

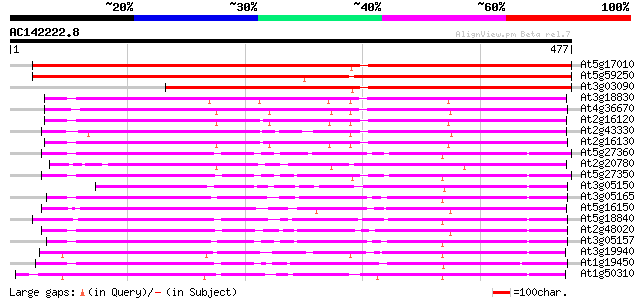
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.8 - phase: 0
(477 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g17010 sugar transporter - like protein 649 0.0
At5g59250 D-xylose-H+ symporter - like protein 537 e-153
At3g03090 hypothetical protein 459 e-129
At3g18830 sugar transporter like protein 231 5e-61
At4g36670 sugar transporter like protein 229 3e-60
At2g16120 putative sugar transporter 215 5e-56
At2g43330 membrane transporter like protein 214 6e-56
At2g16130 putative sugar transporter 213 1e-55
At5g27360 sugar-porter family protein 2 (SFP2) 210 2e-54
At2g20780 putative sugar transporter 203 1e-52
At5g27350 sugar transporter like protein 201 6e-52
At3g05150 putative sugar transporter 201 6e-52
At3g05165 sugar transporter like protein 195 5e-50
At5g16150 sugar transporter like protein 192 3e-49
At5g18840 sugar transporter - like protein 192 4e-49
At2g48020 sugar transporter like protein 191 6e-49
At3g05157 sugar transporter like protein 188 5e-48
At3g19940 putative monosaccharide transport protein, STP4 182 5e-46
At1g19450 integral membrane protein, putative 181 1e-45
At1g50310 hexose transporter, putative 179 3e-45
>At5g17010 sugar transporter - like protein
Length = 503
Score = 649 bits (1675), Expect = 0.0
Identities = 321/461 (69%), Positives = 392/461 (84%), Gaps = 9/461 (1%)
Query: 20 LRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGS 79
L FLFPA GGLL+GY+IGATS ATIS+QS SLSGI+WY+L +V++GL+TSGSLYGAL GS
Sbjct: 49 LPFLFPALGGLLYGYEIGATSCATISLQSPSLSGISWYNLSSVDVGLVTSGSLYGALFGS 108
Query: 80 VLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMY 139
++AF IAD +GRR+EL++AAL+YLVGAL+TA AP + +L+IGR+++G+ +GLAMHAAPMY
Sbjct: 109 IVAFTIADVIGRRKELILAALLYLVGALVTALAPTYSVLIIGRVIYGVSVGLAMHAAPMY 168
Query: 140 IAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGM 199
IAETAP+PIRGQLVSLKEFFIV+G+V GYG+GSL V+ +GWRYM+ S P+AVIMG GM
Sbjct: 169 IAETAPSPIRGQLVSLKEFFIVLGMVGGYGIGSLTVNVHSGWRYMYATSVPLAVIMGIGM 228
Query: 200 WWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGE 259
WWLPASPRW+LLR IQ KG+++ ++ AI+SLC L+G F DSA +QV+EI+AE +++GE
Sbjct: 229 WWLPASPRWLLLRVIQGKGNVENQREAAIKSLCCLRGPAFVDSAAEQVNEILAELTFVGE 288
Query: 260 ENDVTLGEMFRGKCRKALVISAGLVLFQQI---QTVTDHGPTKCSLLRCINPTGFSLAAD 316
+ +VT GE+F+GKC KAL+I GLVLFQQI +V + P+ + GFS A D
Sbjct: 289 DKEVTFGELFQGKCLKALIIGGGLVLFQQITGQPSVLYYAPS------ILQTAGFSAAGD 342
Query: 317 ATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLA 376
ATRVSILLG+ KLIMTGVAVVV+DRLGRRPLLLGGV G+V+SLFLLGSYY+F + V+A
Sbjct: 343 ATRVSILLGLLKLIMTGVAVVVIDRLGRRPLLLGGVGGMVVSLFLLGSYYLFFSASPVVA 402
Query: 377 VVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDL 436
VV LLLYVGCYQ+SFGP+GWLMI+EIFPL+LRG+GLS+AVLVNF ANALVTFAFSPLK+L
Sbjct: 403 VVALLLYVGCYQLSFGPIGWLMISEIFPLKLRGRGLSLAVLVNFGANALVTFAFSPLKEL 462
Query: 437 LGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
LGAGILF F I V SLVFI+FIVPETKGLTLEEIEAKCL
Sbjct: 463 LGAGILFCGFGVICVLSLVFIFFIVPETKGLTLEEIEAKCL 503
>At5g59250 D-xylose-H+ symporter - like protein
Length = 558
Score = 537 bits (1383), Expect = e-153
Identities = 271/461 (58%), Positives = 351/461 (75%), Gaps = 6/461 (1%)
Query: 20 LRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGS 79
L F+FPA GGLLFGYDIGATS AT+S+QS +LSG TW++ V++GL+ SGSLYGAL+GS
Sbjct: 100 LPFIFPALGGLLFGYDIGATSGATLSLQSPALSGTTWFNFSPVQLGLVVSGSLYGALLGS 159
Query: 80 VLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMY 139
+ + +ADFLGRRREL++AA++YL+G+LIT AP+ +L++GRL++G GIGLAMH AP+Y
Sbjct: 160 ISVYGVADFLGRRRELIIAAVLYLLGSLITGCAPDLNILLVGRLLYGFGIGLAMHGAPLY 219
Query: 140 IAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGM 199
IAET P+ IRG L+SLKE FIV+GI+ G+ +GS +D V GWRYM+G +PVA++MG GM
Sbjct: 220 IAETCPSQIRGTLISLKELFIVLGILLGFSVGSFQIDVVGGWRYMYGFGTPVALLMGLGM 279
Query: 200 WWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQ-VDE--IMAEFSY 256
W LPASPRW+LLRA+Q KG LQ K+ A+ +L +L+GR D ++ VD+ + + +Y
Sbjct: 280 WSLPASPRWLLLRAVQGKGQLQEYKEKAMLALSKLRGRPPGDKISEKLVDDAYLSVKTAY 339
Query: 257 LGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAAD 316
E++ E+F+G KAL I GLVLFQQI T + GFS AAD
Sbjct: 340 EDEKSGGNFLEVFQGPNLKALTIGGGLVLFQQI---TGQPSVLYYAGSILQTAGFSAAAD 396
Query: 317 ATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLA 376
ATRVS+++GVFKL+MT VAV VD LGRRPLL+GGVSGI +SLFLL +YY FL ++A
Sbjct: 397 ATRVSVIIGVFKLLMTWVAVAKVDDLGRRPLLIGGVSGIALSLFLLSAYYKFLGGFPLVA 456
Query: 377 VVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDL 436
V LLLYVGCYQISFGP+ WLM++EIFPLR RG+G+S+AVL NF +NA+VTFAFSPLK+
Sbjct: 457 VGALLLYVGCYQISFGPISWLMVSEIFPLRTRGRGISLAVLTNFGSNAIVTFAFSPLKEF 516
