

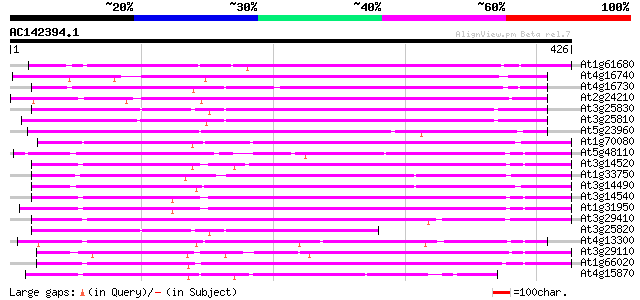
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142394.1 - phase: 0 /pseudo
(426 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g61680 linalool synthase 307 6e-84
At4g16740 limonene cyclase like protein 202 4e-52
At4g16730 limonene cyclase like protein 187 1e-47
At2g24210 myrcene/ocimene synthase (TPS10) 178 6e-45
At3g25830 limonene cyclase, putative 173 2e-43
At3g25810 terpene synthase-related protein 171 7e-43
At5g23960 beta-caryophyllene/alpha-humulene synthase 163 1e-40
At1g70080 unknown protein 152 3e-37
At5g48110 terpene synthase 129 3e-30
At3g14520 unknown protein 127 2e-29
At1g33750 terpene synthase, putative 126 2e-29
At3g14490 terpene synthase-related protein, putative 123 2e-28
At3g14540 unknown protein 118 7e-27
At1g31950 hypothetical protein 118 7e-27
At3g29410 terpene synthase, putative 114 1e-25
At3g25820 terpene synthase-related protein 110 1e-24
At4g13300 putative protein 100 2e-21
At3g29110 putative terpene synthase-related protein 100 2e-21
At1g66020 unknown protein 99 3e-21
At4g15870 delta-cadinene synthase like protein 94 1e-19
>At1g61680 linalool synthase
Length = 569
Score = 307 bits (787), Expect = 6e-84
Identities = 169/417 (40%), Positives = 250/417 (59%), Gaps = 16/417 (3%)
Query: 15 PTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQ 74
P+EN +ID+IQ LGI+ +F +EI+ L ++ + F + +L+E+AL FRLLRQ
Sbjct: 90 PSENLEMIDVIQSLGIDLHFRQEIEQTLH----MIYKEGLQF--NGDLHEIALRFRLLRQ 143
Query: 75 GGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQ 134
G+YV +F + K + DVKGL L+EAS+L +EGE+ L+ + L
Sbjct: 144 EGHYVQEIIFKNILDKKGGFKDVVKNDVKGLTELFEASELRVEGEETLDGAREFTYSRLN 203
Query: 135 AWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNK----WI-CLDELAKM 189
+ H Q + +L P H + K F + +K + W+ L +A++
Sbjct: 204 ELCSGRESH-QKQEIMKSLAQPRHKTVRGLTSKR-FTSMIKIAGQEDPEWLQSLLRVAEI 261
Query: 190 NSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELT 249
+S +K + + E + FKWW +LGL K+++ A PLKW+ W M DP + +R++LT
Sbjct: 262 DSIRLKSLTQGEMSQTFKWWTELGLEKDVEKARSQPLKWHTWSMKILQDPTLTEQRLDLT 321
Query: 250 KPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAE 309
KPISLVY+IDDIFDV+G L++LT+FT V RW+ G + LP +M+V +L +T +
Sbjct: 322 KPISLVYVIDDIFDVYGELEELTIFTRVVERWDHKGLKTLPKYMRVCFEALDMITTEISM 381
Query: 310 MVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHA 369
+YK HG+NP L+ SW L AFL EA W NSG LPTTEEY+ NG+VS+GVH+V++HA
Sbjct: 382 KIYKSHGWNPTYALRQSWASLCKAFLVEAKWFNSGYLPTTEEYMKNGVVSSGVHLVMLHA 441
Query: 370 FFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
+ L+ + +TKE + ++ E P I+ S A ILRL DDL K +Q+G DGSY+ C
Sbjct: 442 YILL--GEELTKEKVELI-ESNPGIVSSAATILRLWDDLGSAKDENQDGTDGSYVEC 495
>At4g16740 limonene cyclase like protein
Length = 1024
Score = 202 bits (513), Expect = 4e-52
Identities = 134/416 (32%), Positives = 212/416 (50%), Gaps = 30/416 (7%)
Query: 3 KQVHKKLVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEK---QFLILSSNPIDFVSS 59
++V K L +E E LID +QRLG+ ++F +EIK L + + N ID
Sbjct: 15 QEVSKMLNETEGLLEQLELIDTLQRLGVSYHFEQEIKKTLTNVHVKNVRAHKNRIDRNRW 74
Query: 60 HELYEVALAFRLLRQGGYY---VNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSI 116
+LY AL FRLLRQ + + +L D +D+KG+++LYEAS LS
Sbjct: 75 GDLYATALEFRLLRQHDVFDGNIGVDLDD--------------KDIKGILSLYEASYLST 120
Query: 117 EGEDGLNDLGYLSCELLQAWLPRHQDHIQAI----YVSNTLQYPIHYGLSRFMDKSIFIN 172
+ L + Y + + L+ ++ +++ ++ V + L+ P H + R +
Sbjct: 121 RIDTKLKESIYYTTKRLRKFVEVNKNETKSYTLRRMVIHALEMPYHRRVGRLEARWYIEV 180
Query: 173 DLKAKNKWICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWP 232
+ + L ELAK++ + V+ ++++E + WW GL K + F + Y
Sbjct: 181 YGERHDMNPILLELAKLDFNFVQAIHQDELKSLSSWWSKTGLTKHLDFVRDRITEGYFSS 240
Query: 233 MACFTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNF 292
+ +P F+ R LTK L+ IDDI+D++GTL++L LFT V +W+++ E LPN+
Sbjct: 241 VGVMYEPEFAYHRQMLTKVFMLITTIDDIYDIYGTLEELQLFTTIVEKWDVNRLEELPNY 300
Query: 293 MKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEY 352
MK+ L N V + GFN I LK SW + FLKEA W SG P EEY
