

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.7 + phase: 0
(555 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
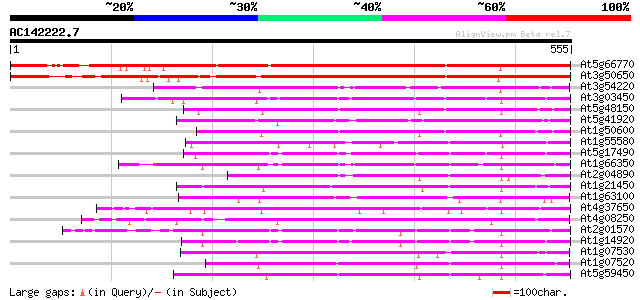
Score E
Sequences producing significant alignments: (bits) Value
At5g66770 SCARECROW gene regulator 594 e-170
At3g50650 scarecrow-like 7 (SCL7) 510 e-145
At3g54220 SCARECROW1 192 5e-49
At3g03450 RGA1-like protein 191 7e-49
At5g48150 SCARECROW gene regulator-like 187 1e-47
At5g41920 SCARECROW gene regulator-like protein 187 1e-47
At1g50600 scarecrow-like 5 (SCL5) 186 4e-47
At1g55580 unknown protein 173 2e-43
At5g17490 RGA-like protein 173 3e-43
At1g66350 gibberellin regulatory protein like 169 5e-42
At2g04890 putative SCARECROW gene regulator 168 6e-42
At1g21450 scarecrow-like 1 (SCL1) 165 5e-41
At1g63100 transcription factor SCARECROW, putative 157 1e-38
At4g37650 SHORT-ROOT (SHR) 156 2e-38
At4g08250 putative protein 156 2e-38
At2g01570 putative RGA1, giberellin repsonse modulation protein 154 1e-37
At1g14920 signal response protein (GAI) 150 1e-36
At1g07530 transcription factor scarecrow-like 14, putative 147 2e-35
At1g07520 transcription factor scarecrow-like 14, putative 145 6e-35
At5g59450 putative SCARECROW transcriptional regulatory protein ... 145 7e-35
>At5g66770 SCARECROW gene regulator
Length = 584
Score = 594 bits (1532), Expect = e-170
Identities = 336/604 (55%), Positives = 409/604 (67%), Gaps = 69/604 (11%)
Query: 1 MAYMCADSGNLMAIAQQVINQKQQQEQHEQQQHQHHHHHQNILGITNPLSVSLHPWHNNN 60
MAYMC DSGNLMAIAQQVI QKQQQEQ +QQ HQ H I GI NPLS L+PW N +
Sbjct: 1 MAYMCTDSGNLMAIAQQVIKQKQQQEQQQQQHHQDHQ----IFGI-NPLS--LNPWPNTS 53
Query: 61 IPVSSSTSLPPLSFPPTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTT----FRFSDF-- 114
+ S S F DPFQV G ++ DP F FP+ H+TT FR SDF
Sbjct: 54 LGFGLSGSA---------FPDPFQVTGGGDSNDPGFPFPNLDHHHATTTGGGFRLSDFGG 104
Query: 115 -------DSDDWMDTLMSAADSHSD------FHLNP---------FSSCPTRLSPQPD-- 150
+SD+WM+TL+S DS +D +H NP F + P+RLS QP
Sbjct: 105 GTGGGEFESDEWMETLISGGDSVADGPDCDTWHDNPDYVIYGPDPFDTYPSRLSVQPSDL 164
Query: 151 -------------LLLPATAPLQIPTQPPTPQRKNDNTTTETTSSFSSSTNPLLKTLTEI 197
L P ++PL IP K D T ++ PLLK + +
Sbjct: 165 NRVIDTSSPLPPPTLWPPSSPLSIPPLTHESPTKEDPETNDSEDDDFDLEPPLLKAIYDC 224
Query: 198 ASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQSSIASSNSSSTT 257
A + ++ PN+A +TL + +S+S+ G+P +RV+FYF++AL+N+++ S SS+SSST
Sbjct: 225 ARISDSD-PNEASKTLLQIRESVSELGDPTERVAFYFTEALSNRLSPNSPATSSSSSST- 282
Query: 258 WEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFA 317
E+L LSYK LNDACPYSKFAHLTANQAILEATE SN IHIVDFGIVQGIQW ALLQA A