Query: 437 LGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
LGA LF +F IA+ SL+F+ +VPETKGL+LEEIE+K L
Sbjct: 517 LGAENLFLLFGGIALVSLLFVILVVPETKGLSLEEIESKIL 557
>At3g03090 hypothetical protein
Length = 342
Score = 459 bits (1182), Expect = e-129
Identities = 230/348 (66%), Positives = 280/348 (80%), Gaps = 9/348 (2%)
Query: 133 MHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVA 192
MHAAPMYIAETAP+ IRG+++SLKEF V+G+V GYG+GSL + ++GWRYM+ P
Sbjct: 1 MHAAPMYIAETAPSQIRGRMISLKEFSTVLGMVGGYGIGSLWITVISGWRYMYATILPFP 60
Query: 193 VIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMA 252
VIMG GM WLPASPRW+LLRA+Q +G+ + L+ AIRSLC+L+G DSA +QV+EI+A
Sbjct: 61 VIMGTGMCWLPASPRWLLLRALQGQGNGENLQQAAIRSLCRLRGSVIADSAAEQVNEILA 120
Query: 253 EFSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQ---TVTDHGPTKCSLLRCINPT 309
E S +GE+ + T GE+FRGKC KAL I+ GLVLFQQI +V + P+ +
Sbjct: 121 ELSLVGEDKEATFGELFRGKCLKALTIAGGLVLFQQITGQPSVLYYAPS------ILQTA 174
Query: 310 GFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFL 369
GFS AADATR+SILLG+ KL+MTGV+V+V+DR+GRRPLLL GVSG+VISLFLLGSYY+F
Sbjct: 175 GFSAAADATRISILLGLLKLVMTGVSVIVIDRVGRRPLLLCGVSGMVISLFLLGSYYMFY 234
Query: 370 DNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFA 429
N +AV LLLYVGCYQ+SFGP+GWLMI+EIFPL+LRG+G+S+AVLVNF ANALVTFA
Sbjct: 235 KNVPAVAVAALLLYVGCYQLSFGPIGWLMISEIFPLKLRGRGISLAVLVNFGANALVTFA 294
Query: 430 FSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
FSPLK+LLGAGILF F I V SL FIY+IVPETKGLTLEEIEAKCL
Sbjct: 295 FSPLKELLGAGILFCAFGVICVVSLFFIYYIVPETKGLTLEEIEAKCL 342
>At3g18830 sugar transporter like protein
Length = 539
Score = 231 bits (590), Expect = 5e-61
Identities = 146/461 (31%), Positives = 231/461 (49%), Gaps = 30/461 (6%)
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
+L GYDIG S A I I+ ++ ++IG+L +LIGS A +D++
Sbjct: 48 ILLGYDIGVMSGAMIYIKRD-------LKINDLQIGILAGSLNIYSLIGSCAAGRTSDWI 100
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR +++A ++ GA++ +PN+ L+ GR + GIG+G A+ AP+Y AE +P R
Sbjct: 101 GRRYTIVLAGAIFFAGAILMGLSPNYAFLMFGRFIAGIGVGYALMIAPVYTAEVSPASSR 160
Query: 150 GQLVSLKEFFIVIGIVAGY--GLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
G L S E FI GI+ GY L + GWR M GI + +VI+ G+ +P SPR
Sbjct: 161 GFLNSFPEVFINAGIMLGYVSNLAFSNLPLKVGWRLMLGIGAVPSVILAIGVLAMPESPR 220
Query: 208 WILL--RAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVTL 265
W+++ R K L D+ + +L+ P + + + S +
Sbjct: 221 WLVMQGRLGDAKRVLDKTSDSPTEATLRLEDIKHAAGIPADCHDDVVQVSRRNSHGEGVW 280
Query: 266 GEMF---RGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADATR 319
E+ R+ ++ + G+ FQQ I V P R G
Sbjct: 281 RELLIRPTPAVRRVMIAAIGIHFFQQASGIDAVVLFSP------RIFKTAGLKTDHQQLL 334
Query: 320 VSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN-------A 372
++ +GV K VA ++DR+GRRPLLL V G+V+SL LG+ +D A
Sbjct: 335 ATVAVGVVKTSFILVATFLLDRIGRRPLLLTSVGGMVLSLAALGTSLTIIDQSEKKVMWA 394
Query: 373 AVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSP 432
V+A+ ++ YV + I GP+ W+ +EIFPLRLR +G S+ V+VN + +++ +F P
Sbjct: 395 VVVAIATVMTYVATFSIGAGPITWVYSSEIFPLRLRSQGSSMGVVVNRVTSGVISISFLP 454
Query: 433 LKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
+ + G FY+F IA + VF Y +PET+G LE+++
Sbjct: 455 MSKAMTTGGAFYLFGGIATVAWVFFYTFLPETQGRMLEDMD 495
Score = 28.5 bits (62), Expect = 8.5
Identities = 36/160 (22%), Positives = 67/160 (41%), Gaps = 16/160 (10%)
Query: 307 NPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYY 366
N F+ A A+ SILLG +M+G + + L L +G ++G + L+GS
Sbjct: 33 NNYAFACAILASMTSILLGYDIGVMSGAMIYIKRDLKINDLQIGILAGSLNIYSLIGSCA 92
Query: 367 I-----FLDNAAVLAVVGLLLYVGCYQISFGP-MGWLMIAEIFPLRLRGKGLSIAVLVNF 420
++ + + G + + G + P +LM + G G+ A+++
Sbjct: 93 AGRTSDWIGRRYTIVLAGAIFFAGAILMGLSPNYAFLMFGRF----IAGIGVGYALMIAP 148
Query: 421 AANALVT------FAFSPLKDLLGAGILFYIFSAIAVASL 454
A V+ F S + + AGI+ S +A ++L
Sbjct: 149 VYTAEVSPASSRGFLNSFPEVFINAGIMLGYVSNLAFSNL 188
>At4g36670 sugar transporter like protein
Length = 493
Score = 229 bits (584), Expect = 3e-60
Identities = 146/462 (31%), Positives = 247/462 (52%), Gaps = 30/462 (6%)
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
++FGYD G S A + I+ + + V+I +LT AL+GS+LA +D +
Sbjct: 29 IIFGYDTGVMSGAMVFIEEDLKT-------NDVQIEVLTGILNLCALVGSLLAGRTSDII 81
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR +++A++++++G+++ + PN+P+L+ GR G+G+G A+ AP+Y AE A R
Sbjct: 82 GRRYTIVLASILFMLGSILMGWGPNYPVLLSGRCTAGLGVGFALMVAPVYSAEIATASHR 141
Query: 150 GQLVSLKEFFIVIGIVAGYGLGSLL--VDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
G L SL I IGI+ GY + + GWR M GI++ ++++ FG+ +P SPR
Sbjct: 142 GLLASLPHLCISIGILLGYIVNYFFSKLPMHIGWRLMLGIAAVPSLVLAFGILKMPESPR 201