Sbjct: 301 MKLCFLCLVNEINQIGYFVLRDKGFNVIPYLKESWADMCTTFLKEAKWYKSGYKPNFEEY 360
Query: 353 LNNGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
+ NG +S+ V +L+H F L ++ +TL IL +++ S A ILRL++DL
Sbjct: 361 MQNGWISSSVPTILLHLFCL------LSDQTLDILGSYNHSVVRSSATILRLANDL 410
>At4g16730 limonene cyclase like protein
Length = 600
Score = 187 bits (474), Expect = 1e-47
Identities = 127/401 (31%), Positives = 206/401 (50%), Gaps = 21/401 (5%)
Query: 17 ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGG 76
E +ID +QRLGI +++ EI L K I + + +L+ +L F LLRQ G
Sbjct: 108 EELEMIDSLQRLGISYHYKHEIHDILRK---IHDQHGEIERETQDLHATSLEFILLRQHG 164
Query: 77 YYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQAW 136
+ V+ + FD R D+KGL++LYEAS S++ E L + + + L +
Sbjct: 165 FDVSQDAFDVFISETGEFRKTLHSDIKGLLSLYEASYFSMDSEFKLKETRIYANKRLSEF 224
Query: 137 LP-------RHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWI-CLDELAK 188
+ R + V L+ P H+ + R ++ +IN + K++ L E A
Sbjct: 225 VAESSKTICREDETYILEMVKRALETPYHWSIRR-LEARWYINVYEKKHEMNPLLLEFAA 283
Query: 189 MNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVEL 248
++ ++++ ++ E K GL K++ F + Y W + F +P F R L
Sbjct: 284 IDFNMLQANHQEE----LKLISSTGLMKQLDFVRDRITESYFWTIGIFYEPEFKYCRKIL 339
Query: 249 TKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFA 308
TK L+ I+DDI+D++GTL++L LFT V +W+++ E LPN+M++ LY N
Sbjct: 340 TKIFMLIVIMDDIYDIYGTLEELELFTNVVEKWDVNHVERLPNYMRMCFLFLYNEINQIG 399
Query: 309 EMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIH 368
V + G N I LK W L FL E+ W +G P+ EEY+ NG++S+ V +L+H
Sbjct: 400 YDVLRDKGLNVIPYLKQVWTDLFKTFLTESKWYKTGHKPSFEEYMQNGVISSSVPTILLH 459
Query: 369 AF-FLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
F L DH I+ +TL+ D + +++ S A ILRL++DL
Sbjct: 460 LFSVLSDH---ISDQTLTD-DSKNHSVVRSCATILRLANDL 496
>At2g24210 myrcene/ocimene synthase (TPS10)
Length = 591
Score = 178 bits (451), Expect = 6e-45
Identities = 117/419 (27%), Positives = 207/419 (48%), Gaps = 22/419 (5%)
Query: 1 MCKQVHKKLVTSEDPT--ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVS 58
+ K+ +K++ E T + ID +Q+LG+ ++F EI L + +N +
Sbjct: 79 LLKEKVRKMLNDEQKTYLDQLEFIDDLQKLGVSYHFEAEIDNILTSSYKKDRTN----IQ 134
Query: 59 SHELYEVALAFRLLRQGGYYVNAELFDCL--KCSKKSLRVKYGEDVKGLIALYEASQLSI 116
+L+ AL FRL RQ G+ V+ ++FD C K +D+ GLI+LYEAS LS
Sbjct: 135 ESDLHATALEFRLFRQHGFNVSEDVFDVFMENCGKFDR-----DDIYGLISLYEASYLST 189
Query: 117 EGEDGLND-LGYLSCELLQAWLPRHQDHI-----QAIYVSNTLQYPIHYGLSRFMDK-SI 169
+ + L + + + L+ ++ H + V L P ++ + R + I
Sbjct: 190 KLDKNLQIFIRPFATQQLRDFVDTHSNEDFGSCDMVEIVVQALDMPYYWQMRRLSTRWYI 249
Query: 170 FINDLKAKNKWICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWY 229
+ + K + + E AK++ +IV+ +++ E V WW + GL K++ FA ++ Y
Sbjct: 250 DVYGKRQNYKNLVVVEFAKIDFNIVQAIHQEELKNVSSWWMETGLGKQLYFARDRIVENY 309
Query: 230 MWPMACFTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENL 289
W + +P + R +TK +L+ IDDI+D++GTL++L LFT A W+++ + L
Sbjct: 310 FWTIGQIQEPQYGYVRQTMTKINALLTTIDDIYDIYGTLEELQLFTVAFENWDINRLDEL 369
Query: 290 PNFMKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTT 349
P +M++ +Y N+ A + + N I LK SW + A+L EA W SG P
Sbjct: 370 PEYMRLCFLVIYNEVNSIACEILRTKNINVIPFLKKSWTDVSKAYLVEAKWYKSGHKPNL 429
Query: 350 EEYLNNGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
EEY+ N +S + +H + + I + L L + N++ + + RL++DL
Sbjct: 430 EEYMQNARISISSPTIFVHFYCVFSDQLSI--QVLETLSQHQQNVVRCSSSVFRLANDL 486
>At3g25830 limonene cyclase, putative
Length = 600
Score = 173 bits (438), Expect = 2e-43
Identities = 119/398 (29%), Positives = 202/398 (49%), Gaps = 12/398 (3%)
Query: 17 ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGG 76
E LID +Q+LG+ ++F EI L L + N +L+ +L FRLLRQ G
Sbjct: 98 EQLELIDDLQKLGVSYHFEIEINDTLTDLHLKMGRNCWKCDKEEDLHATSLEFRLLRQHG 157
Query: 77 YYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLND-LGYLSCELLQA 135
+ V+ +FD + +S K ++ G+I+LYEAS LS + + L+ + + E ++
Sbjct: 158 FDVSENIFDVIIDQIESNTFKTN-NINGIISLYEASYLSTKSDTKLHKVIRPFATEQIRK 216
Query: 136 WLPRHQDHIQAIYVS----NTLQYPIHYGLSRFMDKSIFINDLKAKNKW-ICLDELAKMN 190