Sbjct: 283 -EDLILSYKTLNDACPYSKFAHLTANQAILEATEKSNKIHIVDFGIVQGIQWPALLQALA 341
Query: 318 TRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDE 377
TR+SGKP +R+SGIPA +LG SP S+ ATGNRL +FAK+L LNF+F PILTPI LL+
Sbjct: 342 TRTSGKPTQIRVSGIPAPSLGESPEPSLIATGNRLRDFAKVLDLNFDFIPILTPIHLLNG 401
Query: 378 SSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGF 437
SSF + PDE LAVNFMLQLY LLDE V+ ALRLAKSLNP++VTLGEYE SL RVGF
Sbjct: 402 SSFRVDPDEVLAVNFMLQLYKLLDETPTIVDTALRLAKSLNPRVVTLGEYEVSL-NRVGF 460
Query: 438 VERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV------RERMEDK 491
R + A +++A FESLEPN+ DS ER +VE L GRRI G+IG RERME+K
Sbjct: 461 ANRVKNALQFYSAVFESLEPNLGRDSEERVRVERELFGRRISGLIGPEKTGIHRERMEEK 520
Query: 492 EQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTV 551
EQW+VLMEN GFESV LS+YA+SQAKILLWNY+YS+LYS+VES+P F+SLAW D+PLLT+
Sbjct: 521 EQWRVLMENAGFESVKLSNYAVSQAKILLWNYNYSNLYSIVESKPGFISLAWNDLPLLTL 580
Query: 552 SSWR 555
SSWR
Sbjct: 581 SSWR 584
>At3g50650 scarecrow-like 7 (SCL7)
Length = 542
Score = 510 bits (1313), Expect = e-145
Identities = 296/583 (50%), Positives = 377/583 (63%), Gaps = 69/583 (11%)
Query: 1 MAYMCADSGNLMAIAQQVINQKQQQE-QHEQQQHQHHHHHQNILGITNPLSVSLHPWHNN 59
MAYMC DSGNLMAIAQQ+I QKQQQ+ QH+QQ+ Q + PW N
Sbjct: 1 MAYMCTDSGNLMAIAQQLIKQKQQQQSQHQQQEEQEQEPN---------------PWPNP 45
Query: 60 NIPVSSSTSLPPLSFPPTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTTFRFSDFDSDDW 119
+ + P + F+DPFQV DP F+FP + +FDSD+W
Sbjct: 46 SFG---------FTLPGSGFSDPFQV-----TNDPGFHFPHLEHHQNAAVASEEFDSDEW 91
Query: 120 MDTLMSAADS---HSDFHL---NPFSSCPTRLSPQPDLLLPA----TAPLQIPTQP---- 165
M++L++ D+ + DF + +PF S P+RLS P L +A Q+P P
Sbjct: 92 MESLINGGDASQTNPDFPIYGHDPFVSFPSRLSA-PSYLNRVNKDDSASQQLPPPPASTA 150
Query: 166 ---PTPQRKNDNTTTETTSSFSSSTNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQ 222
P+P F + P+ K + + A +KP +TL + +S+S+
Sbjct: 151 IWSPSPPSPQHPPPPPPQPDFDLN-QPIFKAIHDYA-----RKPETKPDTLIRIKESVSE 204
Query: 223 NGNPNQRVSFYFSQALTNKITAQSSIASSNSSSTTWEELTLSYKALNDACPYSKFAHLTA 282
+G+P QRV +YF++AL++K T + S+SSS++ E+ LSYK LNDACPYSKFAHLTA
Sbjct: 205 SGDPIQRVGYYFAEALSHKETE----SPSSSSSSSLEDFILSYKTLNDACPYSKFAHLTA 260
Query: 283 NQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPV 342
NQAILEAT SNNIHIVDFGI QGIQW+ALLQA ATRSSGKP +RISGIPA +LG SP
Sbjct: 261 NQAILEATNQSNNIHIVDFGIFQGIQWSALLQALATRSSGKPTRIRISGIPAPSLGDSPG 320
Query: 343 SSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDE 402
S+ ATGNRL +FA +L LNFEF P+LTPI+LL+ SSF + PDE L VNFML+LY LLDE
Sbjct: 321 PSLIATGNRLRDFAAILDLNFEFYPVLTPIQLLNGSSFRVDPDEVLVVNFMLELYKLLDE 380
