

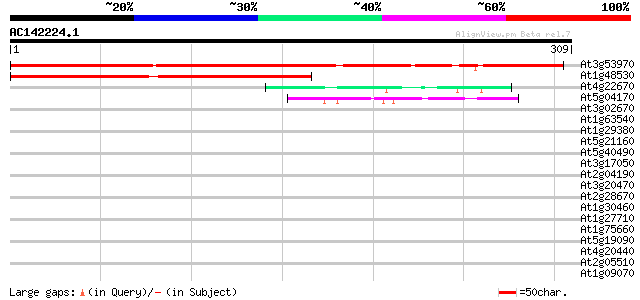
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142224.1 - phase: 0
(309 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g53970 unknown protein 317 7e-87
At1g48530 hypothetical protein 143 1e-34
At4g22670 HSP associated protein like 46 3e-05
At5g04170 EF - hand Calcium binding protein - like 45 5e-05
At3g02670 unknown protein 39 0.003
At1g63540 unknown protein 39 0.003
At1g29380 beta-1,3 glucanase, putative 39 0.003
At5g21160 unknown protein 39 0.005
At5g40490 ribonucleoprotein -like 38 0.006
At3g17050 unknown protein 38 0.006
At2g04190 unknown protein 38 0.006
At3g20470 glycine-rich protein, putative 38 0.008
At2g28670 putative disease resistance response protein 38 0.008
At1g30460 hypothetical protein 38 0.008
At1g27710 unknown protein 38 0.008
At1g75660 Dhp1-like protein 37 0.010
At5g19090 unknown protein 37 0.014
At4g20440 putative snRNP protein 37 0.018
At2g05510 putative glycine-rich protein 37 0.018
At1g09070 unknown protein 37 0.018
>At3g53970 unknown protein
Length = 302
Score = 317 bits (811), Expect = 7e-87
Identities = 169/311 (54%), Positives = 207/311 (66%), Gaps = 17/311 (5%)
Query: 1 MATDKSVLAVIRAARPTFRNQNDKIAFVVHSSFLASGYILTATGPQALSDTALSNPSNDE 60
MA ++V+A+IR ARP FRN +DK+AF +HSSF+ASGYILTATG A +D ALS+ S ++
Sbjct: 1 MANSQTVMAMIRLARPPFRNNHDKVAFAIHSSFVASGYILTATGRPAFADEALSSSSQND 60
Query: 61 VSVDHWNELNDEYAFVYANSEKEGLKKVLVKCLVMNGKLLVHALSEGFKEPLHLEINVAD 120
V ++ WNE EYAFVYAN K+G KK+LVKCL M+ KLLV A+++G EP HLEI V D
Sbjct: 61 VGIEGWNEFEGEYAFVYAN-PKKGSKKILVKCLAMDDKLLVDAIADGGAEPAHLEIKVGD 119
Query: 121 YAGEDGGSNFSQQFKNLDKLVKKIDADILSKLDGTAHASSSTKSSKTSDSTRPETSEPVP 180
YA E ++S QFKNLDKLV + ++I+ KLDG +S S + + P + P
Sbjct: 120 YAEESNEGDYSAQFKNLDKLVTDLQSEIIDKLDGKPKPVASRAQSSSETNEEPRYYDDTP 179
Query: 181 GFGEPAGPNDYHPGFIVPPVPAGSG-SDLFPGPGAGMYPSRGDHDFG-GSMLVGPNDPRW 238
P GP + G +VPP+P G SDLFPGPGAGMYP RG FG GSMLVGP DPR+
Sbjct: 180 ---NPLGPQIHPSGVVVPPIPGNGGYSDLFPGPGAGMYPGRG--GFGDGSMLVGPTDPRF 234
Query: 239 FSGGTGGGIGGGPGFIG----GLPGAPPGARFDPIGPPGVPGFEPNRFARNPRRPGFDAH 294
F G G PGF+G G+P PPGARFDP GPPGVPGFEP RF R P R H
Sbjct: 235 FPFGDG---SDRPGFMGPPHPGMP--PPGARFDPYGPPGVPGFEPGRFTRQPPRGPGGGH 289
Query: 295 PDLQHFRRDTD 305
PDL+HF +D
Sbjct: 290 PDLEHFPGGSD 300
>At1g48530 hypothetical protein
Length = 164
Score = 143 bits (361), Expect = 1e-34
Identities = 80/168 (47%), Positives = 111/168 (65%), Gaps = 6/168 (3%)
Query: 1 MATDKSVLAVIRAARPTFRNQNDKIAFVVHSSFLASGYILTATGPQALSDTALSNPSND- 59
MAT +V+ VIR+ P FRN+NDKIAFVVH+S SG+ILT+TG A + ALS+ +
Sbjct: 1 MATTHAVMGVIRSVMPAFRNKNDKIAFVVHASLAVSGFILTSTGRPAFAHDALSSSTTQC 60
Query: 60 EVSVDHWNELNDEYAFVYANSEKEGLKKVLVKCLVMNGKLLVHALSEGFKEPLHLEINVA 119
V ++ WNE ++EYAFVY K +K+ LVKCL MN KLLV A++E KE HL+I V
Sbjct: 61 LVGIEGWNEFDEEYAFVY----KCPVKRYLVKCLAMNDKLLVDAIAEDGKEFGHLQIEVG 116
Query: 120 DYAGEDG-GSNFSQQFKNLDKLVKKIDADILSKLDGTAHASSSTKSSK 166
+Y E G ++ QFKN DKLV ++ ++IL KL+ + + S+ +K
Sbjct: 117 NYIDESGEEGDYDTQFKNFDKLVTELKSEILCKLNTVSTTTKSSSETK 164
>At4g22670 HSP associated protein like