++ + + I V + L+ P H+ + R +D +I+ + K+ + L E AK++
Sbjct: 217 FVD--DEDTKNIEVREKAYHALEMPYHWRMRR-LDTRWYIDAYEKKHDMNLVLIEFAKID 273
Query: 191 SSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTK 250
+IV+ ++ + V +WW+D L ++ F ++ Y W + +P F R +T
Sbjct: 274 FNIVQAAHQEDLKYVSRWWKDTCLTNQLPFVRDRIVENYFWTVGLIYEPQFGYIRRIMTI 333
Query: 251 PISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEM 310
+LV IDDI+D++GTL++L LFT V W+++ LP +M++ LY N
Sbjct: 334 VNALVTTIDDIYDIYGTLEELELFTSMVENWDVNRLGELPEYMRLCFLILYNEINGIGCD 393
Query: 311 VYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAF 370
+ K + I LK SW L +L EA W G P+ EEY+ N +S +LIH
Sbjct: 394 ILKYKKIDVIPYLKKSWADLCRTYLVEAKWYKRGYKPSLEEYMQNAWISISAPTILIH-- 451
Query: 371 FLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
F + I+ + L L + +I+ A +LRL++DL
Sbjct: 452 FYCVFSDQISVQNLETLSQHRQHIVRCSATVLRLANDL 489
>At3g25810 terpene synthase-related protein
Length = 511
Score = 171 bits (433), Expect = 7e-43
Identities = 114/404 (28%), Positives = 205/404 (50%), Gaps = 9/404 (2%)
Query: 10 VTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAF 69
V ++ E LID +Q+LG+ ++F +EI L L N +L+ AL F
Sbjct: 5 VETKSRLEKLELIDDLQKLGVSYHFEQEINNILTNFHLENGKNIWKCDKEEDLHATALEF 64
Query: 70 RLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLND-LGYL 128
RLLRQ G+ V+ ++FD + +S K +++ +I LYEAS LS + + L+ +
Sbjct: 65 RLLRQHGFGVSEDIFDVIIDKIESNTFK-SDNITSIITLYEASYLSTKSDTKLHKVIRPF 123
Query: 129 SCELLQAWLPRHQDHIQAIY---VSNTLQYPIHYGLSRFMDKSIFINDLKAKNKW-ICLD 184
+ E ++ ++ + + + L+ P H+ + R ++ +I+ + K+ + L
Sbjct: 124 ATEQIRNFVDDESETYNIMLREMAIHALEIPYHWRMRR-LETRWYIDAYEKKHDMNLFLA 182
Query: 185 ELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNE 244
E AK++ +IV+ ++ + V WW++ GL ++ F ++ Y W + +P F
Sbjct: 183 EFAKIDFNIVQTAHQEDVKYVSCWWKETGLGSQLHFVRDRIVENYFWTVGMIYEPQFGYI 242
Query: 245 RVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVT 304
R + +L+ +IDDI+D++GT ++L LFT V W+++ + LP +MK+ +L+
Sbjct: 243 RRIVAIVAALITVIDDIYDIYGTPEELELFTAMVQNWDINRLDELPEYMKLCFLTLFNEI 302
Query: 305 NNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHV 364
N V K + I K SW L A+L EA W G P+ EEY+ N +S
Sbjct: 303 NAMGCDVLKCKNIDVIPYFKKSWADLCKAYLVEAKWYKGGYKPSVEEYMQNAWISISAPT 362
Query: 365 VLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
+LIH F + I+ + L L ++ +++ A +LRL++DL
Sbjct: 363 MLIH--FYCAFSGQISVQILESLVQQQQDVVRCSATVLRLANDL 404
>At5g23960 beta-caryophyllene/alpha-humulene synthase
Length = 547
Score = 163 bits (413), Expect = 1e-40
Identities = 110/400 (27%), Positives = 203/400 (50%), Gaps = 12/400 (3%)
Query: 14 DPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLR 73
+P EN ID + RLG+ ++F ++I L+K F L + +LY V + F++ R
Sbjct: 55 NPIENIKFIDALCRLGVSYHFEKDIVEQLDKSFDCLDFPQMVRQEGCDLYTVGIIFQVFR 114
Query: 74 QGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELL 133
Q G+ ++A++F+ K + D G+++LYEA+Q GED +++ S L
Sbjct: 115 QFGFKLSADVFEKFKDENGKFKGHLVTDAYGMLSLYEAAQWGTHGEDIIDEALAFSRSHL 174
Query: 134 QAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNSSI 193
+ R H+ AI + N L++P H G+SR + + ++ L E AK++ ++
Sbjct: 175 EEISSRSSPHL-AIRIKNALKHPYHKGISRIETRQYISYYEEEESCDPTLLEFAKIDFNL 233
Query: 194 VKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKPIS 253
++ +++ E V +W ++ ++ + + + Y+W + + +P +S RV T +
Sbjct: 234 LQILHREELACVTRWHHEMEFKSKVTYTRHRITEAYLWSLGTYFEPQYSQARVITTMALI 293
Query: 254 LVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMV-- 311
L +DD++D +GT+++L LFT+A++ W + +P + S+ +Y VT F + +
Sbjct: 294 LFTALDDMYDAYGTMEELELFTDAMDEWLPVVPDEIP--IPDSMKFIYNVTVEFYDKLDE 351
Query: 312 -YKKHGFNPID-TLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHA 369
+K G + LK S + N +++EA WL + T +EY N I+S+G + ++
Sbjct: 352 ELEKEGRSGCGFHLKKSLQKTANGYMQEAKWLKKDYIATFDEYKENAILSSGYYALIAMT 411
Query: 370 FFLM-DHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
F M D AK E LS P I + I R +DD+
Sbjct: 412 FVRMTDVAKLDAFEWLS----SHPKIRVASEIISRFTDDI 447
>At1g70080 unknown protein
Length = 611
Score = 152 bits (384), Expect = 3e-37