Query: 208 WILLRAIQKKGD--LQTLKDTAIRSLCQLQG-RTFHDSAPQQVDEIMAEFSYLGEENDVT 264
W++++ K+G L+ + ++ + + Q + P+ VD+++ V
Sbjct: 202 WLIMQGRLKEGKEILELVSNSPEEAELRFQDIKAAAGIDPKCVDDVVKMEGKKTHGEGVW 261
Query: 265 LGEMFRGK--CRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADATR 319
+ R R+ L+ + G+ FQ I+ V +GP R G +
Sbjct: 262 KELILRPTPAVRRVLLTALGIHFFQHASGIEAVLLYGP------RIFKKAGITTKDKLFL 315
Query: 320 VSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAA------ 373
V+I +G+ K A +++D++GRR LLL V G+VI+L +LG NA
Sbjct: 316 VTIGVGIMKTTFIFTATLLLDKVGRRKLLLTSVGGMVIALTMLGFGLTMAQNAGGKLAWA 375
Query: 374 -VLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSP 432
VL++V +V + I GP+ W+ +E+FPL+LR +G S+ V VN NA V+ +F
Sbjct: 376 LVLSIVAAYSFVAFFSIGLGPITWVYSSEVFPLKLRAQGASLGVAVNRVMNATVSMSFLS 435
Query: 433 LKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
L + G F++F+ +A + F +F++PETKG +LEEIEA
Sbjct: 436 LTSAITTGGAFFMFAGVAAVAWNFFFFLLPETKGKSLEEIEA 477
>At2g16120 putative sugar transporter
Length = 511
Score = 215 bits (547), Expect = 5e-56
Identities = 147/464 (31%), Positives = 230/464 (48%), Gaps = 35/464 (7%)
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
++ GYDIG S A+I I+ L V++ +L +L+GS A +D+L
Sbjct: 38 IILGYDIGVMSGASIFIKDD-------LKLSDVQLEILMGILNIYSLVGSGAAGRTSDWL 90
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR +++A + GAL+ FA N+P +++GR V GIG+G AM AP+Y AE AP R
Sbjct: 91 GRRYTIVLAGAFFFCGALLMGFATNYPFIMVGRFVAGIGVGYAMMIAPVYTAEVAPASSR 150
Query: 150 GQLVSLKEFFIVIGIVAGYGLGSLL--VDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
G L S E FI IGI+ GY + GWR+M G+ + +V + G+ +P SPR
Sbjct: 151 GFLTSFPEIFINIGILLGYVSNYFFSKLPEHLGWRFMLGVGAVPSVFLAIGVLAMPESPR 210
Query: 208 WILLRAIQKKGD----LQTLKDTAIRSLCQLQGRTFHDSAPQQV-DEIMAEFSYLGEEND 262
W++L+ + GD L +T ++ +L P + D+++ +
Sbjct: 211 WLVLQG--RLGDAFKVLDKTSNTKEEAISRLDDIKRAVGIPDDMTDDVIVVPNKKSAGKG 268
Query: 263 VTLGEMFR--GKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADA 317
V + R R L+ G+ QQ I V + PT + G D
Sbjct: 269 VWKDLLVRPTPSVRHILIACLGIHFAQQASGIDAVVLYSPT------IFSKAGLKSKNDQ 322
Query: 318 TRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN------ 371
++ +GV K + V VVDR GRR LLL + G+ +SL LG+ ++
Sbjct: 323 LLATVAVGVVKTLFIVVGTCVVDRFGRRALLLTSMGGMFLSLTALGTSLTVINRNPGQTL 382
Query: 372 --AAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFA 429
A LAV ++ +V + I GP+ W+ +EIFP+RLR +G S+ V++N + ++
Sbjct: 383 KWAIGLAVTTVMTFVATFSIGAGPVTWVYCSEIFPVRLRAQGASLGVMLNRLMSGIIGMT 442
Query: 430 FSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
F L L G F +F+ +A A+ VF + +PET+G+ LEE+E
Sbjct: 443 FLSLSKGLTIGGAFLLFAGVAAAAWVFFFTFLPETRGIPLEEME 486
Score = 29.3 bits (64), Expect = 5.0
Identities = 41/183 (22%), Positives = 75/183 (40%), Gaps = 43/183 (23%)
Query: 311 FSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYI--- 367
F+ A A+ SI+LG +M+G ++ + D L + L + GI+ L+GS
Sbjct: 27 FACAILASMTSIILGYDIGVMSGASIFIKDDLKLSDVQLEILMGILNIYSLVGSGAAGRT 86
Query: 368 --FLDNAAVLAVVGLLLYVGCYQISFGP---------------MGWLMI------AEIFP 404
+L + + G + G + F +G+ M+ AE+ P
Sbjct: 87 SDWLGRRYTIVLAGAFFFCGALLMGFATNYPFIMVGRFVAGIGVGYAMMIAPVYTAEVAP 146
Query: 405 LRLRG-------KGLSIAVLVNFAANALVTFAFSPLKD------LLGAGILFYIFSAIAV 451
RG ++I +L+ + +N + FS L + +LG G + +F AI V
Sbjct: 147 ASSRGFLTSFPEIFINIGILLGYVSN----YFFSKLPEHLGWRFMLGVGAVPSVFLAIGV 202
Query: 452 ASL 454
++
Sbjct: 203 LAM 205
>At2g43330 membrane transporter like protein
Length = 509
Score = 214 bits (546), Expect = 6e-56
Identities = 147/462 (31%), Positives = 233/462 (49%), Gaps = 42/462 (9%)
Query: 28 GGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIG-----LLTSGSLYGALIGSVLA 82
GGLLFGYD G S A + I+ D + V+ + S +L GA+IG+
Sbjct: 41 GGLLFGYDTGVISGALLYIKD---------DFEVVKQSSFLQETIVSMALVGAMIGAAAG 91
Query: 83 FNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAE 142
I D+ GR++ L A +++ GA++ A AP+ +L+ GRL+ G+G+G+A AP+YIAE
Sbjct: 92 GWINDYYGRKKATLFADVVFAAGAIVMAAAPDPYVLISGRLLVGLGVGVASVTAPVYIAE 151
Query: 143 TAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWL 202
+P+ +RG LVS I G Y + S WR+M G+S AVI M ++
Sbjct: 152 ASPSEVRGGLVSTNVLMITGGQFLSYLVNSAFTQVPGTWRWMLGVSGVPAVIQFILMLFM 211
Query: 203 PASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEEND 262
P SPRW+ ++ +K +Q L T + +L+ H SA ++ ++ +
Sbjct: 212 PESPRWLFMKN-RKAEAIQVLART--YDISRLEDEIDHLSAAEEEEK--------QRKRT 260
Query: 263 VTLGEMFRGK-CRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADAT 318
V ++FR K R A + AGL FQQ I TV + PT + GF A
Sbjct: 261 VGYLDVFRSKELRLAFLAGAGLQAFQQFTGINTVMYYSPT------IVQMAGFHSNQLAL 314
Query: 319 RVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAV---- 374
+S+++ T V + +D GR+ L L + G++ISL +L + +
