRGPPLIISGSDDGTA 163
Query: 251 ---DLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHS 307
D++ + + V FS ++ I +G D +KVW +R + LE H
Sbjct: 164 KLWDMRQRGAIQTFPDKYQITAVSFSDAADKIFTGGVDNDVKVWDLRKGEATMTLE-GHQ 222
Query: 308 QGVTSVSFSRDGSQLLSTSFDSTARIHGLK----SGKMLKEFRGH----TSYVNDATFTN 359
+T +S S DGS LL+ D+ + ++ + +K F GH + +++
Sbjct: 223 DTITGMSLSPDGSYLLTNGMDNKLCVWDMRPYAPQNRCVKIFEGHQHNFEKNLLKCSWSP 282
Query: 360 DGTRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIV-VCNK 418
DGT+V SSD V +WD TT +K P G SVN P TE I+ C+
Sbjct: 283 DGTKVTAGSSDRMVHIWD--TTSRRTIYKLP----GHTGSVNECVFHP--TEPIIGSCSS 334
Query: 419 TSSIYI 424
+IY+
Sbjct: 335 DKNIYL 340
Score = 52.0 bits (123), Expect = 8e-07
Identities = 39/175 (22%), Positives = 72/175 (40%), Gaps = 15/175 (8%)
Query: 211 FGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQYQAEETFMMHDEPVLCV 270
F K FS + + +D ++VWD G +A T H + + +
Sbjct: 177 FPDKYQITAVSFSDAADKIFTGGVDNDVKVWDLRKG--------EATMTLEGHQDTITGM 228
Query: 271 DFSRDSEMIASGSTDGKIKVWRIRTA----QCLRRLE---HAHSQGVTSVSFSRDGSQLL 323
S D + + D K+ VW +R +C++ E H + + S+S DG+++
Sbjct: 229 SLSPDGSYLLTNGMDNKLCVWDMRPYAPQNRCVKIFEGHQHNFEKNLLKCSWSPDGTKVT 288
Query: 324 STSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASSDCTVKVWDL 378
+ S D I S + + + GHT VN+ F + + SSD + + ++
Sbjct: 289 AGSSDRMVHIWDTTSRRTIYKLPGHTGSVNECVFHPTEPIIGSCSSDKNIYLGEI 343
>At4g29830 unknown protein
Length = 321
Score = 75.9 bits (185), Expect = 5e-14
Identities = 62/198 (31%), Positives = 92/198 (46%), Gaps = 12/198 (6%)
Query: 196 DVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYL-VSCSIDGFIEVWDYISGKLKKDLQY 254
DVD TN + + S +F P G L V+ +++WD S +L L
Sbjct: 88 DVD----TNATIAVLEAPPSEVWGMQFEPKGTILAVAGGSSASVKLWDTASWRLISTLSI 143
Query: 255 QAEETFMMHDEP-----VLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQG 309
+ D+ VL V +S + + +A GS DG I V+ + ++ L +LE H+
Sbjct: 144 PRPDAPKPSDKTSSKKFVLSVAWSPNGKRLACGSMDGTICVFDVDRSKLLHQLE-GHNMP 202
Query: 310 VTSVSFSR-DGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITAS 368
V S+ FS D L S S D +H + +L GHTS+V + DG + T S
Sbjct: 203 VRSLVFSPVDPRVLFSGSDDGHVNMHDAEGKTLLGSMSGHTSWVLSVDASPDGGAIATGS 262
Query: 369 SDCTVKVWDLKTTDCLQT 386
SD TV++WDLK +QT
Sbjct: 263 SDRTVRLWDLKMRAAIQT 280
Score = 52.8 bits (125), Expect = 5e-07
Identities = 51/242 (21%), Positives = 99/242 (40%), Gaps = 11/242 (4%)
Query: 278 MIASGSTDGKIKVWRIRTAQCLRRLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLK 337
++ +GS D +K+WR L R HS GV +++ G S+S DS R+ +
Sbjct: 32 LLLTGSLDETVKLWRPDELD-LVRTNTGHSLGVAALAAHPSGIIAASSSIDSFVRVFDVD 90
Query: 338 SGKMLKEFRGHTSYVNDATFTNDGTRVITAS-SDCTVKVWDLKTTDCLQTFKPPPP--LR 394
+ + S V F GT + A S +VK+WD + + T P P +
Sbjct: 91 TNATIAVLEAPPSEVWGMQFEPKGTILAVAGGSSASVKLWDTASWRLISTLSIPRPDAPK 150
Query: 395 GGDASVNSVFI----FPKNTEHIVVCNKTSSIYIMTL-QGQVVKSFSSGKREGGDFVAAC 449
D + + F+ + N + + + +I + + + +++ V +
Sbjct: 151 PSDKTSSKKFVLSVAWSPNGKRLACGSMDGTICVFDVDRSKLLHQLEGHNMPVRSLVFSP 210
Query: 450 ISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMK 509
+ P+ ++ +D ++ + L M+ H ++ V P +AT S D T++
Sbjct: 211 VDPRV--LFSGSDDGHVNMHDAEGKTLLGSMSGHTSWVLSVDASPDGGAIATGSSDRTVR 268
Query: 510 LW 511
LW
Sbjct: 269 LW 270
Score = 42.0 bits (97), Expect = 8e-04
Identities = 49/211 (23%), Positives = 84/211 (39%), Gaps = 21/211 (9%)
Query: 299 LRRLEHAHSQGV---TSVSFSRDGSQLLST-SFDSTARIHGLKSGKMLKEFRGHTSYVND 354
L+ +E+AH V T V + D LL T S D T ++ +++ GH+ V