Identities = 110/412 (26%), Positives = 198/412 (47%), Gaps = 14/412 (3%)
Query: 22 IDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGGYYVNA 81
I + LG+ ++F E+I L+ F + ID +LY V++ FR+ R G+Y+
Sbjct: 123 IHTLVSLGVSYHFEEKIVEFLKDAFENIEDMIID-CKEDDLYTVSIIFRVFRLYGHYITP 181
Query: 82 ELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQAWL---- 137
++F+ K + + +DV+G+++ YEAS ED L + + + L+ +L
Sbjct: 182 DIFNRFKGDDGNFKKCLNDDVRGMLSFYEASHFGTTTEDILEEAMSFTQKHLELFLVGEK 241
Query: 138 PRHQDHIQAIYVSNTLQYPIHYGLSRFMDKS-IFINDLKAKNKWICLDELAKMNSSIVKF 196
+H HI + + L P ++ L + + I +L+ + + L +LAK+N ++
Sbjct: 242 AKHYPHITKL-IQAALYIPQNFNLEILVAREYIDFYELETDHNEMLL-KLAKLNFRFLQL 299
Query: 197 MNKNESVEVFKWWEDLGLAKEMKFAGYNPL-KWYMWPMACFTDPCFSNERVELTKPISLV 255
+ + WW++L L ++ L + Y W + +P +S R+ L K I LV
Sbjct: 300 QYIQDLKTLTTWWKELDLVSKIPVYFRERLAEPYFWATGIYYEPQYSAARIMLAKSIILV 359
Query: 256 YIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVYKKH 315
I+D+ FDV+GT+D++ +A+ RW+ D + LP+++KV + + + E V +
Sbjct: 360 DIVDNTFDVYGTIDEVKSLVQAIERWDSDAVDVLPDYLKVVFRTTFDLFKELEEYVSSEA 419
Query: 316 GFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFFLMDH 375
+ L+ +L+EA W N G LP+ EEY+ G+ ST V+L F M
Sbjct: 420 RSFTMQYAYEQLRILMKGYLQEAEWSNRGHLPSHEEYIEVGVASTAGEVLLAMTFIPMGD 479
Query: 376 AKGI-TKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
A G+ E L P + +++ RL DD+ K + G + +NC
Sbjct: 480 AAGVGVYEWL----RSRPKLTHALFVKSRLRDDIATYKEEMKRGDVCNGINC 527
>At5g48110 terpene synthase
Length = 575
Score = 129 bits (324), Expect = 3e-30
Identities = 110/427 (25%), Positives = 194/427 (44%), Gaps = 33/427 (7%)
Query: 4 QVHKKLVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDF-VSSHEL 62
+V +KLV D E HLI ++ LG+ ++F ++I+ L+ F N D + ++
Sbjct: 97 KVSEKLVCM-DVKERIHLIHLLVSLGVAYHFEKQIEEFLKVDF----ENVEDMNLGEEDM 151
Query: 63 YEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGL 122
Y +++ FR+ R + +++++F+ K + +D K L + + + G
Sbjct: 152 YSISVIFRVFRLYRHKLSSDVFNRFKEENGDFKKCLLDDKKSLTKQWASRGNTWNYFVGG 211
Query: 123 NDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWIC 182
++ +LS + Q++ + + +Q+ Y D+++
Sbjct: 212 SNEEHLSGHIKNVLYLSQQENAEVVMSREYIQF---YEQETHHDETLL------------ 256
Query: 183 LDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGY---NPLKWYMWPMACFTDP 239
+ AK+N ++ E + KWW++L L E K Y ++ W MA + +P
Sbjct: 257 --KFAKINFKFMQLHYVQELQTIVKWWKELDL--ESKIPNYYRVRAVECLYWAMAVYMEP 312
Query: 240 CFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSS 299
+S R+ L+K + L IIDD++D + TL + FTE + RWE D A ++P+ MKV L S
Sbjct: 313 QYSVARIILSKSLVLWTIIDDLYDAYCTLPEAIAFTENMERWETD-AIDMPDHMKVLLRS 371
Query: 300 LYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVS 359
L + +F V + ++ W RL A L + W +G +P +EY+ GIV+
Sbjct: 372 LIDLMEDFKGEVRSEGRLYSVEYGIDEWKRLFRADLTISKWARTGYIPNYDEYMEVGIVT 431
Query: 360 TGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGL 419
GV V + AF M A KE + P I ++ RL DD+ K G
Sbjct: 432 GGVDVTVAFAFIGMGEA---GKEAFDWIRSR-PKFIQTIDLKSRLRDDVATYKDEMARGE 487
Query: 420 DGSYLNC 426
+ +NC
Sbjct: 488 IATGINC 494
>At3g14520 unknown protein
Length = 605
Score = 127 bits (318), Expect = 2e-29
Identities = 93/420 (22%), Positives = 193/420 (45%), Gaps = 23/420 (5%)
Query: 17 ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGG 76
E LI ++ LG ++F +EI+ L+ F L V +L +++ F + R G
Sbjct: 114 EKIRLIHLLVSLGSSYHFDKEIQDILKHSFTKLDDI---IVGEDDLETISIMFEVFRLYG 170
Query: 77 YYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQAW 136
+ ++ + FD + + +DV+G++ L+E + L ED +++ + L +W
Sbjct: 171 HKMSCDAFDRFRGEDGRFKESLAKDVRGMLQLFEVAHLGTPSEDIMDEASSFAQNHLDSW 230
Query: 137 L--------PRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSI--FINDLKAKNKWICLDEL 186
+ P HIQ N+L P + + + + + + NK L +
Sbjct: 231 IGGNVSGATPHLLKHIQ-----NSLYIPRYCNIEVLVAREYISYYEQEEGHNK--ILLKF 283
Query: 187 AKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERV 246
AK+N + +F E + KWW+DL LA ++ + ++ ++ + + +P +S R+
Sbjct: 284 AKLNFNFCQFHYIQELKTLTKWWKDLDLASKLPYIRDRLVESHLGGLGPYFEPHYSLGRI 343
Query: 247 ELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNN 306