Query: 403 NTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALD 462
+V ALRLA+SLNP+IVTLGEYE SL RV F R + + +++A FESLEPN+ D
Sbjct: 381 TATTVGTALRLARSLNPRIVTLGEYEVSL-NRVEFANRVKNSLRFYSAVFESLEPNLDRD 439
Query: 463 SPERFQVESLLLGRRIDGVI---------GVR-ERMEDKEQWKVLMENCGFESVGLSHYA 512
S ER +VE +L GRRI ++ G R ME+KEQW+VLME GFE V S+YA
Sbjct: 440 SKERLRVERVLFGRRIMDLVRSDDDNNKPGTRFGLMEEKEQWRVLMEKAGFEPVKPSNYA 499
Query: 513 ISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+SQAK+LLWNY+YS+LYSLVES+P F+SLAW +VPLLTVSSWR
Sbjct: 500 VSQAKLLLWNYNYSTLYSLVESEPGFISLAWNNVPLLTVSSWR 542
>At3g54220 SCARECROW1
Length = 653
Score = 192 bits (487), Expect = 5e-49
Identities = 138/421 (32%), Positives = 215/421 (50%), Gaps = 27/421 (6%)
Query: 143 TRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTTTETTSSFSSSTNPLLKTLTEIASLIE 202
T PQP+ + +Q T +RK + + LL L + A +
Sbjct: 247 TDAPPQPETVTATVPAVQTNTAEALRERKEEIKRQKQDEEGLH----LLTLLLQCAEAVS 302
Query: 203 TQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQ-----SSIASSNSSSTT 257
+A + L +++ + G QRV+ YFS+A++ ++ +++ S T
Sbjct: 303 ADNLEEANKLLLEISQLSTPYGTSAQRVAAYFSEAMSARLLNSCLGIYAALPSRWMPQTH 362
Query: 258 WEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFA 317
++ +++ N P KF+H TANQAI EA E +++HI+D I+QG+QW L A
Sbjct: 363 SLKMVSAFQVFNGISPLVKFSHFTANQAIQEAFEKEDSVHIIDLDIMQGLQWPGLFHILA 422
Query: 318 TRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDE 377
+R G P+ VR++G LGTS + ++ ATG RLS+FA LGL FEF P+ + LD
Sbjct: 423 SRPGGPPH-VRLTG-----LGTS-MEALQATGKRLSDFADKLGLPFEFCPLAEKVGNLDT 475
Query: 378 SSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGF 437
++ EA+AV++ L + L + T S L L + L PK+VT+ E L+ F
Sbjct: 476 ERLNVRKREAVAVHW---LQHSLYDVTGSDAHTLWLLQRLAPKVVTV--VEQDLSHAGSF 530
Query: 438 VERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV----RERMEDKEQ 493
+ RF A +Y++A F+SL + +S ER VE LL + I V+ V R E
Sbjct: 531 LGRFVEAIHYYSALFDSLGASYGEESEERHVVEQQLLSKEIRNVLAVGGPSRSGEVKFES 590
Query: 494 WKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSS 553
W+ M+ CGF+ + L+ A +QA +LL + S Y+LV+ L L WKD+ LLT S+
Sbjct: 591 WREKMQQCGFKGISLAGNAATQATLLLGMFP-SDGYTLVDDN-GTLKLGWKDLSLLTASA 648
Query: 554 W 554
W
Sbjct: 649 W 649
>At3g03450 RGA1-like protein
Length = 547
Score = 191 bits (486), Expect = 7e-49
Identities = 143/470 (30%), Positives = 241/470 (50%), Gaps = 38/470 (8%)
Query: 111 FSDFDSDDWMDTLMSAADSHSDFHLNPFSSCPTRLSPQPDL-LLPATAPL----QIPTQP 165
++ D +W+++++S ++ + L+ SC R + DL +P + ++ +
Sbjct: 89 YNPSDLSNWVESMLSELNNPASSDLDTTRSCVDR--SEYDLRAIPGLSAFPKEEEVFDEE 146
Query: 166 PTPQR--------KNDNTTTETTSSFSSSTNP-LLKTLTEIASLIETQKPNQAIETLTHL 216