Sbjct: 315 FLSLIVAAMNAAGTVVGIYFIDHCGRKKLALSSLFGVIISLLILSVSFFKQSETSSDGGL 374
Query: 375 ---LAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFS 431
LAV+GL LY+ + GP+ W + +EI+P + RG ++ VN+ +N +V F
Sbjct: 375 YGWLAVLGLALYIVFFAPGMGPVPWTVNSEIYPQQYRGICGGMSATVNWISNLIVAQTFL 434
Query: 432 PLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
+ + G G+ F I + IAV +++F+ VPET+GLT E+E
Sbjct: 435 TIAEAAGTGMTFLILAGIAVLAVIFVIVFVPETQGLTFSEVE 476
>At2g16130 putative sugar transporter
Length = 511
Score = 213 bits (543), Expect = 1e-55
Identities = 147/465 (31%), Positives = 229/465 (48%), Gaps = 35/465 (7%)
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
++ GYDIG S A I I+ L V++ +L +LIGS A +D++
Sbjct: 38 IILGYDIGVMSGAAIFIKDD-------LKLSDVQLEILMGILNIYSLIGSGAAGRTSDWI 90
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR +++A + GAL+ FA N+P +++GR V GIG+G AM AP+Y E AP R
Sbjct: 91 GRRYTIVLAGFFFFCGALLMGFATNYPFIMVGRFVAGIGVGYAMMIAPVYTTEVAPASSR 150
Query: 150 GQLVSLKEFFIVIGIVAGYGLGSLL--VDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
G L S E FI IGI+ GY + GWR+M GI + +V + G+ +P SPR
Sbjct: 151 GFLSSFPEIFINIGILLGYVSNYFFAKLPEHIGWRFMLGIGAVPSVFLAIGVLAMPESPR 210
Query: 208 WILLRAIQKKGD----LQTLKDTAIRSLCQLQGRTFHDSAPQQV-DEIMAEFSYLGEEND 262
W++++ + GD L +T ++ +L P + D+++ +
Sbjct: 211 WLVMQG--RLGDAFKVLDKTSNTKEEAISRLNDIKRAVGIPDDMTDDVIVVPNKKSAGKG 268
Query: 263 VTLGEMFR--GKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADA 317
V + R R L+ G+ QQ I V + PT + G D
Sbjct: 269 VWKDLLVRPTPSVRHILIACLGIHFSQQASGIDAVVLYSPT------IFSRAGLKSKNDQ 322
Query: 318 TRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN------ 371
++ +GV K + V +VDR GRR LLL + G+ SL LG+ +D
Sbjct: 323 LLATVAVGVVKTLFIVVGTCLVDRFGRRALLLTSMGGMFFSLTALGTSLTVIDRNPGQTL 382
Query: 372 --AAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFA 429
A LAV ++ +V + + GP+ W+ +EIFP+RLR +G S+ V++N + ++
Sbjct: 383 KWAIGLAVTTVMTFVATFSLGAGPVTWVYASEIFPVRLRAQGASLGVMLNRLMSGIIGMT 442
Query: 430 FSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
F L L G F +F+ +AVA+ VF + +PET+G+ LEEIE+
Sbjct: 443 FLSLSKGLTIGGAFLLFAGVAVAAWVFFFTFLPETRGVPLEEIES 487
>At5g27360 sugar-porter family protein 2 (SFP2)
Length = 478
Score = 210 bits (534), Expect = 2e-54
Identities = 144/456 (31%), Positives = 237/456 (51%), Gaps = 37/456 (8%)
Query: 28 GGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIAD 87
G FG +G TS A I I DL + S S GA IG++ + +A
Sbjct: 45 GSFSFGVSLGYTSGAEIGIMKD-------LDLSIAQFSAFASLSTLGAAIGALFSGKMAI 97
Query: 88 FLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTP 147
LGRR+ + V+ L+ ++G AFA + L GR+ GIG+GL + P+YIAE +P
Sbjct: 98 ILGRRKTMWVSDLLCIIGWFSIAFAKDVMWLNFGRISSGIGLGLISYVVPVYIAEISPKH 157
Query: 148 IRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
+RG + G+ Y G+ L WR + + + I G++++P SPR
Sbjct: 158 VRGTFTFTNQLLQNSGLAMVYFSGNFL-----NWRILALLGALPCFIQVIGLFFVPESPR 212
Query: 208 WILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVTLGE 267
W + K G + L++ SL +L+G S E+M + + ++ + +
Sbjct: 213 W-----LAKVGSDKELEN----SLLRLRGGNADISREASDIEVMTKM--VENDSKSSFCD 261
Query: 268 MFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSILLGVF 327
+F+ K R LV+ GL+L QQ + ++LR GFS+ +T LLG+F
Sbjct: 262 LFQRKYRYTLVVGIGLMLIQQFSGSSAVLSYASTILR---KAGFSVTIGST----LLGLF 314
Query: 328 KLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYY------IFLDNAAVLAVVGLL 381
+ + V++VD+ GRRPLLL VSG+ I+ L+G + + + V + +
Sbjct: 315 MIPKAMIGVILVDKWGRRPLLLTSVSGMCITSMLIGVAFTLQKMQLLPELTPVFTFICVT 374
Query: 382 LYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGI 441
LY+G Y I G + W++++EIFP+ ++ SI LV+++++++VT+AF+ L + G
Sbjct: 375 LYIGTYAIGLGGLPWVIMSEIFPMNIKVTAGSIVTLVSWSSSSIVTYAFNFLLEWSTQG- 433
Query: 442 LFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
FY+F A+ +L+FI+ +VPETKGL+LEEI+A +
Sbjct: 434 TFYVFGAVGGLALLFIWLLVPETKGLSLEEIQASLI 469
>At2g20780 putative sugar transporter
Length = 547
Score = 203 bits (517), Expect = 1e-52
Identities = 138/447 (30%), Positives = 224/447 (49%), Gaps = 29/447 (6%)
Query: 35 DIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFLGRRRE 94
D+G S A + IQ IT + + IG L+ SL+G+L G +D +GR+
Sbjct: 93 DVGVMSGAVLFIQQDLK--ITEVQTEVL-IGSLSIISLFGSLAGG----RTSDSIGRKWT 145
Query: 95 LLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIRGQLVS 154
+ +AAL++ GA + A AP+F +L+IGR + GIGIGL + AP+YIAE +PT RG S
Sbjct: 146 MALAALVFQTGAAVMAVAPSFEVLMIGRTLAGIGIGLGVMIAPVYIAEISPTVARGFFTS 205
Query: 155 LKEFFIVIGIVAGYGLGSLL--VDTVAGWRYMFGISSPVAVIMGFGMWWLPASPRWILLR 212
E FI +GI+ GY + WR M + +V +GF + +P SPRW+++
Sbjct: 206 FPEIFINLGILLGYVSNYAFSGLSVHISWRIMLAVGILPSVFIGFALCVIPESPRWLVM- 264