+ K I + ++DD +D H T+ ++ + TE + R + + LP++++ L S+++V
Sbjct: 344 IVAKIIMTMVVVDDTYDAHATVPEVAVLTECLQRLNIGADDKLPDYLRTVLESVFEVMGE 403
Query: 307 FAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVL 366
+ + K + + + + A + W +G +P+ +EY+ G+V+ G+
Sbjct: 404 IEQEMRPKGRSYGVKQVLERFKNVAKADKQLTEWARTGDVPSFDEYMKVGLVTAGMDGYA 463
Query: 367 IHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
+ F M+ KE L P II ++ + RL++D+ ++ G + LNC
Sbjct: 464 GYCFIGMEDVS--EKEAFEWLSSN-PLIIQALNVMFRLANDVGTYETEINRGEVANGLNC 520
>At1g33750 terpene synthase, putative
Length = 603
Score = 126 bits (317), Expect = 2e-29
Identities = 96/418 (22%), Positives = 200/418 (46%), Gaps = 22/418 (5%)
Query: 17 ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGG 76
E LI ++ LG HYF +EI+ LE+ F L + F +L A+ F + R G
Sbjct: 117 ERICLIHLLICLGTFHYFEKEIEEILEQAFRKLD---MLFTDEDDLETTAIMFEVFRLYG 173
Query: 77 YYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLND-LGYLSCEL--- 132
+ ++ ++FD K + DV+G++ LYEA+ L+ E L++ L + L
Sbjct: 174 HKISCDVFDRFKGVDAKFKEHLVSDVRGMLQLYEAAHLATPFETILDEALSFTRYHLESL 233
Query: 133 --LQAWLPRHQDHI-QAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKM 189
QA P HI A+Y L+ I + +I+ + + L + AK+
Sbjct: 234 AGQQATAPHISRHILNALYKPRFLKMEI-------IAAREYIHFYQKEGHDETLLKFAKL 286
Query: 190 NSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELT 249
N + + E + KWW+D+ L ++ + L+ ++ MA + +P +S R+ T
Sbjct: 287 NFNFCQLHYVRELKTLTKWWKDIDLPYKLPYIRDRLLETFIGVMAVYLEPHYSLGRIIAT 346
Query: 250 KPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAE 309
K ++ ++DD D +GT ++ +++ RW+ + LP+ +++ + S+ + + E
Sbjct: 347 KVSQVIVVMDDTCDAYGTFSEVRSLIDSLERWDPGAIDKLPSCLRIVIQSIVETMEDI-E 405
Query: 310 MVYKKHGFNPIDTLKISWVRLL-NAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIH 368
K G + + ++++ A+ + + W +G +PT ++Y+ G+ S+G+ ++
Sbjct: 406 REMKPRGRSSSVQDTVEEIKIMGRAYAEISKWARAGHVPTFDDYIELGLDSSGIRCFAMY 465
Query: 369 AFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
+F M+ + + + + P ++ +++ I RL++D+ G + + G + +NC
Sbjct: 466 SFISMEDCE---ENQTNAWFKSKPKMLRALSVIFRLTNDIAGFEEEMRRGEVVNGVNC 520
>At3g14490 terpene synthase-related protein, putative
Length = 601
Score = 123 bits (309), Expect = 2e-28
Identities = 92/416 (22%), Positives = 190/416 (45%), Gaps = 16/416 (3%)
Query: 17 ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSS-HELYEVALAFRLLRQG 75
E LI ++ LGI ++F EI L + F N D ++ ++L ++ F + R
Sbjct: 112 ERIRLIHLLMNLGIAYHFEIEIDEILGQAF----GNLDDIIAKENDLETISTMFEVFRLR 167
Query: 76 GYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQA 135
GYY+ F+ K + ED++G++ LYEA+ L ED +++ + L++
Sbjct: 168 GYYMPCYAFNRFKGEDGRFKESLAEDIRGMLQLYEAAHLGTPSEDIMDEALSFTRYRLES 227
Query: 136 WLPRH----QDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNS 191
H H+ ++ N L ++ L + + + ++ L + AK+N
Sbjct: 228 LTSNHTATASPHLSK-HIQNALYRARYHNLEILVAREYISFYEQEEDHDETLLKFAKLNF 286
Query: 192 SIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKP 251
+ + E ++ KWW++L LA ++ + ++ Y +A + +P +S R+ +TK
Sbjct: 287 NYCQLHYIQELKDLTKWWKELDLASKLPYIRDRIVEVYFGALALYFEPRYSLGRIIVTKI 346
Query: 252 ISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMV 311
+V + +D D +GTL ++T ++ RW++ E LP+++K+ +++ E
Sbjct: 347 TMIVTVFNDTCDAYGTLPEVTSLVDSFQRWDLGDIEKLPSYVKIVFRGVFETLEEI-EQE 405
Query: 312 YKKHGFNPIDTLKISWVRLL-NAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAF 370
+ G + I + + ++ L A+L + W + +PT EEY+ G+ ++ H
Sbjct: 406 MRPQGRSRIVQVAVDEIKKLGKAYLAISKWARASHVPTFEEYMEFGMQTSMDHFAAYSFI 465
Query: 371 FLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
+ D + T E + P ++ ++ + R+ +D+ + G LNC
Sbjct: 466 AMEDCDENQTCEWY----KSRPKMMEALNGVFRIKNDINTFEQEMSRGEVAKGLNC 517
>At3g14540 unknown protein
Length = 602
Score = 118 bits (295), Expect = 7e-27
Identities = 90/418 (21%), Positives = 188/418 (44%), Gaps = 19/418 (4%)
Query: 17 ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGG 76
E LI ++ LG ++F +EI+ L+ F L V +L +++ F + R G
Sbjct: 111 EKIRLIHLLVSLGSSYHFDKEIQDILKHSFTKLDGI---IVEEDDLETISIMFEVFRLYG 167