Length = 441
Score = 45.8 bits (107), Expect = 3e-05
Identities = 49/147 (33%), Positives = 57/147 (38%), Gaps = 34/147 (23%)
Query: 142 KKIDADILSKLDGTAHASSSTKSSKTSDSTRPETSEPVPGFGEPAGPNDYHPGFIVPPVP 201
KK + D L + A K + S S+RP G G P G PG + P
Sbjct: 257 KKAERDRLRRRAEAQAAYDKAKKEEQSSSSRPS------GGGFPGGMPGGFPGGMPGGFP 310
Query: 202 AGSGS--DLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGG----GIGGG-PGFI 254
G G FPG GM G M P F GG GG G+GGG PG
Sbjct: 311 GGMGGMPGGFPGGMGGM----------GGM------PGGFPGGMGGGMPAGMGGGMPGMG 354
Query: 255 GGLP-----GAPPGARFDPIGPPGVPG 276
GG+P G PGA G G+PG
Sbjct: 355 GGMPAGMGGGGMPGAGGGMPGGGGMPG 381
Score = 38.1 bits (87), Expect = 0.006
Identities = 31/95 (32%), Positives = 37/95 (38%), Gaps = 15/95 (15%)
Query: 201 PAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGA 260
P+G G FPG G +P F G M GG GG GG G +GG+PG
Sbjct: 288 PSGGG---FPGGMPGGFPGGMPGGFPGGM-----------GGMPGGFPGGMGGMGGMPGG 333
Query: 261 PPGARFDPIGPPGVPGFEPNRFARNPRRPGFDAHP 295
PG + P G+ G P P G P
Sbjct: 334 FPGGMGGGM-PAGMGGGMPGMGGGMPAGMGGGGMP 367
>At5g04170 EF - hand Calcium binding protein - like
Length = 354
Score = 45.1 bits (105), Expect = 5e-05
Identities = 49/147 (33%), Positives = 62/147 (41%), Gaps = 30/147 (20%)
Query: 154 GTAHASSSTKSSKTSDSTR---PETSEPV---PG-FGEPAGPNDYHPGFIVPPVPAGS-- 204
G + SS+ S+ T S+ P S P PG + +P Y G+ PP P+GS
Sbjct: 51 GASKPQSSSSSAPTYGSSSYGAPPPSAPYAPSPGDYNKPPKEKPYGGGYGAPP-PSGSSD 109
Query: 205 ----GSDLFP------GPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFI 254
G+ P G G G P G D+G G PR S G GGG GG P
Sbjct: 110 YGSYGAGPRPSQPSGHGGGYGATPPHGVSDYGS---YGGAPPRPASSGHGGGYGGYP--- 163
Query: 255 GGLPGAPPGARFDPIGPPG-VPGFEPN 280
P A G+ F + P G PG +PN
Sbjct: 164 ---PQASYGSPFASLIPSGFAPGTDPN 187
>At3g02670 unknown protein
Length = 217
Score = 39.3 bits (90), Expect = 0.003
Identities = 56/181 (30%), Positives = 70/181 (37%), Gaps = 30/181 (16%)
Query: 139 KLVKKIDADILSKLDGTAHASSSTKSSKTSDSTRPETSEP----------VPGFGEPAGP 188
K V ++ ++ + A S S ++S T P P PG G P P
Sbjct: 34 KPVDRVVTTTTNEANNLAFVSDPFSSLQSSPPTSPIPGSPGFRLPFPFPSSPG-GNPGIP 92
Query: 189 NDYHPGFIVP-PVPAGSGSDLFPG-PGAGMYPSR-GDHDFGGSMLVGPNDPRWFSGGTGG 245
PGF +P P P+ G + PG PG P G G L P P GG+
Sbjct: 93 GS--PGFRLPFPFPSSPGGN--PGIPGIPGIPGLPGIPGSPGFRLPFPF-PSSPGGGSIP 147
Query: 246 GIGGGPGFI---------GGLPGAP-PGARFDPIGPPGVP-GFEPNRFARNPRRPGFDAH 294
GI G PGF GG+PG P P P+ PG+P F P P PGF
Sbjct: 148 GIPGSPGFRLPFPFPPSGGGIPGLPLPFPPLPPVTIPGLPLPFPPLPPVTIPSFPGFRFP 207
Query: 295 P 295
P
Sbjct: 208 P 208
>At1g63540 unknown protein
Length = 635
Score = 39.3 bits (90), Expect = 0.003
Identities = 44/166 (26%), Positives = 66/166 (39%), Gaps = 27/166 (16%)
Query: 154 GTAHASSSTKSSKTSDSTRPETSEP-----VPGFGEPAGPNDYHPGFIVPPVPAGSGSDL 208
G + +SS+ S++ +T P P + + + P VPP P+ SGS L
Sbjct: 289 GISPVASSSTSTEIFGATPASLFSPFGPMQAPVQASASSTSTFPPFGCVPPSPS-SGSSL 347
Query: 209 FPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGF--IGGLPGAPPGARF 266
F + FG L P+ +F + +G P +G LPG+P + F