Sbjct: 6 LKSIENAHEDSVWAATWVPATEDRPALLLTGSLDETVKLWRPDELDLVRTNTGHSLGVAA 65
Query: 355 ATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIV 414
G ++S D V+V+D+ T + + PP + V + PK T V
Sbjct: 66 LAAHPSGIIAASSSIDSFVRVFDVDTNATIAVLEAPP------SEVWGMQFEPKGTILAV 119
Query: 415 VCNKTSSIYIM-TLQGQVVKSFS---------SGKREGGDFV-AACISPKGEWIYCVGED 463
++S+ + T +++ + S S K FV + SP G+ + C D
Sbjct: 120 AGGSSASVKLWDTASWRLISTLSIPRPDAPKPSDKTSSKKFVLSVAWSPNGKRLACGSMD 179
Query: 464 RNMYCFSYQSGKLEHLMTVHDKEIIGVTHHP 494
+ F KL H + H+ + + P
Sbjct: 180 GTICVFDVDRSKLLHQLEGHNMPVRSLVFSP 210
Score = 39.3 bits (90), Expect = 0.005
Identities = 40/166 (24%), Positives = 67/166 (40%), Gaps = 42/166 (25%)
Query: 210 KFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLKKDLQ---------------- 253
K +K +SP+G+ L S+DG I V+D KL L+
Sbjct: 154 KTSSKKFVLSVAWSPNGKRLACGSMDGTICVFDVDRSKLLHQLEGHNMPVRSLVFSPVDP 213
Query: 254 --------------YQAEETFMM-----HDEPVLCVDFSRDSEMIASGSTDGKIKVWRIR 294
+ AE ++ H VL VD S D IA+GS+D +++W ++
Sbjct: 214 RVLFSGSDDGHVNMHDAEGKTLLGSMSGHTSWVLSVDASPDGGAIATGSSDRTVRLWDLK 273
Query: 295 TAQCLRRLEHAHSQGVTSVSFSRDG------SQLLSTSFDSTARIH 334
++ + + H+ V SV+F G +L S S D + ++
Sbjct: 274 MRAAIQTMSN-HNDQVWSVAFRPPGGTGVRAGRLASVSDDKSVSLY 318
>At5g60940 cleavage stimulation factor 50K chain
Length = 429
Score = 73.9 bits (180), Expect = 2e-13
Identities = 89/400 (22%), Positives = 168/400 (41%), Gaps = 37/400 (9%)
Query: 143 AQIAQAIAAEVTV-----VPPSRLMALIGQALKWQQH--------QGLLPPGTQFDLFRG 189
+Q+A A+A+ VPP+RL+ L+ + L + + LLP
Sbjct: 34 SQVASAVASATMTPLNIEVPPNRLLELVAKGLAAENNGTLRGVSSSVLLPSSYGSITTPR 93
Query: 190 TAAMKQDVDDM------YPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDY 243
TA++ V+ P + S T+ KS CARFSPDG + + D I++++
Sbjct: 94 TASIDFSVNHAKGSSKTIPKHESKTLS-EHKSVVRCARFSPDGMFFATGGADTSIKLFEV 152
Query: 244 ISGKLKKDLQYQAE---ETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLR 300
K QA TF H EP+ +DF S ++ S + D IK + R
Sbjct: 153 PKVKQMISGDTQARPLIRTFYDHAEPINDLDFHPRSTILISSAKDNCIKFFDFSKTTAKR 212
Query: 301 RLE-HAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFR----GHTSYVNDA 355
+ + V S+SF G LL+ + ++ + + + G + +N
Sbjct: 213 AFKVFQDTHNVRSISFHPSGEFLLAGTDHPIPHLYDVNTYQCFLPSNFPDSGVSGAINQV 272
Query: 356 TFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVV 415
+++ G+ ITAS D ++++D + C+++ G + V S +F K+ ++
Sbjct: 273 RYSSTGSIYITASKDGAIRLFDGVSAKCVRSIGN----AHGKSEVTSA-VFTKDQRFVLS 327
Query: 416 CNKTSSIYIMTL-QGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRN--MYCFSYQ 472
K S++ + + G++VK + KR A + E++ + E N + +
Sbjct: 328 SGKDSTVKLWEIGSGRMVKEYLGAKRVKLR-SQAIFNDTEEFVISIDEASNEVVTWDART 386
Query: 473 SGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLWK 512
+ K+ + H+ + H P ++ T D +++ WK
Sbjct: 387 ADKVAKWPSNHNGAPRWIEHSPVESVFVTCGIDRSIRFWK 426
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,602,326
Number of Sequences: 26719
Number of extensions: 490183
Number of successful extensions: 2816
Number of sequences better than 10.0: 214
Number of HSP's better than 10.0 without gapping: 159
Number of HSP's successfully gapped in prelim test: 55
Number of HSP's that attempted gapping in prelim test: 1190
Number of HSP's gapped (non-prelim): 943
length of query: 513
length of database: 11,318,596
effective HSP length: 104
effective length of query: 409
effective length of database: 8,539,820
effective search space: 3492786380
effective search space used: 3492786380
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC142394.6