Query: 77 YYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGL--------NDLGYL 128
+ ++ + FD + + +DV+G++ L+E + L ED + N L L
Sbjct: 168 HKMSCDAFDRFRGGDGRFKESLAKDVRGMLQLFEVAHLGTLSEDIMDEALRFTRNHLESL 227
Query: 129 SCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAK 188
+ + + P HIQ N+L P + + + + + + L + AK
Sbjct: 228 TSGNVSSASPHILKHIQ-----NSLYIPRYCNIEVLVAREYISYYEQEEGYNEILLKFAK 282
Query: 189 MNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVEL 248
+N + + E + KWW+DL LA ++ + ++ ++ + + +P +S R+ +
Sbjct: 283 LNFNFCQCHYIQEIKTLTKWWKDLDLASKLPYIRDRSVESHLGGLGPYFEPQYSLGRIIV 342
Query: 249 TKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFA 308
K I ++ + DD +D H T+ + T+ TE R + + L ++++ L S+++V
Sbjct: 343 AKTIMIIVVADDTYDAHATIPEATVLTEYFQRLNIGADDKLSGYLRIVLESVFEVMGEIE 402
Query: 309 EMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIH 368
+ + K + + + + A+ + W G +PT +EY+ G+V+ G+ +
Sbjct: 403 QEMSPKGRSYSVKQVLERFKIIAKAYKQLTEWARKGHVPTFDEYMKVGLVTAGMGDYAGY 462
Query: 369 AFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
F M+ KE L+ P +I ++ + R+++D+ ++ G + LNC
Sbjct: 463 CFIGMEDIN--EKEAFEWLNSN-PLLIDALNVLFRIANDVGTYETEINRGEVANGLNC 517
>At1g31950 hypothetical protein
Length = 608
Score = 118 bits (295), Expect = 7e-27
Identities = 94/428 (21%), Positives = 192/428 (43%), Gaps = 20/428 (4%)
Query: 8 KLVTSEDPT-ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVA 66
KL++SE E LI ++ +GI ++F +EI+ L+ F L V +L ++
Sbjct: 107 KLMSSESSDKEKIRLIHLLVSMGISYHFDKEIQDILKHSFTKLDDI---IVGEDDLETIS 163
Query: 67 LAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGL---- 122
+ F + + G+ ++ + FD + + + D +G++ L+E + L E +
Sbjct: 164 IMFEVFKLYGHKMSCDAFDRFRGNDGRFKESLVRDFRGMLQLFEVAHLGTPCEVIMDEAL 223
Query: 123 ----NDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKN 178
N L L+ P HIQ N+L P + + + + + +
Sbjct: 224 SFTRNHLESLTSGNASTASPHLLKHIQ-----NSLYIPRYCNIEVLVAREYISYYEQEEG 278
Query: 179 KWICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTD 238
L + AK+N + +F E + KWW DL LA ++ + ++ ++ + + +
Sbjct: 279 HDEILLKFAKLNFNFCQFHYVQELKTLTKWWRDLDLASKLPYIRDRLVESHLVALGPYFE 338
Query: 239 PCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLS 298
P +S R+ + K ++ ++DD +D + TL Q+ TE + RW ++ ++ LP+++++ L
Sbjct: 339 PHYSLGRIIVAKINMIMVVVDDTYDAYATLPQVKALTECLQRWSIEVSDKLPDYLRIVLG 398
Query: 299 SLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIV 358
SL+ V + + + + A+ + A W +G + T +EY+ G++
Sbjct: 399 SLFDVMGEIEREMRPLGRLYRVKQVVEKIKIITKAYQEIAKWARTGHVSTFDEYMKVGVL 458
Query: 359 STGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNG 418
+ G+ + F M+ KE L+ P II + + RL++D+ ++ G
Sbjct: 459 TAGMADYAAYCFIGMEDIN--EKEAFEWLNSN-PLIIKHLTAMFRLANDVGTYETEINRG 515
Query: 419 LDGSYLNC 426
+ LNC
Sbjct: 516 EVANGLNC 523
>At3g29410 terpene synthase, putative
Length = 603
Score = 114 bits (284), Expect = 1e-25
Identities = 93/415 (22%), Positives = 189/415 (45%), Gaps = 15/415 (3%)
Query: 17 ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGG 76
E LI ++ LGI +Y+ EI+ L K + L+ D +L +A+ F + R G
Sbjct: 116 ERIRLIHLLISLGIAYYYENEIEEILHKAYGKLACLISD---EDDLETIAIMFEVFRLYG 172
Query: 77 YYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQAW 136
+ + ++F+ K + DV+GL+ LYEA+ L ED +++ + L+
Sbjct: 173 HKMPCDVFERFKSEDGKFKESLVGDVRGLLQLYEAAHLGAPSEDIMDEALSFARYHLEPL 232
Query: 137 LPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNSSIVKF 196
+V N L ++ + + + + +++ L +K+N + +
Sbjct: 233 AGTETSSNLFKHVENVLYRARYHSIEILVARQYISFYDQEEDQDETLLRFSKLNFNFCQM 292
Query: 197 MNKNESVEVFKWWEDLGLAKEMKFA-GYNPLKWYMWPMACFTDPCFSNERVELTKPISLV 255
E V +WW++LG+A ++ ++ ++ Y+ + +P +S R+ L K ++
Sbjct: 293 HYVKELKIVTRWWKELGIASKLPYSIRERNVETYLGGLGVLFEPRYSLARIFLAKLTLIM 352
Query: 256 YIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNF-AEMVYK- 313
++DD D + TL ++ +A +RW++ E LP +M++ S+++ + EM+ +
Sbjct: 353 TVVDDTCDAYATLPEVQSLHDAFHRWDLRAMEELPRYMRIIYQSVFETVEDIDREMIARG 412
Query: 314 KHG--FNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFF 371
KHG ID +K L+ +L A W S +P+ E+Y+ G S+ + + F