+ +R +D +T S T L+ L A I + N A + +
Sbjct: 147 ASSKRIRLGSWCESSDESTRSVVLVDSQETGVRLVHALVACAEAIHQENLNLADALVKRV 206
Query: 217 NKSISQNGNPNQRVSFYFSQALTNKI----TAQSSIASSNSSSTTWEELTLSYKALNDAC 272
+V+ YF+QAL +I TA++ + ++ + S +EE+ + ++C
Sbjct: 207 GTLAGSQAGAMGKVATYFAQALARRIYRDYTAETDVCAAVNPS--FEEVLEMH--FYESC 262
Query: 273 PYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGI 332
PY KFAH TANQAILEA + +H++D G+ QG+QW AL+QA A R G P S R++GI
Sbjct: 263 PYLKFAHFTANQAILEAVTTARRVHVIDLGLNQGMQWPALMQALALRPGGPP-SFRLTGI 321
Query: 333 PAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILT-PIELLDESSFCIQPD-EALAV 390
T S+ G +L++FA+ +G+ FEF + + L+ F +P+ E L V
Sbjct: 322 GPPQ--TENSDSLQQLGWKLAQFAQNMGVEFEFKGLAAESLSDLEPEMFETRPESETLVV 379
Query: 391 NFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAA 450
N + +L+ LL + S+EK L K++ P IVT+ E EA+ + F++RF A +Y+++
Sbjct: 380 NSVFELHRLL-ARSGSIEKLLNTVKAIKPSIVTVVEQEAN-HNGIVFLDRFNEALHYYSS 437
Query: 451 FFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFES 505
F+SLE + +L S +R E + LGR+I V+ ER E QW++ M++ GF+
Sbjct: 438 LFDSLEDSYSLPSQDRVMSE-VYLGRQILNVVAAEGSDRVERHETAAQWRIRMKSAGFDP 496
Query: 506 VGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+ L A QA +LL Y+ Y VE L + W+ PL+T S+W+
Sbjct: 497 IHLGSSAFKQASMLLSLYATGDGYR-VEENDGCLMIGWQTRPLITTSAWK 545
>At5g48150 SCARECROW gene regulator-like
Length = 490
Score = 187 bits (475), Expect = 1e-47
Identities = 131/399 (32%), Positives = 191/399 (47%), Gaps = 21/399 (5%)
Query: 173 DNTTTETTSSFSS-----STNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPN 227
D+T ++ + + S S L L A + A + L + +S +G P
Sbjct: 97 DSTASQEINGWRSTLEAISRRDLRADLVSCAKAMSENDLMMAHSMMEKLRQMVSVSGEPI 156
Query: 228 QRVSFYFSQALTNKITAQSSI---ASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQ 284
QR+ Y + L ++ + S A + EL L + CPY KF +++AN
Sbjct: 157 QRLGAYLLEGLVAQLASSGSSIYKALNRCPEPASTELLSYMHILYEVCPYFKFGYMSANG 216
Query: 285 AILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSS 344
AI EA + N +HI+DF I QG QW L+QAFA R G P +RI+GI M +
Sbjct: 217 AIAEAMKEENRVHIIDFQIGQGSQWVTLIQAFAARPGGPPR-IRITGIDDMTSAYARGGG 275
Query: 345 ISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENT 404
+S GNRL++ AK + FEF + + + + ++P EALAVNF L+++ DE+
Sbjct: 276 LSIVGNRLAKLAKQFNVPFEFNSVSVSVSEVKPKNLGVRPGEALAVNFAFVLHHMPDESV 335
Query: 405 ---NSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMAL 461
N ++ LR+ KSL+PK+VTL E E++ T F RF NY+AA FES++ +
Sbjct: 336 STENHRDRLLRMVKSLSPKVVTLVEQESN-TNTAAFFPRFMETMNYYAAMFESIDVTLPR 394
Query: 462 DSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFESVGLSHYAISQA 516