Query: 213 AIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYL-GEENDVTLGEMFRG 271
KG + + ++ +++ D A +++ EI ++ G E+ E+
Sbjct: 265 ----KGRVDSAREVLMKT------NERDDEAEERLAEIQLAAAHTEGSEDRPVWRELLSP 314
Query: 272 K--CRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSILLGVFKL 329
RK L++ G+ FQQI T T + G ++ +GV K
Sbjct: 315 SPVVRKMLIVGFGIQCFQQI---TGIDATVYYSPEILKEAGIQDETKLLAATVAVGVTKT 371
Query: 330 IMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLAVVGLLLYVG---C 386
+ A ++D +GR+PLL G+ + LF L FL + + LL G
Sbjct: 372 VFILFATFLIDSVGRKPLLYVSTIGMTLCLFCLSFTLTFLGQGTLGITLALLFVCGNVAF 431
Query: 387 YQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYIF 446
+ I GP+ W++ +EIFPLRLR + ++ + N + LV +F + + G F++F
Sbjct: 432 FSIGMGPVCWVLTSEIFPLRLRAQASALGAVGNRVCSGLVAMSFLSVSRAITVGGTFFVF 491
Query: 447 SAIAVASLVFIYFIVPETKGLTLEEIE 473
S ++ S++F+Y +VPET G +LE+IE
Sbjct: 492 SLVSALSVIFVYVLVPETSGKSLEQIE 518
>At5g27350 sugar transporter like protein
Length = 474
Score = 201 bits (512), Expect = 6e-52
Identities = 137/459 (29%), Positives = 231/459 (49%), Gaps = 43/459 (9%)
Query: 28 GGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIAD 87
G FG G TS A + DL + S + GA IG++ N+A
Sbjct: 41 GSFSFGVATGYTSGAETGVMKD-------LDLSIAQFSAFGSFATLGAAIGALFCGNLAM 93
Query: 88 FLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTP 147
+GRR + V+ + + G L AFA LL GR++ GIG GL + P+YIAE P
Sbjct: 94 VIGRRGTMWVSDFLCITGWLSIAFAKEVVLLNFGRIISGIGFGLTSYVVPVYIAEITPKH 153
Query: 148 IRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
+RG + G+ Y G+ + WR + + + I G++++P SPR
Sbjct: 154 VRGTFTFSNQLLQNAGLAMIYFCGNFIT-----WRTLALLGALPCFIQVIGLFFVPESPR 208
Query: 208 WILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVTLGE 267
W + K G + L++ SL +L+GR S ++ EI + ++ + +
Sbjct: 209 W-----LAKVGSDKELEN----SLFRLRGRDADIS--REASEIQVMTKMVENDSKSSFSD 257
Query: 268 MFRGKCRKALVISAGLVLFQQIQ---TVTDHGPTKCSLLRCINPTGFSLAADATRVSILL 324
+F+ K R LV+ GL+L QQ V + T GFS+A T +L
Sbjct: 258 LFQRKYRYTLVVGIGLMLIQQFSGSAAVISYAST------IFRKAGFSVAIGTT----ML 307
Query: 325 GVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYY------IFLDNAAVLAVV 378
G+F + + +++VD+ GRRPLL+ G+ ++ LLG + + + +L+ +
Sbjct: 308 GIFVIPKAMIGLILVDKWGRRPLLMTSAFGMSMTCMLLGVAFTLQKMQLLSELTPILSFI 367
Query: 379 GLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLG 438
+++Y+ Y I G + W++++EIFP+ ++ SI LV+F+++++VT+AF+ L +
Sbjct: 368 CVMMYIATYAIGLGGLPWVIMSEIFPINIKVTAGSIVTLVSFSSSSIVTYAFNFLFEWST 427
Query: 439 AGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
G F+IF+ I A+L+FI+ +VPETKGL+LEEI+ +
Sbjct: 428 QG-TFFIFAGIGGAALLFIWLLVPETKGLSLEEIQVSLI 465
>At3g05150 putative sugar transporter
Length = 463
Score = 201 bits (512), Expect = 6e-52
Identities = 125/408 (30%), Positives = 221/408 (53%), Gaps = 31/408 (7%)
Query: 74 GALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAM 133
GA++G++ + I+DF+GR+ + +++++ +G LI A L GR + G G G
Sbjct: 77 GAVLGAITSGKISDFIGRKGAMRLSSVISAIGWLIIYLAKGDVPLDFGRFLTGYGCGTLS 136
Query: 134 HAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAV 193
P++IAE +P +RG L +L + FIVIG+ + + L+ V WR + V
Sbjct: 137 FVVPVFIAEISPRKLRGALATLNQLFIVIGLASMF-----LIGAVVNWRTLALTGVAPCV 191
Query: 194 IMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAE 253
++ FG W++P SPRW L + + D + A++ L Q ++ ++ E +A
Sbjct: 192 VLFFGTWFIPESPRW--LEMVGRHSDF----EIALQKLRGPQANITREAG--EIQEYLAS 243
Query: 254 FSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSL 313
++L + TL ++ K + +++ GL+ FQQ + ++ S
Sbjct: 244 LAHLPK---ATLMDLIDKKNIRFVIVGVGLMFFQQFVGIN-------GVIFYAQQIFVSA 293
Query: 314 AADATRVSILLGVFKLIMTGV-AVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIF---- 368
A T SIL + ++++T + A +++DRLGRRPLL+ G++I L+G+ ++
Sbjct: 294 GASPTLGSILYSIEQVVLTALGATLLIDRLGRRPLLMASAVGMLIGCLLIGNSFLLKAHG 353
Query: 369 --LDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALV 426
LD LAV G+L+Y+G + I G + W++++EIFP+ L+G + +VN+ ++ LV
Sbjct: 354 LALDIIPALAVSGVLVYIGSFSIGMGAIPWVIMSEIFPINLKGTAGGLVTVVNWLSSWLV 413
Query: 427 TFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
+F F+ L G FY++ + V +++FI +VPETKG TLEEI+A
Sbjct: 414 SFTFNFLMIWSPHG-TFYVYGGVCVLAIIFIAKLVPETKGRTLEEIQA 460
>At3g05165 sugar transporter like protein
Length = 467
Score = 195 bits (495), Expect = 5e-50
Identities = 134/449 (29%), Positives = 219/449 (47%), Gaps = 37/449 (8%)
Query: 32 FGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFLGR 91
+G G TS A +I DL + S G +G++ + +A LGR
Sbjct: 44 YGCAAGYTSGAETAIMKE-------LDLSMAQFSAFGSFLNVGGAVGALFSGQLAVILGR 96
Query: 92 RRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIRGQ 151
RR L + G L AFA N L +GR+ GIG+GL + P+YIAE P +RG