Sbjct: 413 KHGRLQLTIDEIK----SLMIWYLGIAKWARSDQVPSFEDYMEIGTPSSALDDFASYGFI 468
Query: 372 LMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
MD ++ L P I +++ + R+ +D+ + G + +NC
Sbjct: 469 AMDDC---DQKQLKEWFYSKPKIFHALNALFRIRNDIVTFEQEMSRGEVANGVNC 520
>At3g25820 terpene synthase-related protein
Length = 363
Score = 110 bits (276), Expect = 1e-24
Identities = 80/270 (29%), Positives = 142/270 (51%), Gaps = 10/270 (3%)
Query: 17 ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGG 76
E LID +Q+LG+ ++F EI L L + N +L+ +L FRLLRQ G
Sbjct: 66 EQLELIDDLQKLGVSYHFEIEINDTLTDLHLKMGRNCWKCDKEEDLHATSLEFRLLRQHG 125
Query: 77 YYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLND-LGYLSCELLQA 135
+ V+ +FD + +S K ++ G+I+LYEAS LS + + L+ + + E ++
Sbjct: 126 FDVSENIFDVIIDQIESNTFKTN-NINGIISLYEASYLSTKSDTKLHKVIRPFATEQIRK 184
Query: 136 WLPRHQDHIQAIYVS----NTLQYPIHYGLSRFMDKSIFINDLKAKNKW-ICLDELAKMN 190
++ + + I V + L+ P H+ + R +D +I+ + K+ + L E AK++
Sbjct: 185 FVD--DEDTKNIEVREKAYHALEMPYHWRMRR-LDTRWYIDAYEKKHDMNLVLIEFAKID 241
Query: 191 SSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTK 250
+IV+ ++ + V +WW+D L ++ F ++ Y W + +P F R +T
Sbjct: 242 FNIVQAAHQEDLKYVSRWWKDTCLTNQLPFVRDRIVENYFWTVGLIYEPQFGYIRRIMTI 301
Query: 251 PISLVYIIDDIFDVHGTLDQLTLFTEAVNR 280
+LV IDDI+D++GTL++L LFT V +
Sbjct: 302 VNALVTTIDDIYDIYGTLEELELFTSMVEK 331
>At4g13300 putative protein
Length = 471
Score = 100 bits (249), Expect = 2e-21
Identities = 105/414 (25%), Positives = 180/414 (43%), Gaps = 24/414 (5%)
Query: 7 KKLVTSEDPTENFH----LIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHEL 62
+KL++S+ E LI + LG+ +F EIK LE F + D ++L
Sbjct: 51 RKLLSSKGDDETSKRKVLLIQSLLSLGLAFHFENEIKDILEDAFRRIDDITGD---ENDL 107
Query: 63 YEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGL 122
+++ FR+ R G+ + + +F ED KG+++LYEA+ L + L
Sbjct: 108 STISIMFRVFRTYGHNLPSSVFKRFTGDDGKFERSLTEDAKGILSLYEAAHLGTTTDYIL 167
Query: 123 NDLGYLSCELLQAWLPRH--QDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKW 180
++ + L++ L + HI + + NTL P + + + + + ++
Sbjct: 168 DEALEFTSSHLKSLLVGGMCRPHILRL-IRNTLYLPQRWNMEAVIAREYISFYEQEEDHD 226
Query: 181 ICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEM--KFAGYNPLKWYMWPMACFTD 238
L LAK+N +++ E KWW +LGL + +F W M F +
Sbjct: 227 KMLLRLAKLNFKLLQLHYIKELKTFIKWWMELGLTSKWPSQFRERIVEAWLAGLMMYF-E 285
Query: 239 PCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLS 298
P FS RV K L+ I+DD D + ++ +LT + V RW DG L + ++
Sbjct: 286 PQFSGGRVIAAKFNYLLTILDDACDHYFSIPELTRLVDCVERWNHDGIHTLEDISRIIFK 345
Query: 299 SLYKVTNNFAEMVYKK----HGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLN 354
V ++ V K + ++ LKI L+ A L W LP+ EE++
Sbjct: 346 LALDVFDDIGRGVRSKGCSYYLKEMLEELKI----LVRANLDLVKWARGNQLPSFEEHVE 401
Query: 355 NGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
G ++ + L+++F M A G KE + P +I S+A RL DD+
Sbjct: 402 VGGIALTTYATLMYSFVGMGEAVG--KEAYEWVRSR-PRLIKSLAAKGRLMDDI 452
>At3g29110 putative terpene synthase-related protein
Length = 569
Score = 100 bits (249), Expect = 2e-21
Identities = 96/424 (22%), Positives = 189/424 (43%), Gaps = 39/424 (9%)
Query: 21 LIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHE--LYEVALAFRLLRQGGYY 78
+I ++ LG+ ++F EEI+ +L+ F ID + + E LY ++ F + R GY
Sbjct: 86 MIYLLVSLGLAYHFEEEIEKSLKDGF-----EKIDEIIAGEDDLYTISTIFWVFRTYGYN 140
Query: 79 VNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLND-LGYLSCELL---- 133
+++++F K + ED +G+++LYEA+ L + L++ L + S L+
Sbjct: 141 MSSDVFRRFKEENGKFKESLIEDARGMLSLYEAAHLGTTTDYILDEALDFASNNLVSLAE 200
Query: 134 QAWLPRHQDHIQAIYVSNTLQYPIHYGLS--------RFMDKSIFINDLKAKNKWICLDE 185
P H + ++ N L H+ + RF ++ + +++ K
Sbjct: 201 DGMCPSH----LSTHIRNALSISQHWNMEIIVAVQYIRFYEQEVGHDEMLLK-------- 248
Query: 186 LAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPL--KWYMWPMACFTDPCFSN 243
AK+N ++V+ + E + KW++D + ++ Y P+ + + + A F +P FS+
Sbjct: 249 FAKLNFNLVQRLYLQEVKILTKWYKDQDIHSKLP-PYYRPVVTEMHFFSTATFFEPQFSH 307
Query: 244 ERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGA-ENLPNFMKVSLSSLYK 302
R+ TK ++DD D + T ++ ++ RW D A + P+++KV +