Sbjct: 348 F------------NSAFGS--LPAPSSSNFFGQSSSNLLGQNPSTTGVGYLPGSPLNSSF 393
Query: 267 DPIGPPGVPGFEPNRFARNPRRPG---FDAHPDLQHFRRDTDSDYI 309
G +PG N F NP G A P QHF + D+ +
Sbjct: 394 PGFGVGYLPGSSSNLFRSNPPNFGGGSIGAGP--QHFGFNGDASVL 437
>At1g29380 beta-1,3 glucanase, putative
Length = 228
Score = 39.3 bits (90), Expect = 0.003
Identities = 36/114 (31%), Positives = 43/114 (37%), Gaps = 8/114 (7%)
Query: 150 SKLDGTAHASSSTKSSKTSDSTRPETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLF 209
S LD A + T P S P +P+ P+ PG P P G +
Sbjct: 22 SPLDVIAQGQGGQGDIPVVNPTAPGGSTTTPTITQPSPPSLTFPG---PTTPTGG----Y 74
Query: 210 PGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPG 263
P P G P+ G G+ G D GG GGG GG G GG GA G
Sbjct: 75 P-PLDGTTPTGGYPPLYGTTPPGGGDVGGGGGGYGGGTPGGGGGGGGDTGAGAG 127
>At5g21160 unknown protein
Length = 826
Score = 38.5 bits (88), Expect = 0.005
Identities = 34/112 (30%), Positives = 40/112 (35%), Gaps = 17/112 (15%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSR------GDHDFG 226
P PVP E + P +PA G P P + R G +FG
Sbjct: 158 PPYPVPVPPVTESGNEKQVQASPLPPVLPAPQGDPGKPWPHQRGFDPRNMPQGAGPRNFG 217
Query: 227 GSMLVGPNDPRWFSGGTGGGIGGGPGFIGG---LPGAPPGARFDPIGPPGVP 275
+GP G +G GPGF G LPG PPGA P P P
Sbjct: 218 RPPFMGP--------APGFLVGPGPGFPGPVYYLPGPPPGAIRGPYPPRFAP 261
>At5g40490 ribonucleoprotein -like
Length = 423
Score = 38.1 bits (87), Expect = 0.006
Identities = 27/68 (39%), Positives = 31/68 (44%), Gaps = 5/68 (7%)
Query: 211 GPGAGMYPSRGDHDFGGSMLVGPNDPRW----FSGGTGGGIGGGPG-FIGGLPGAPPGAR 265
G G G+ P RG+ G S G + +S G GGG GGGPG GG G P G
Sbjct: 279 GYGGGVGPYRGEPALGYSGRYGGGGGGYNRGGYSMGGGGGYGGGPGDMYGGSYGEPGGGY 338
Query: 266 FDPIGPPG 273
P G G
Sbjct: 339 GGPSGSYG 346
Score = 35.8 bits (81), Expect = 0.030
Identities = 34/99 (34%), Positives = 41/99 (41%), Gaps = 14/99 (14%)
Query: 177 EPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDP 236
EP G+G P+G Y G+ + G G G G Y G +D GG VG
Sbjct: 333 EPGGGYGGPSG--SYGGGYGSSGIGGYGGG--MGGAGGGGYRGGGGYDMGG---VGGGGA 385
Query: 237 RWFSGGTGGGIGGGP-----GFIGGLPGAPPGARFDPIG 270
+ G GGG GGG G GG G G R+ P G
Sbjct: 386 GGYGAG-GGGNGGGSFYGGGGGRGGYGGGGSG-RYHPYG 422
Score = 31.2 bits (69), Expect = 0.75
Identities = 22/67 (32%), Positives = 28/67 (40%), Gaps = 9/67 (13%)
Query: 211 GPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIG 270
G G G Y +RG + GG + GG G GG G GG G P G+ G
Sbjct: 300 GGGGGGY-NRGGYSMGGG--------GGYGGGPGDMYGGSYGEPGGGYGGPSGSYGGGYG 350
Query: 271 PPGVPGF 277
G+ G+
Sbjct: 351 SSGIGGY 357
Score = 28.5 bits (62), Expect = 4.8
Identities = 17/41 (41%), Positives = 19/41 (45%), Gaps = 1/41 (2%)
Query: 232 GPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARF-DPIGP 271
GP P GG GGG GG G GG G G + +GP
Sbjct: 246 GPGGPYKSGGGYGGGRSGGYGGYGGEFGGYGGGGYGGGVGP 286
Score = 28.1 bits (61), Expect = 6.3
Identities = 22/58 (37%), Positives = 25/58 (42%), Gaps = 2/58 (3%)
Query: 209 FPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARF 266
+ GPG G Y S G + G S G F G GGG GGG G G P R+
Sbjct: 244 YGGPG-GPYKSGGGYGGGRSGGYGGYGGE-FGGYGGGGYGGGVGPYRGEPALGYSGRY 299