Sbjct: 97 RRTLWACDFFCVFGWLSIAFAKNVFWLDLGRISLGIGVGLISYVVPVYIAEITPKHVRGA 156
Query: 152 LVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPRWILL 211
+ + G+ Y G TV WR M I + ++ G++++P SPRW+
Sbjct: 157 FTASNQLLQNSGVSLIYFFG-----TVINWRVMAVIGAIPCILQTIGIFFIPESPRWLAK 211
Query: 212 RAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVTLGEMFRG 271
+ K+ + SL +L+G+ S + EI L E++ + +MF+
Sbjct: 212 IRLSKEVE---------SSLHRLRGKDTDVSG--EAAEIQVMTKMLEEDSKSSFSDMFQK 260
Query: 272 KCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSILLGVFKLIM 331
K R+ LV+ GL+L QQ+ + ++ R GFS S++ GVF +
Sbjct: 261 KYRRTLVVGIGLMLIQQLSGASGITYYSNAIFR---KAGFSERLG----SMIFGVFVIPK 313
Query: 332 TGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYY------IFLDNAAVLAVVGLLLYVG 385
V +++VDR GRRPLLL G+ I L+G + + + + V +L+Y G
Sbjct: 314 ALVGLILVDRWGRRPLLLASAVGMSIGSLLIGVSFTLQQMNVLPELIPIFVFVNILVYFG 373
Query: 386 CYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYI 445
C+ G + W++++EIFP+ ++ +I L ++ + V++AF+ + + G FYI
Sbjct: 374 CFAFGIGGLPWVIMSEIFPINIKVSAGTIVALTSWTSGWFVSYAFNFMFEWSAQG-TFYI 432
Query: 446 FSAIAVASLVFIYFIVPETKGLTLEEIEA 474
F+A+ S +FI+ +VPETKG +LEE++A
Sbjct: 433 FAAVGGMSFIFIWMLVPETKGQSLEELQA 461
>At5g16150 sugar transporter like protein
Length = 546
Score = 192 bits (488), Expect = 3e-49
Identities = 138/452 (30%), Positives = 232/452 (50%), Gaps = 33/452 (7%)
Query: 28 GGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIAD 87
G +LFGY +G + A + GI + V G + S L GA +GS +AD
Sbjct: 115 GAILFGYHLGVVNGALEYLAKDL--GIAE---NTVLQGWIVSSLLAGATVGSFTGGALAD 169
Query: 88 FLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTP 147
GR R + A+ +GA + A A + +++GRL+ GIGIG++ P+YI+E +PT
Sbjct: 170 KFGRTRTFQLDAIPLAIGAFLCATAQSVQTMIVGRLLAGIGIGISSAIVPLYISEISPTE 229
Query: 148 IRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
IRG L S+ + FI IGI+A G L WR MFG++ +V++ GM + P SPR
Sbjct: 230 IRGALGSVNQLFICIGILAALIAGLPLAANPLWWRTMFGVAVIPSVLLAIGMAFSPESPR 289
Query: 208 WILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGE---ENDVT 264
W++ Q A +++ L G+ ++V E++ + S G+ E +
Sbjct: 290 WLV---------QQGKVSEAEKAIKTLYGK-------ERVVELVRDLSASGQGSSEPEAG 333
Query: 265 LGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSILL 324
++F + K + + A L LFQQ+ + S+ R + +D S L+
Sbjct: 334 WFDLFSSRYWKVVSVGAALFLFQQLAGINAVVYYSTSVFR-----SAGIQSDVA-ASALV 387
Query: 325 GVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAA---VLAVVGLL 381
G + T VA ++D++GR+ LLL G+ +S+ LL + + AA LAVVG +
Sbjct: 388 GASNVFGTAVASSLMDKMGRKSLLLTSFGGMALSMLLLSLSFTWKALAAYSGTLAVVGTV 447
Query: 382 LYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGI 441
LYV + + GP+ L++ EIF R+R K +++++ +++ +N ++ F + G
Sbjct: 448 LYVLSFSLGAGPVPALLLPEIFASRIRAKAVALSLGMHWISNFVIGLYFLSVVTKFGISS 507
Query: 442 LFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
++ F+ + V ++++I V ETKG +LEEIE
Sbjct: 508 VYLGFAGVCVLAVLYIAGNVVETKGRSLEEIE 539
>At5g18840 sugar transporter - like protein
Length = 482
Score = 192 bits (487), Expect = 4e-49
Identities = 131/461 (28%), Positives = 229/461 (49%), Gaps = 32/461 (6%)
Query: 20 LRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGS 79
L LF F + ++ G+ + QSS + +L E + S GA++G+
Sbjct: 42 LMVLFSTFVAVCGSFEFGSCVGYSAPTQSSIRQDL---NLSLAEFSMFGSILTIGAMLGA 98
Query: 80 VLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMY 139
V++ I+DF GR+ + +A + G L F LL +GR G GIG+ + P+Y
Sbjct: 99 VMSGKISDFSGRKGAMRTSACFCITGWLAVFFTKGALLLDVGRFFTGYGIGVFSYVVPVY 158
Query: 140 IAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGM 199
IAE +P +RG L +L + IVIG + +GSL+ W+ + +++ FG+
Sbjct: 159 IAEISPKNLRGGLTTLNQLMIVIGSSVSFLIGSLI-----SWKTLALTGLAPCIVLLFGL 213
Query: 200 WWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGE 259
++P SPRW+ +K+ + +L +L+G+ + D I L
Sbjct: 214 CFIPESPRWLAKAGHEKEFRV---------ALQKLRGK--DADITNEADGIQVSIQALEI 262
Query: 260 ENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATR 319
+ ++ K ++++I L++FQQ + G GF+ T
Sbjct: 263 LPKARIQDLVSKKYGRSVIIGVSLMVFQQFVGINGIG---FYASETFVKAGFTSGKLGT- 318
Query: 320 VSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYY------IFLDNAA 373
I + ++ +T + +++D+ GRRPL++ GI + L G+ + + L+
Sbjct: 319 --IAIACVQVPITVLGTILIDKSGRRPLIMISAGGIFLGCILTGTSFLLKGQSLLLEWVP 376
Query: 374 VLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPL 433
LAV G+L+YV + I GP+ W++++EIFP+ ++G S+ VLVN++ V++ F+ L
Sbjct: 377 SLAVGGVLIYVAAFSIGMGPVPWVIMSEIFPINVKGIAGSLVVLVNWSGAWAVSYTFNFL 436
Query: 434 KDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
G FY++SA A A+++F+ +VPETKG TLEEI+A
Sbjct: 437 MSWSSPG-TFYLYSAFAAATIIFVAKMVPETKGKTLEEIQA 476
>At2g48020 sugar transporter like protein
Length = 463
Score = 191 bits (486), Expect = 6e-49