Sbjct: 308 ARILQTKLFMAELLVDDTCDRYATFSEVESLINSLQRWAPDDAMDTHPDYLKVVFKFILN 367
Query: 303 VTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGV 362
+ + + ++ K + R + L A +G +P+ EEY+ G G
Sbjct: 368 AFEECEKELRPQGRSYSLEQTKEEYKRFAKSNLDLAKLAQAGNVPSFEEYMEVGKDEIGA 427
Query: 363 HVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGS 422
V++ + MD+ + E L S A+I+R +DL G + + G +
Sbjct: 428 FVIVAGSLMGMDNIDAV--EAYDFLKSR-SKFSQSSAEIVRYLNDLAGFEDDMRRGCVST 484
Query: 423 YLNC 426
LNC
Sbjct: 485 GLNC 488
>At1g66020 unknown protein
Length = 598
Score = 99.4 bits (246), Expect = 3e-21
Identities = 93/413 (22%), Positives = 180/413 (43%), Gaps = 17/413 (4%)
Query: 21 LIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGGYYVN 80
+I ++ LG+ H+F EEI L F + D +L V++ F R+ G+Y++
Sbjct: 113 MIYMLVSLGLAHHFEEEIYETLRDGFGKIEEMMED---EDDLCTVSIIFWAFRRYGHYIS 169
Query: 81 AELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLND-LGYLSCELLQ----A 135
+++F K S + + KG+++LYEA+ L + L + L + S L
Sbjct: 170 SDVFRRFKGSNGNFKESLTGYAKGMLSLYEAAHLGTTKDYILQEALSFTSSHLESLAACG 229
Query: 136 WLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNSSIVK 195
P H ++++ N L P H+ + + + K+ L + AK++ +++
Sbjct: 230 TCPPHL----SVHIQNVLSVPQHWNMEILVPVEYIPFYEQEKDHDEILLKFAKLSFKLLQ 285
Query: 196 FMNKNESVEVFKWWEDLGLAKEMK-FAGYNPLKWYMWPMACFTDPCFSNERVELTKPISL 254
+ V KW+++L A ++ + N + Y + +A P S ER+ LT+ +
Sbjct: 286 LQYIQDLKIVTKWYKELEFASKLPPYFRDNIVVNYFYVLAVIYTPQHSYERIMLTQYFTC 345
Query: 255 VYIIDDIFDVHGTLDQLTLFTEAVNRW-EMDGAENLPNFMKVSLSSLYKVTNNFAEMVYK 313
+ I+DD FD + +L + ++ RW D + P+++K+ L+ + K F + +
Sbjct: 346 LAILDDTFDRYASLPEAISLANSLERWAPNDAMDKQPDYLKIVLNFILKTFEVFQKELEP 405
Query: 314 KHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFFLM 373
+ + + + A W ++ +P+ EEY+ G V L F M
Sbjct: 406 EGRSYTVKATIEEFKTVTKGNFDLAKWAHAVHVPSFEEYMEVGEEEISVCSTLAGIFMCM 465
Query: 374 DHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
+ + TKE L P I ++ RL +D+ G + G + +NC
Sbjct: 466 E--QKATKEDYEWLKSR-PKFIQTLCARCRLKNDITGFEDDMSRGYVTNAVNC 515
>At4g15870 delta-cadinene synthase like protein
Length = 582
Score = 94.0 bits (232), Expect = 1e-19
Identities = 84/363 (23%), Positives = 156/363 (42%), Gaps = 24/363 (6%)
Query: 13 EDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLL 72
E + I ++ LG+ +F +EI+ +LE+ F + +LY +++ F +
Sbjct: 117 ESTKKRILFIYMLVSLGVAFHFEDEIEQSLEEGFEKIQEM---IAGEDDLYTISIMFWVF 173
Query: 73 RQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCEL 132
R GY ++ ++F K + DVKG+++LYEA+ L ED L + +
Sbjct: 174 RTYGYNMSTDVFKRFKGENEKFMESITSDVKGMVSLYEAAHLRTTREDILEEALSFTTRN 233
Query: 133 LQ--AWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSI--FINDLKAKNKWICLDELAK 188
L+ A HI + + N L P HY + F + NK + AK
Sbjct: 234 LESLARAGASSPHI-LMRIRNALCMPQHYNAEMIFAREYISFYEQEEDHNKMLL--RFAK 290
Query: 189 MNSSIVKFMNKNESVEVFKWWEDLGLAKEM-KFAGYNPLKWYMWPMACFTDPCFSNERVE 247
+N ++ E + KWW+ LA ++ + ++ Y++ + + +P FS RV
Sbjct: 291 INFKFLQLNWIQELKTLTKWWKQQDLASKLPPYFRDRLIECYLFAIMIYFEPQFSLGRVS 350
Query: 248 LTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNF 307
L K ++ ++DD D +G + T + + RW+ D ++LP++MK +
Sbjct: 351 LAKINTVFTLVDDTCDRYGNVSD-TNISVCMYRWDPDCMDSLPDYMKTVFKFAWNTFEEC 409
Query: 308 AEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLI 367
+ G + + L+E W ++PT +EYL G V ++V +
Sbjct: 410 ENAGIMEEGLS----------YDVQGALEE--WEQGDVVPTFDEYLEIGGVEVTMYVSIA 457
Query: 368 HAF 370
+F
Sbjct: 458 CSF 460
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,854,435
Number of Sequences: 26719
Number of extensions: 421647
Number of successful extensions: 1187
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 33
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 1062
Number of HSP's gapped (non-prelim): 51
length of query: 426
length of database: 11,318,596
effective HSP length: 102
effective length of query: 324
effective length of database: 8,593,258
effective search space: 2784215592
effective search space used: 2784215592
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC142394.1