Score = 27.7 bits (60), Expect = 8.2
Identities = 13/23 (56%), Positives = 13/23 (56%)
Query: 239 FSGGTGGGIGGGPGFIGGLPGAP 261
F GG G G GGG G G PG P
Sbjct: 228 FGGGYGDGYGGGHGGGYGGPGGP 250
>At3g17050 unknown protein
Length = 349
Score = 38.1 bits (87), Expect = 0.006
Identities = 29/83 (34%), Positives = 31/83 (36%), Gaps = 11/83 (13%)
Query: 181 GFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFS 240
G G G H G I G+G L G G G+ G GG
Sbjct: 94 GGGAGGGLGGGHGGGIGGGAGGGAGGGLGGGHGGGIGGGAGGGSGGG-----------LG 142
Query: 241 GGTGGGIGGGPGFIGGLPGAPPG 263
GG GGG GGG G GGL G G
Sbjct: 143 GGIGGGAGGGAGGGGGLGGGHGG 165
Score = 37.7 bits (86), Expect = 0.008
Identities = 32/90 (35%), Positives = 34/90 (37%), Gaps = 7/90 (7%)
Query: 181 GFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHD-FGGSMLVGPNDPRWF 239
G G G H G I GSG L G G G G GG +G F
Sbjct: 172 GGGAGGGLGGGHGGGIGGGAGGGSGGGLGGGIGGGAGGGAGGGGGAGGGGGLGGGHGGGF 231
Query: 240 SGGTGGGIGGGP------GFIGGLPGAPPG 263
GG GGG+GGG GF GG G G
Sbjct: 232 GGGAGGGLGGGAGGGTGGGFGGGAGGGAGG 261
Score = 36.2 bits (82), Expect = 0.023
Identities = 22/60 (36%), Positives = 27/60 (44%), Gaps = 3/60 (5%)
Query: 211 GPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIG 270
G G G +G FGG + G GG GGG+GGG G GG+ G G +G
Sbjct: 38 GLGGGFGGGKG---FGGGIGAGGGFGGGAGGGAGGGLGGGAGGGGGIGGGAGGGAGGGLG 94
Score = 35.8 bits (81), Expect = 0.030
Identities = 26/64 (40%), Positives = 27/64 (41%), Gaps = 5/64 (7%)
Query: 202 AGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGP--GFIGGLPG 259
AG G L G G G G GG+ G F GG GGG GGG GF GG G
Sbjct: 217 AGGGGGLGGGHGGGFGGGAGGGLGGGA---GGGTGGGFGGGAGGGAGGGAGGGFGGGAGG 273
Query: 260 APPG 263
G
Sbjct: 274 GAGG 277
Score = 35.4 bits (80), Expect = 0.040
Identities = 31/90 (34%), Positives = 34/90 (37%), Gaps = 11/90 (12%)
Query: 181 GFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFS 240
GFG AG G + G+G G G G G FGG G F
Sbjct: 230 GFGGGAG------GGLGGGAGGGTGGGFGGGAGGGAGGGAGG-GFGGG--AGGGAGGGFG 280
Query: 241 GGTGGGIGGGPGFIGGLPGAPPGARFDPIG 270
GG GGG GGG G GG G G +G
Sbjct: 281 GGAGGGAGGGAG--GGFGGGAGGGHGGGVG 308
Score = 33.9 bits (76), Expect = 0.11
Identities = 29/90 (32%), Positives = 33/90 (36%), Gaps = 1/90 (1%)
Query: 181 GFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFS 240
G G G H G I G+G L G G G+ G GG + G
Sbjct: 152 GAGGGGGLGGGHGGGIGGGAGGGAGGGLGGGHGGGIGGGAGGGS-GGGLGGGIGGGAGGG 210
Query: 241 GGTGGGIGGGPGFIGGLPGAPPGARFDPIG 270
G GGG GGG G GG G G +G
Sbjct: 211 AGGGGGAGGGGGLGGGHGGGFGGGAGGGLG 240
Score = 32.3 bits (72), Expect = 0.33
Identities = 23/61 (37%), Positives = 26/61 (41%), Gaps = 7/61 (11%)
Query: 203 GSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPP 262
G+G G G G G GG + G GG GGG+GGG G GGL G
Sbjct: 54 GAGGGFGGGAGGGAGGGLGGGAGGGGGIGGGA-----GGGAGGGLGGGAG--GGLGGGHG 106
Query: 263 G 263
G
Sbjct: 107 G 107
Score = 31.6 bits (70), Expect = 0.57
Identities = 27/76 (35%), Positives = 28/76 (36%), Gaps = 9/76 (11%)
Query: 203 GSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGP--GFIGGLPGA 260