D +R VE L R + +I ER E +W+ GF LS S
Sbjct: 395 DHKQRINVEQHCLARDVVNIIACEGADRVERHELLGKWRSRFGMAGFTPYPLSPLVNSTI 454
Query: 517 KILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
K LL N YS Y L E A L L W L+ +W+
Sbjct: 455 KSLLRN--YSDKYRLEERDGA-LYLGWMHRDLVASCAWK 490
>At5g41920 SCARECROW gene regulator-like protein
Length = 405
Score = 187 bits (475), Expect = 1e-47
Identities = 127/399 (31%), Positives = 207/399 (51%), Gaps = 23/399 (5%)
Query: 166 PTPQRKNDNTTTETTSSFSSSTNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGN 225
P+ ++ ET + ++ LL L + A + T +A L+ +++ S G+
Sbjct: 16 PSSAKRRIEFPEETLENDGAAAIKLLSLLLQCAEYVATDHLREASTLLSEISEICSPFGS 75
Query: 226 PNQRVSFYFSQALTNKITAQSSIASSNSSSTTWEELTL--------SYKALNDACPYSKF 277
+RV YF+QAL ++ SS S S + + LT+ + + N P KF
Sbjct: 76 SPERVVAYFAQALQTRVI--SSYLSGACSPLSEKPLTVVQSQKIFSALQTYNSVSPLIKF 133
Query: 278 AHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMAL 337
+H TANQAI +A +G +++HI+D ++QG+QW AL A+R K S+RI+G + +
Sbjct: 134 SHFTANQAIFQALDGEDSVHIIDLDVMQGLQWPALFHILASRPR-KLRSIRITGFGSSS- 191
Query: 338 GTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPI-ELLDESSFCIQPDEALAVNFMLQL 396
+++TG RL++FA L L FEF PI I L+D S + EA+ V++M
Sbjct: 192 -----DLLASTGRRLADFASSLNLPFEFHPIEGIIGNLIDPSQLATRQGEAVVVHWMQ-- 244
Query: 397 YNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLE 456
+ L D N++E L + + L P ++T+ E E S F+ RF A +Y++A F++L
Sbjct: 245 HRLYDVTGNNLE-TLEILRRLKPNLITVVEQELSYDDGGSFLGRFVEALHYYSALFDALG 303
Query: 457 PNMALDSPERFQVESLLLGRRIDGVIGVRERMEDKEQWKVLMENCGFESVGLSHYAISQA 516
+ +S ERF VE ++LG I ++ + +WK + GF V L +QA
Sbjct: 304 DGLGEESGERFTVEQIVLGTEIRNIVAHGGGRRKRMKWKEELSRVGFRPVSLRGNPATQA 363
Query: 517 KILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+LL ++ Y+LVE + L L WKD+ LLT S+W+
Sbjct: 364 GLLLGMLPWNG-YTLVE-ENGTLRLGWKDLSLLTASAWK 400
>At1g50600 scarecrow-like 5 (SCL5)
Length = 597
Score = 186 bits (471), Expect = 4e-47
Identities = 129/380 (33%), Positives = 187/380 (48%), Gaps = 15/380 (3%)
Query: 186 STNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQ 245
S L L E A +E ++ L + +S +G P QR+ Y + L ++ +
Sbjct: 223 SRGDLKGVLYECAKAVENYDLEMTDWLISQLQQMVSVSGEPVQRLGAYMLEGLVARLASS 282
Query: 246 SS--IASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGI 303
S + T EL L +ACPY KF + +AN AI EA + + +HI+DF I
Sbjct: 283 GSSIYKALRCKDPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHIIDFQI 342
Query: 304 VQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNF 363
QG QW +L++A R G PN VRI+GI + + G RL + A++ G+ F
Sbjct: 343 SQGGQWVSLIRALGARPGGPPN-VRITGIDDPRSSFARQGGLELVGQRLGKLAEMCGVPF 401