Identities = 139/454 (30%), Positives = 232/454 (50%), Gaps = 39/454 (8%)
Query: 28 GGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIAD 87
G FG G +S A +I++ L E L S +GA+IG++ + IAD
Sbjct: 38 GSFAFGSCAGYSSPAQAAIRND-------LSLTIAEFSLFGSLLTFGAMIGAITSGPIAD 90
Query: 88 FLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTP 147
+GR+ + V++ +VG L FA L +GRL G G+G + P++IAE AP
Sbjct: 91 LVGRKGAMRVSSAFCVVGWLAIIFAKGVVALDLGRLATGYGMGAFSYVVPIFIAEIAPKT 150
Query: 148 IRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
RG L +L + I G+ + +G+L+ WR + I G++++P SPR
Sbjct: 151 FRGALTTLNQILICTGVSVSFIIGTLVT-----WRVLALIGIIPCAASFLGLFFIPESPR 205
Query: 208 WILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVTLGE 267
W+ K T + A+R +L+G+ S ++ EI L + +
Sbjct: 206 WL------AKVGRDTEFEAALR---KLRGKKADIS--EEAAEIQDYIETLERLPKAKMLD 254
Query: 268 MFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVS-ILLGV 326
+F+ + ++++I+ GL++FQQ + C I F A TR+ I+ V
Sbjct: 255 LFQRRYIRSVLIAFGLMVFQQFGGING----ICFYTSSI----FEQAGFPTRLGMIIYAV 306
Query: 327 FKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLG-SYYIFLDNAA-----VLAVVGL 380
++++T + +VDR GR+PLLL +G+VI + S+Y+ + + A VLAVVG+
Sbjct: 307 LQVVITALNAPIVDRAGRKPLLLVSATGLVIGCLIAAVSFYLKVHDMAHEAVPVLAVVGI 366
Query: 381 LLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAG 440
++Y+G + G M W++++EIFP+ ++G +A LVN+ V++ F+ L G
Sbjct: 367 MVYIGSFSAGMGAMPWVVMSEIFPINIKGVAGGMATLVNWFGAWAVSYTFNFLMSWSSYG 426
Query: 441 ILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
F I++AI ++VF+ IVPETKG TLE+I+A
Sbjct: 427 -TFLIYAAINALAIVFVIAIVPETKGKTLEQIQA 459
>At3g05157 sugar transporter like protein
Length = 458
Score = 188 bits (478), Expect = 5e-48
Identities = 138/449 (30%), Positives = 220/449 (48%), Gaps = 37/449 (8%)
Query: 32 FGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFLGR 91
+G G TS A +I DL + S G +G++ + +A LGR
Sbjct: 35 YGCANGYTSGAETAIMKE-------LDLSMAQFSAFGSFLNLGGAVGALFSGQLAVILGR 87
Query: 92 RRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIRGQ 151
RR L L + G L AFA N L +GR+ GIG+GL + P+YIAE P +RG
Sbjct: 88 RRTLWACDLFCIFGWLSIAFAKNVLWLDLGRISLGIGVGLTSYVVPVYIAEITPKHVRGA 147
Query: 152 LVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPRWILL 211
+ GI Y G TV WR + I + I G++++P SPRW
Sbjct: 148 FSASTLLLQNSGISLIYFFG-----TVINWRVLAVIGALPCFIPVIGIYFIPESPRW--- 199
Query: 212 RAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVTLGEMFRG 271
+ K G ++ +++ SL +L+G+ S + EI L E++ + +MF+
Sbjct: 200 --LAKIGSVKEVEN----SLHRLRGKDADVS--DEAAEIQVMTKMLEEDSKSSFCDMFQK 251
Query: 272 KCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSILLGVFKLIM 331
K R+ LV+ GL+L QQ+ + ++ R GFS S++ GVF +
Sbjct: 252 KYRRTLVVGIGLMLIQQLSGASGITYYSNAIFR---KAGFSERLG----SMIFGVFVIPK 304
Query: 332 TGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYY------IFLDNAAVLAVVGLLLYVG 385
V +++VDR GRRPLLL G+ I L+G + +F + V + +L+Y G
Sbjct: 305 ALVGLILVDRWGRRPLLLASAVGMSIGSLLIGVSFTLQEMNLFPEFIPVFVFINILVYFG 364
Query: 386 CYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYI 445
+ I G + W++++EIFP+ ++ SI L ++ V++ F+ + + G FYI
Sbjct: 365 FFAIGIGGLPWIIMSEIFPINIKVSAGSIVALTSWTTGWFVSYGFNFMFEWSAQG-TFYI 423
Query: 446 FSAIAVASLVFIYFIVPETKGLTLEEIEA 474
F+ + SL+FI+ +VPETKG +LEE++A
Sbjct: 424 FAMVGGLSLLFIWMLVPETKGQSLEELQA 452
>At3g19940 putative monosaccharide transport protein, STP4
Length = 514
Score = 182 bits (461), Expect = 5e-46
Identities = 147/478 (30%), Positives = 224/478 (46%), Gaps = 56/478 (11%)
Query: 26 AFGGLLFGYDIGATSSAT------------ISIQSSSLSGITWY-DLDAVEIGLLTSGSL 72
A GGLLFGYD+G + T + Q T Y D + L TS
Sbjct: 33 AMGGLLFGYDLGISGGVTSMEEFLTKFFPQVESQMKKAKHDTAYCKFDNQMLQLFTSSLY 92
Query: 73 YGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLA 132
AL+ S +A I GR+ + + L +L+GAL AFA N +L+IGRL+ G+G+G A
Sbjct: 93 LAALVASFMASVITRKHGRKVSMFIGGLAFLIGALFNAFAVNVSMLIIGRLLLGVGVGFA 152
Query: 133 MHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVA----GYGLGSLLVDTVAGWRYMFGIS 188
+ P+Y++E AP IRG L + I IGI+ YG + GWR G++
Sbjct: 153 NQSTPVYLSEMAPAKIRGALNIGFQMAITIGILVANLINYGTSKM---AQHGWRVSLGLA 209
Query: 189 SPVAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQ-V 247
+ AV+M G + LP +P +L R + A + L +++G D Q +
Sbjct: 210 AVPAVVMVIGSFILPDTPNSMLERG---------KNEEAKQMLKKIRGADNVDHEFQDLI 260
Query: 248 DEIMAEFSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLR 304
D + A ++ + + K R AL+ + + FQQ I + + P L
Sbjct: 261 DAVEA-----AKKVENPWKNIMESKYRPALIFCSAIPFFQQITGINVIMFYAPVLFKTL- 314
Query: 305 CINPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGS 364
GF A A +++ GV ++ T V++ VDR GRR L L G + I L+GS
Sbjct: 315 -----GFGDDA-ALMSAVITGVVNMLSTFVSIYAVDRYGRRLLFLEGGIQMFICQLLVGS 368