G+G G G G G FGG G GG GGG GGG GF GG G
Sbjct: 274 GAGGGFGGGAGGGAGGGAGG-GFGGGAGGG------HGGGVGGGFGGGSGGGFGGGAGGG 326
Query: 261 PPGARFDPIGPPGVPG 276
G G G G
Sbjct: 327 AGGGAGGGFGGGGGAG 342
Score = 30.8 bits (68), Expect = 0.97
Identities = 12/17 (70%), Positives = 13/17 (75%)
Query: 241 GGTGGGIGGGPGFIGGL 257
GG GGG GGG GF GG+
Sbjct: 37 GGLGGGFGGGKGFGGGI 53
Score = 30.4 bits (67), Expect = 1.3
Identities = 31/93 (33%), Positives = 35/93 (37%), Gaps = 9/93 (9%)
Query: 181 GFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFS 240
G G G H G I GSG L G G G G GG L G +
Sbjct: 114 GGGAGGGLGGGHGGGIGGGAGGGSGGGLGGGIGGGAGGGAG----GGGGLGGGHGGGIGG 169
Query: 241 GGTG---GGIGGGPGFIGGLPGAPPGARFDPIG 270
G G GG+GGG G GG+ G G +G
Sbjct: 170 GAGGGAGGGLGGGHG--GGIGGGAGGGSGGGLG 200
Score = 30.0 bits (66), Expect = 1.7
Identities = 22/57 (38%), Positives = 23/57 (39%), Gaps = 7/57 (12%)
Query: 203 GSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPG 259
G+G G G G G FGG G GG GGG GGG G GG G
Sbjct: 298 GAGGGHGGGVGGGFGGGSGG-GFGGGAGGGAG------GGAGGGFGGGGGAGGGFGG 347
Score = 28.1 bits (61), Expect = 6.3
Identities = 25/76 (32%), Positives = 26/76 (33%), Gaps = 21/76 (27%)
Query: 181 GFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFS 240
GFG AG H G + GSG G G G G
Sbjct: 294 GFGGGAGGG--HGGGVGGGFGGGSGGGFGGGAGGGAGGGAG------------------- 332
Query: 241 GGTGGGIGGGPGFIGG 256
GG GGG G G GF GG
Sbjct: 333 GGFGGGGGAGGGFGGG 348
Score = 27.7 bits (60), Expect = 8.2
Identities = 26/75 (34%), Positives = 29/75 (38%), Gaps = 2/75 (2%)
Query: 202 AGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAP 261
AG G L G G G+ G GG L G + G GG GG G IGG G
Sbjct: 153 AGGGGGLGGGHGGGIGGGAGGGAGGG--LGGGHGGGIGGGAGGGSGGGLGGGIGGGAGGG 210
Query: 262 PGARFDPIGPPGVPG 276
G G G+ G
Sbjct: 211 AGGGGGAGGGGGLGG 225
>At2g04190 unknown protein
Length = 412
Score = 38.1 bits (87), Expect = 0.006
Identities = 29/86 (33%), Positives = 33/86 (37%), Gaps = 1/86 (1%)
Query: 180 PGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWF 239
PG G GP + PG+ + G F G GAG P G G G D R +
Sbjct: 39 PGRGYGGGPRVHGPGYGIGSRGPDPGPGFFFG-GAGPGPGYGGGGGHGPGYGGGGDGRGY 97
Query: 240 SGGTGGGIGGGPGFIGGLPGAPPGAR 265
TGGG G G L PP R
Sbjct: 98 GSETGGGNHGPEGVTRTLRDEPPSHR 123
Score = 28.9 bits (63), Expect = 3.7
Identities = 26/68 (38%), Positives = 28/68 (40%), Gaps = 13/68 (19%)
Query: 203 GSGSDLFPGPGAGMYPSRGDHDFGGSMLV-------GPNDPRWFSGGTGGGIGGGPGFIG 255
G G F G G G P RG +GG V G P G GG G GPG+ G
Sbjct: 26 GGGGPAFGGRGGG--PGRG---YGGGPRVHGPGYGIGSRGPDPGPGFFFGGAGPGPGY-G 79
Query: 256 GLPGAPPG 263
G G PG
Sbjct: 80 GGGGHGPG 87
Score = 28.9 bits (63), Expect = 3.7
Identities = 20/45 (44%), Positives = 21/45 (46%), Gaps = 6/45 (13%)
Query: 239 FSGGTGG----GIGGGP--GFIGGLPGAPPGARFDPIGPPGVPGF 277
F GG GG G GGGP G+ GG PG GP PGF
Sbjct: 23 FGGGGGGPAFGGRGGGPGRGYGGGPRVHGPGYGIGSRGPDPGPGF 67
>At3g20470 glycine-rich protein, putative
Length = 174
Score = 37.7 bits (86), Expect = 0.008
Identities = 31/78 (39%), Positives = 33/78 (41%), Gaps = 20/78 (25%)