Query: 364 EFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKSLNPK 420
EF ++ ++ EALAVNF L L+++ DE+ N ++ LRL K L+P
Sbjct: 402 EFHGAALCCTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKHLSPN 461
Query: 421 IVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDG 480
+VTL E EA+ T F+ RF N++ A FES++ +A D ER VE L R +
Sbjct: 462 VVTLVEQEAN-TNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAREVVN 520
Query: 481 VIGV-----RERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQ 535
+I ER E +W+ GF+ LS Y + K LL SYS Y+L E
Sbjct: 521 LIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLL--ESYSEKYTLEERD 578
Query: 536 PAFLSLAWKDVPLLTVSSWR 555
A L L WK+ PL+T +WR
Sbjct: 579 GA-LYLGWKNQPLITSCAWR 597
>At1g55580 unknown protein
Length = 445
Score = 173 bits (439), Expect = 2e-43
Identities = 139/439 (31%), Positives = 222/439 (49%), Gaps = 69/439 (15%)
Query: 175 TTTE--------TTSSFSSSTNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNP 226
TTTE ++S+ +S+++ L + L A+ + A L+ L+ + S +G+
Sbjct: 18 TTTENPPPLCIASSSAATSASHHLRRLLFTAANFVSQSNFTAAQNLLSILSLNSSPHGDS 77
Query: 227 NQRVSFYFSQALTNKITAQSSIASSNSSST-TWEELTLS--------------------- 264
+R+ F++AL+ +I Q ++ + +T T E+T+S
Sbjct: 78 TERLVHLFTKALSVRINRQQQDQTAETVATWTTNEMTMSNSTVFTSSVCKEQFLFRTKNN 137
Query: 265 --------YKALNDACPYSKFAHLTANQAILEATEGSNN--IHIVDFGIVQGIQWAALLQ 314
Y LN P+ +F HLTANQAIL+ATE ++N +HI+D I QG+QW L+Q
Sbjct: 138 NSDFESCYYLWLNQLTPFIRFGHLTANQAILDATETNDNGALHILDLDISQGLQWPPLMQ 197
Query: 315 AFATRS---SGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEF------ 365
A A RS S P S+RI+G V+ ++ TG+RL+ FA LGL F+F
Sbjct: 198 ALAERSSNPSSPPPSLRITGC------GRDVTGLNRTGDRLTRFADSLGLQFQFHTLVIV 251
Query: 366 ----TPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKI 421
+L I LL S+ E +AVN + L+ + +++ + + L KSLN +I
Sbjct: 252 EEDLAGLLLQIRLLALSAV---QGETIAVNCVHFLHKIFNDDGDMIGHFLSAIKSLNSRI 308
Query: 422 VTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGV 481
VT+ E EA+ F+ RF A +++ A F+SLE + +S ER +E G+ I V
Sbjct: 309 VTMAEREANHGDH-SFLNRFSEAVDHYMAIFDSLEATLPPNSRERLTLEQRWFGKEILDV 367
Query: 482 IGVRE-----RMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQP 536
+ E R E W+ +M+ GF +V + +A+SQAK+LL + S Y+L +
Sbjct: 368 VAAEETERKQRHRRFEIWEEMMKRFGFVNVPIGSFALSQAKLLLRLHYPSEGYNL-QFLN 426
Query: 537 AFLSLAWKDVPLLTVSSWR 555
L L W++ PL +VSSW+
Sbjct: 427 NSLFLGWQNRPLFSVSSWK 445
>At5g17490 RGA-like protein
Length = 523
Score = 173 bits (438), Expect = 3e-43
Identities = 126/393 (32%), Positives = 204/393 (51%), Gaps = 23/393 (5%)
Query: 173 DNTTTETTSS---FSSSTNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQR 229