Query: 365 YY----------IFLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSI 414
+ A + + +YV + S+GP+GWL+ +EI PL +R G +I
Sbjct: 369 FIGARFGTSGTGTLTPATADWILAFICVYVAGFAWSWGPLGWLVPSEICPLEIRPAGQAI 428
Query: 415 AVLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEI 472
V VN L+ F + + G LFY F+++ VFIYF++PETKG+ +EE+
Sbjct: 429 NVSVNMFFTFLIGQFFLTMLCHMKFG-LFYFFASMVAIMTVFIYFLLPETKGVPIEEM 485
>At1g19450 integral membrane protein, putative
Length = 488
Score = 181 bits (458), Expect = 1e-45
Identities = 133/462 (28%), Positives = 236/462 (50%), Gaps = 40/462 (8%)
Query: 23 LFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLA 82
L A G + FG+ G +S +I L E + S S GA++G++ +
Sbjct: 54 LIVALGPIQFGFTCGYSSPTQAAITKD-------LGLTVSEYSVFGSLSNVGAMVGAIAS 106
Query: 83 FNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAE 142
IA+++GR+ L++AA+ ++G L +FA + L +GRL+ G G+G+ + P+YIAE
Sbjct: 107 GQIAEYVGRKGSLMIAAIPNIIGWLSISFAKDTSFLYMGRLLEGFGVGIISYTVPVYIAE 166
Query: 143 TAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWL 202
AP +RG L S+ + + IGI+ Y LG + WR + + ++ G++++
Sbjct: 167 IAPQTMRGALGSVNQLSVTIGIMLAYLLGLFV-----PWRILAVLGVLPCTLLIPGLFFI 221
Query: 203 PASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEEND 262
P SPRW + K G L D SL L+G F +V+EI + + +
Sbjct: 222 PESPRW-----LAKMG----LTDDFETSLQVLRG--FETDITVEVNEIKRSVASSSKRSA 270
Query: 263 VTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSI 322
V ++ R + L++ GL+ QQ+ + ++ T ++A +
Sbjct: 271 VRFVDLKRRRYYFPLMVGIGLLALQQLGGINGVLFYSSTIFESAGVTSSNVA------TF 324
Query: 323 LLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLG-SYYI---------FLDNA 372
+GV +++ TG+A +VD+ GRR LL+ G+ ISL ++ ++Y+ +
Sbjct: 325 GVGVVQVVATGIATWLVDKAGRRLLLMISSIGMTISLVIVAVAFYLKEFVSPDSNMYNIL 384
Query: 373 AVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSP 432
++++VVG++ V + GP+ WL+++EI P+ ++G SIA L+N+ + LVT +
Sbjct: 385 SMVSVVGVVAMVISCSLGMGPIPWLIMSEILPVNIKGLAGSIATLLNWFVSWLVTMTANM 444
Query: 433 LKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
L +G F +++ + ++VF+ VPETKG TLEEI+A
Sbjct: 445 LL-AWSSGGTFTLYALVCGFTVVFVSLWVPETKGKTLEEIQA 485
>At1g50310 hexose transporter, putative
Length = 517
Score = 179 bits (454), Expect = 3e-45
Identities = 146/498 (29%), Positives = 232/498 (46%), Gaps = 61/498 (12%)
Query: 6 GALLVDLPKTCLKNLRFLFPAFGGLLFGYDIGATSSAT------------ISIQSSSLSG 53
G + V + TC+ A GGLLFGYD+G + T + Q
Sbjct: 19 GGVTVFVIMTCI------VAAMGGLLFGYDLGISGGVTSMEEFLSKFFPEVDKQMHEARR 72
Query: 54 ITWY-DLDAVEIGLLTSGSLYGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFA 112
T Y D + L TS AL S +A + GR+ + V + +L+G+L AFA
Sbjct: 73 ETAYCKFDNQLLQLFTSSLYLAALASSFVASAVTRKYGRKISMFVGGVAFLIGSLFNAFA 132
Query: 113 PNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGI----VAGY 168
N +L++GRL+ G+G+G A + P+Y++E AP IRG L + I IGI + Y
Sbjct: 133 TNVAMLIVGRLLLGVGVGFANQSTPVYLSEMAPAKIRGALNIGFQMAITIGILIANLINY 192
Query: 169 GLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAI 228
G + + GWR G+++ AVIM G + LP +P +L R ++ A
Sbjct: 193 GTSQMAKN---GWRVSLGLAAVPAVIMVIGSFVLPDTPNSMLERGKYEQ---------AR 240
Query: 229 RSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQ 288
L +++G D+ ++ ++ + ++ + K R ALV + + FQQ
Sbjct: 241 EMLQKIRGA---DNVDEEFQDLCDACEAAKKVDNPWKNIFQQAKYRPALVFCSAIPFFQQ 297
Query: 289 IQTVTDHGPTKCSLLRCINPTGF---SLAADATRVS-ILLGVFKLIMTGVAVVVVDRLGR 344
I T +++ P F A DA+ +S ++ G ++ T V++ VDR GR
Sbjct: 298 I--------TGINVIMFYAPVLFKTLGFADDASLISAVITGAVNVVSTLVSIYAVDRYGR 349
Query: 345 RPLLLGGVSGIVISLFLLGSYY----------IFLDNAAVLAVVGLLLYVGCYQISFGPM 394
R L L G +++S ++G+ A + + LYV + S+GP+
Sbjct: 350 RILFLEGGIQMIVSQIVVGTLIGMKFGTTGSGTLTPATADWILAFICLYVAGFAWSWGPL 409
Query: 395 GWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASL 454
GWL+ +EI PL +R G +I V VN L+ F + + G LFY F +
Sbjct: 410 GWLVPSEICPLEIRPAGQAINVSVNMFFTFLIGQFFLTMLCHMKFG-LFYFFGGMVAVMT 468
Query: 455 VFIYFIVPETKGLTLEEI 472
VFIYF++PETKG+ +EE+
Sbjct: 469 VFIYFLLPETKGVPIEEM 486
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.145 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,675,963
Number of Sequences: 26719
Number of extensions: 394524
Number of successful extensions: 1549
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 68
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 1221
Number of HSP's gapped (non-prelim): 134
length of query: 477
length of database: 11,318,596
effective HSP length: 103
effective length of query: 374
effective length of database: 8,566,539
effective search space: 3203885586
effective search space used: 3203885586
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC142222.8