Query: 202 AGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGL---P 258
AGSG L G GAG FGG GG+GGG GGG G GGL
Sbjct: 97 AGSGGGLGGGGGAG-------GGFGGGA----------GGGSGGGFGGGAGAGGGLGGGG 139
Query: 259 GAPPGARFDPIGPPGVPG 276
GA G F G G+ G
Sbjct: 140 GAGGGGGFGGGGGSGIGG 157
Score = 35.0 bits (79), Expect = 0.052
Identities = 25/57 (43%), Positives = 25/57 (43%), Gaps = 5/57 (8%)
Query: 203 GSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPG 259
G GS L G G G G GG L G GG GGG GGG G GGL G
Sbjct: 54 GGGSGLGGGGGFGGGGGLGGGAGGGGGLGGGA-----GGGAGGGFGGGAGSGGGLGG 105
Score = 33.9 bits (76), Expect = 0.11
Identities = 24/55 (43%), Positives = 25/55 (44%), Gaps = 11/55 (20%)
Query: 202 AGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGG 256
AG+G L G GAG G FGG G GG GGG G G GF GG
Sbjct: 129 AGAGGGLGGGGGAG-----GGGGFGGGGGSG------IGGGFGGGAGAGGGFGGG 172
Score = 33.1 bits (74), Expect = 0.20
Identities = 21/55 (38%), Positives = 23/55 (41%), Gaps = 3/55 (5%)
Query: 209 FPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPG 263
F PG G+ G GG +G GG GGG GGG G GG G G
Sbjct: 40 FHHPGGGL---GGGGGIGGGSGLGGGGGFGGGGGLGGGAGGGGGLGGGAGGGAGG 91
Score = 27.7 bits (60), Expect = 8.2
Identities = 29/85 (34%), Positives = 30/85 (35%), Gaps = 12/85 (14%)
Query: 181 GFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFS 240
GFG AG G G+G G G G FGG G
Sbjct: 92 GFGGGAGSGGGLGG------GGGAGGGFGGGAGGG-----SGGGFGGGAGAGGGLGGGGG 140
Query: 241 GGTGGGIGGGPGF-IGGLPGAPPGA 264
G GGG GGG G IGG G GA
Sbjct: 141 AGGGGGFGGGGGSGIGGGFGGGAGA 165
>At2g28670 putative disease resistance response protein
Length = 447
Score = 37.7 bits (86), Expect = 0.008
Identities = 32/90 (35%), Positives = 38/90 (41%), Gaps = 8/90 (8%)
Query: 181 GFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSR----GDHDFGGS-MLVGPND 235
GFG AG G + AG+GS G G G+ P+ G GGS L
Sbjct: 62 GFGFGAGSGSSGSGSTGSGLGAGTGSIPSSGSGPGLLPTASSVPGSLAGGGSGSLPTTGS 121
Query: 236 PRWFSGGTGGGIGGGPG---FIGGLPGAPP 262
GTG +GGGPG +GG GA P
Sbjct: 122 ATGAGAGTGSALGGGPGAGSALGGGAGAGP 151
Score = 37.4 bits (85), Expect = 0.010
Identities = 37/115 (32%), Positives = 42/115 (36%), Gaps = 7/115 (6%)
Query: 154 GTAHASSSTKSSKTSDSTRPETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPG 213
G + S SS + P T+ VPG G AG+GS L GPG
Sbjct: 80 GLGAGTGSIPSSGSGPGLLP-TASSVPGSLAGGGSGSLPTTGSATGAGAGTGSALGGGPG 138
Query: 214 AGMYPSRG---DHDFGGSMLVGPNDPRWFSGGT---GGGIGGGPGFIGGLPGAPP 262
AG G GG GP G+ GGG G GP GG GA P
Sbjct: 139 AGSALGGGAGAGPALGGGAGAGPALGGGAGAGSALGGGGAGAGPALGGGGAGAGP 193
>At1g30460 hypothetical protein
Length = 678
Score = 37.7 bits (86), Expect = 0.008
Identities = 35/100 (35%), Positives = 42/100 (42%), Gaps = 17/100 (17%)
Query: 178 PVPGFGE----PAGPNDYHP----GFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGG-- 227
P+PG G GP D P G+ P P G G F G Y R DF G
Sbjct: 507 PMPGMGGFPLGVMGPGDAFPYGPGGYNGMPDPFGMGPRPF-----GPYGPRFGGDFRGPV 561
Query: 228 -SMLVGPNDPRWFSGGTGGGIGGGPG-FIGGLPGAPPGAR 265
M+ P+ F G G +GGG G +GG+ AP G R