D+ T+E+T S + L++ L A ++ + + A + + + +
Sbjct: 137 DSVTSESTRSVVLIEETGVRLVQALVACAEAVQLENLSLADALVKRVGLLAASQAGAMGK 196
Query: 230 VSFYFSQALTNKITAQSSIASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEA 289
V+ YF++AL +I A++ S E L +++ D+CPY KFAH TANQAILEA
Sbjct: 197 VATYFAEALARRIYRIHPSAAAIDPSFE-EILQMNFY---DSCPYLKFAHFTANQAILEA 252
Query: 290 TEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATG 349
S +H++D G+ QG+QW AL+QA A R G P S R++G+ S I G
Sbjct: 253 VTTSRVVHVIDLGLNQGMQWPALMQALALRPGGPP-SFRLTGVG----NPSNREGIQELG 307
Query: 350 NRLSEFAKLLGLNFEFTPILTP-IELLDESSFCIQPD-EALAVNFMLQLYNLLDENTNSV 407
+L++ A+ +G+ F+F + T + L+ F + + E L VN + +L+ +L + S+
Sbjct: 308 WKLAQLAQAIGVEFKFNGLTTERLSDLEPDMFETRTESETLVVNSVFELHPVLSQ-PGSI 366
Query: 408 EKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERF 467
EK L K++ P +VT+ E EA+ V F++RF A +Y+++ F+SLE + + S +R
Sbjct: 367 EKLLATVKAVKPGLVTVVEQEANHNGDV-FLDRFNEALHYYSSLFDSLEDGVVIPSQDRV 425
Query: 468 QVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWN 522
E + LGR+I ++ ER E QW+ M + GF+ V L A QA +LL
Sbjct: 426 MSE-VYLGRQILNLVATEGSDRIERHETLAQWRKRMGSAGFDPVNLGSDAFKQASLLLAL 484
Query: 523 YSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
Y VE L LAW+ PL+ S+W+
Sbjct: 485 SGGGDGYR-VEENDGSLMLAWQTKPLIAASAWK 516
>At1g66350 gibberellin regulatory protein like
Length = 511
Score = 169 bits (427), Expect = 5e-42
Identities = 137/469 (29%), Positives = 224/469 (47%), Gaps = 55/469 (11%)
Query: 108 TFRFSDFDSDDWMDTLMSAADSHSDFHLNPFSSCPTRLSPQPDLLLPATAPLQIPTQPPT 167
T ++ D W+++++S D PTR+ +PD A P
Sbjct: 72 TVHYNPSDLSGWVESMLSDLD-------------PTRIQEKPDSEYDLRAIPGSAVYPRD 118
Query: 168 PQRKNDNTTTETTSSFSSSTNP-----------LLKTLTEIASLIETQKPNQAIETLTHL 216
+ T S SS+ + L+ L A ++ A + H+
Sbjct: 119 EHVTRRSKRTRIESELSSTRSVVVLDSQETGVRLVHALLACAEAVQQNNLKLADALVKHV 178
Query: 217 NKSISQNGNPNQRVSFYFSQALTNKITA---QSSIASSNSSSTTWEELTLSYKALNDACP 273
S ++V+ YF++ L +I + +A S+ S T L + + ++CP
Sbjct: 179 GLLASSQAGAMRKVATYFAEGLARRIYRIYPRDDVALSSFSDT----LQIHFY---ESCP 231
Query: 274 YSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIP 333
Y KFAH TANQAILE + +H++D G+ G+QW AL+QA A R +G P+ R++GI
Sbjct: 232 YLKFAHFTANQAILEVFATAEKVHVIDLGLNHGLQWPALIQALALRPNGPPD-FRLTGI- 289
Query: 334 AMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPI-LTPIELLDESSFCIQPD-EALAVN 391
G S ++ I G +L + A +G+NFEF I L + L I+P E++AVN
Sbjct: 290 ----GYS-LTDIQEVGWKLGQLASTIGVNFEFKSIALNNLSDLKPEMLDIRPGLESVAVN 344
Query: 392 FMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAF 451
+ +L+ LL + S++K L KS+ P I+T+ E EA+ V F++RF + +Y+++