Sbjct: 562 PGMMFPGRPPQQFPHGGYGMMGGGRGPHMGGMGNAPRGGR 601
>At1g27710 unknown protein
Length = 212
Score = 37.7 bits (86), Expect = 0.008
Identities = 23/54 (42%), Positives = 27/54 (49%), Gaps = 7/54 (12%)
Query: 210 PGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPG 263
PG G G Y G GG +++G F GG G G GGG G+ GG G PG
Sbjct: 112 PGYGGGGYGPGGG---GGGVVIGGG----FGGGAGYGSGGGLGWDGGNGGGGPG 158
Score = 35.4 bits (80), Expect = 0.040
Identities = 38/103 (36%), Positives = 45/103 (42%), Gaps = 10/103 (9%)
Query: 156 AHASSSTKSSKTSDSTRPETSEPVPGFGEPA-GPNDYHPGFIVPPVPAGSGSDLFPGPGA 214
AH+S KS + + E + G G P G Y PG V G G G GA
Sbjct: 83 AHSSEIGKSIRHVTGFKGELTAGGYGGGGPGYGGGGYGPGGGGGGVVIGGGF----GGGA 138
Query: 215 GMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGL 257
G Y S G + G G P + SGG GGIGGG G GG+
Sbjct: 139 G-YGSGGGLGWDGGN--GGGGPGYGSGG--GGIGGGGGIGGGV 176
>At1g75660 Dhp1-like protein
Length = 1020
Score = 37.4 bits (85), Expect = 0.010
Identities = 31/110 (28%), Positives = 42/110 (38%), Gaps = 14/110 (12%)
Query: 152 LDGTAHASSSTKSSKTSDSTRPETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPG 211
L AH S +D + T P PG+G Y+P VPP+P G + P
Sbjct: 859 LGNAAHRLVSNSLQMGTDRYQTPTDVPAPGYG-------YNPPQYVPPIPYQHGGYMAP- 910
Query: 212 PGA------GMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIG 255
PGA Y +RG + G P++P G G G+ G
Sbjct: 911 PGAQGYAQPAPYQNRGGYQPRGPSGRFPSEPYQSQSREGQHASRGGGYSG 960
>At5g19090 unknown protein
Length = 587
Score = 37.0 bits (84), Expect = 0.014
Identities = 26/75 (34%), Positives = 31/75 (40%), Gaps = 4/75 (5%)
Query: 203 GSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGF----IGGLP 258
G G + GP G G GG + G P + G GGG GGGP +GG
Sbjct: 371 GGGVQMNGGPNGGKKGGGGGGGGGGGPMSGGLPPGFRPMGGGGGGGGGPQSMSMPMGGAM 430
Query: 259 GAPPGARFDPIGPPG 273
G P G+ G PG
Sbjct: 431 GGPMGSLPQMGGGPG 445
Score = 36.2 bits (82), Expect = 0.023
Identities = 32/78 (41%), Positives = 35/78 (44%), Gaps = 8/78 (10%)
Query: 203 GSGSDLFPGPGAGMYPSRGDHDFGG-SMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAP 261
G G L G G G+ GG M GPN + GG GGG GGG GGL P
Sbjct: 349 GGGHPLDGKMGGGGGGPNGNKGGGGVQMNGGPNGGK--KGGGGGGGGGGGPMSGGL---P 403
Query: 262 PGARFDPIGPPGVPGFEP 279
PG F P+G G G P
Sbjct: 404 PG--FRPMGGGGGGGGGP 419
Score = 33.1 bits (74), Expect = 0.20
Identities = 35/117 (29%), Positives = 44/117 (36%), Gaps = 29/117 (24%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVP--------------PVPAGS---GSDL-FP---- 210
P +P+ G G PN+ P ++P P PAG G + FP
Sbjct: 254 PNKMKPMMGGGNMIMPNNMMPNMMMPNAQQMLNAHKNGGGPGPAGGKIEGKGMPFPVQMG 313
Query: 211 GPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGG----PGFIGGLPGAPPG 263
G G G +G GG + N GG GG GGG G +GG G P G
Sbjct: 314 GGGGGPGGKKGGPGGGGGNMGNQNQG---GGGKNGGKGGGGHPLDGKMGGGGGGPNG 367
Score = 27.7 bits (60), Expect = 8.2
Identities = 15/33 (45%), Positives = 16/33 (48%)
Query: 226 GGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLP 258
GG N + GG GGG GGG G GG P
Sbjct: 109 GGGGPANNNKGQKIGGGGGGGGGGGGGGGGGPP 141
>At4g20440 putative snRNP protein
Length = 257
Score = 36.6 bits (83), Expect = 0.018
Identities = 43/138 (31%), Positives = 50/138 (36%), Gaps = 16/138 (11%)
Query: 172 RPETSEPVPGFGEPAGPN-----DYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFG 226
+P S PV G G PA P PP+ G L P P G P G
Sbjct: 125 QPGLSGPVRGVGGPAPGMMQPQISRPPQLSAPPIIRPPGQMLPPPPFGGQGPPMGRGPPP 184
Query: 227 GSMLVGPNDPRWFSGGTGGGIGGGP------GFIGGLPGAPPGARFDPIGPPGVPGFEPN 280
+ P P+ FSG G P G + G P P G + P PG+P P
Sbjct: 185 PYGMRPP--PQQFSGPPPPQYGQRPMIPPPGGMMRGPPPPPHGMQGPPPPRPGMPP-APG 241
Query: 281 RFARNPRRPGFDAHPDLQ 298
FA P RPG H Q
Sbjct: 242 GFA--PPRPGMPPHNQQQ 257
>At2g05510 putative glycine-rich protein
Length = 127
Score = 36.6 bits (83), Expect = 0.018
Identities = 32/117 (27%), Positives = 41/117 (34%), Gaps = 21/117 (17%)
Query: 160 SSTKSSKTSDSTRPETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPS 219
S +++ S +PE+ V P YH G G G G +
Sbjct: 19 SEVSAARQSGMVKPESEATVQ-------PEGYHGGH--------------GGHGGGGHYG 57
Query: 220 RGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPG 276
G H GG G + + GG GG GGG G GG G G + G G G
Sbjct: 58 GGGHGHGGHNGGGGHGLDGYGGGHGGHYGGGGGHYGGGGGHGGGGHYGGGGHHGGGG 114
>At1g09070 unknown protein
Length = 324
Score = 36.6 bits (83), Expect = 0.018
Identities = 39/140 (27%), Positives = 55/140 (38%), Gaps = 32/140 (22%)
Query: 182 FGEPAGPNDYHPGFI--------VPPVPAGSGS-DLFPGPGAGM---YPSRGDHDFGGSM 229
+G +GP+ P + V P G G+ +P P AG YP +G HD
Sbjct: 140 YGSSSGPHAPVPSAMDHKTMDQPVTAYPPGHGAPSAYPAPPAGPSSGYPPQG-HDDKHDG 198
Query: 230 LVGPNDPRWFSGGTGG--------GIGGGPGFI----GGLPGAPPGARFD-------PIG 270
+ G + GTGG GG PG+ GG PG PP + P G
Sbjct: 199 VYGYPQQAGYPAGTGGYPPPGAYPQQGGYPGYPPQQQGGYPGYPPQGPYGYPQQGYPPQG 258
Query: 271 PPGVPGFEPNRFARNPRRPG 290
P G P + + + P++ G
Sbjct: 259 PYGYPQQQAHGKPQKPKKHG 278
Score = 28.5 bits (62), Expect = 4.8
Identities = 41/150 (27%), Positives = 50/150 (33%), Gaps = 56/150 (37%)
Query: 161 STKSSKTSDSTRPETSEPVPGFGEP-------AGPN---------DYHPGFIVPPVPAG- 203
S KT D +P T+ P PG G P AGP+ D H G P AG
Sbjct: 152 SAMDHKTMD--QPVTAYP-PGHGAPSAYPAPPAGPSSGYPPQGHDDKHDGVYGYPQQAGY 208
Query: 204 -SGSDLFPGPGA----GMYPSRGDHDFGGSMLVGPNDPRWF------------------- 239
+G+ +P PGA G YP GG P P +
Sbjct: 209 PAGTGGYPPPGAYPQQGGYPGYPPQQQGGYPGYPPQGPYGYPQQGYPPQGPYGYPQQQAH 268
Query: 240 ------------SGGTGGGIGGGPGFIGGL 257
G G G+G G G +GGL
Sbjct: 269 GKPQKPKKHGKAGAGMGLGLGLGAGLLGGL 298
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,459,150
Number of Sequences: 26719
Number of extensions: 466578
Number of successful extensions: 3044
Number of sequences better than 10.0: 139
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 91
Number of HSP's that attempted gapping in prelim test: 1501
Number of HSP's gapped (non-prelim): 685
length of query: 309
length of database: 11,318,596
effective HSP length: 99
effective length of query: 210
effective length of database: 8,673,415
effective search space: 1821417150
effective search space used: 1821417150
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC142224.1