Sbjct: 345 SVFELHRLL-AHPGSIDKFLSTIKSIRPDIMTVVEQEANHNGTV-FLDRFTESLHYYSSL 402
Query: 452 FESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFESV 506
F+SLE + D + L LGR+I ++ ER E QW+ GF+ V
Sbjct: 403 FDSLEGPPSQDR----VMSELFLGRQILNLVACEGEDRVERHETLNQWRNRFGLGGFKPV 458
Query: 507 GLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+ A QA +LL Y+ + Y++ E++ L L W+ PL+ S+WR
Sbjct: 459 SIGSNAYKQASMLLALYAGADGYNVEENEGCLL-LGWQTRPLIATSAWR 506
>At2g04890 putative SCARECROW gene regulator
Length = 413
Score = 168 bits (426), Expect = 6e-42
Identities = 119/350 (34%), Positives = 176/350 (50%), Gaps = 22/350 (6%)
Query: 216 LNKSISQNGNPNQRVSFYFSQALTNKITAQ-SSIASSNSSSTTWEELTLSYK-ALNDACP 273
L +S +G P QR+ Y + L ++ A SSI S S LSY L++ CP
Sbjct: 76 LRGMVSISGEPIQRLGAYMLEGLVARLAASGSSIYKSLQSREPESYEFLSYVYVLHEVCP 135
Query: 274 YSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIP 333
Y KF +++AN AI EA + IHI+DF I QG QW AL+QAFA R G PN +RI+G+
Sbjct: 136 YFKFGYMSANGAIAEAMKDEERIHIIDFQIGQGSQWIALIQAFAARPGGAPN-IRITGV- 193
Query: 334 AMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQPDEALAVNFM 393
S + RL + AK + F F + P ++ + ++ EAL VNF
Sbjct: 194 ------GDGSVLVTVKKRLEKLAKKFDVPFRFNAVSRPSCEVEVENLDVRDGEALGVNFA 247
Query: 394 LQLYNLLDENT---NSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAA 450
L++L DE+ N ++ LR+ KSL+PK+VTL E E + T F+ RF +Y+ A
Sbjct: 248 YMLHHLPDESVSMENHRDRLLRMVKSLSPKVVTLVEQECNTNTS-PFLPRFLETLSYYTA 306
Query: 451 FFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR--ERMEDKE---QWKVLMENCGFES 505
FES++ + + ER +E + R + +I ER+E E +WK GFE
Sbjct: 307 MFESIDVMLPRNHKERINIEQHCMARDVVNIIACEGAERIERHELLGKWKSRFSMAGFEP 366
Query: 506 VGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
LS + + LL +YS +E + L L W D L++ +W+
Sbjct: 367 YPLSSIISATIRALLRDYSNG---YAIEERDGALYLGWMDRILVSSCAWK 413
>At1g21450 scarecrow-like 1 (SCL1)
Length = 593
Score = 165 bits (418), Expect = 5e-41
Identities = 114/400 (28%), Positives = 192/400 (47%), Gaps = 15/400 (3%)
Query: 166 PTPQRKNDNTTTETTSSFSSSTNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGN 225
P D+ + ++ S P + L A + K +A+ + L + +S G+
Sbjct: 199 PKESSSADSNSHVSSKEVVSQATPK-QILISCARALSEGKLEEALSMVNELRQIVSIQGD 257
Query: 226 PNQRVSFYFSQALTNKITAQSSIA--SSNSSSTTWEELTLSYKALNDACPYSKFAHLTAN 283
P+QR++ Y + L ++ A + +E + + L + CP KF L AN
Sbjct: 258 PSQRIAAYMVEGLAARMAASGKFIYRALKCKEPPSDERLAAMQVLFEVCPCFKFGFLAAN 317
Query: 284 QAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVS 343
AILEA +G +HI+DF I QG Q+ L+++ A GK +R++GI +
Sbjct: 318 GAILEAIKGEEEVHIIDFDINQGNQYMTLIRSIA-ELPGKRPRLRLTGIDDPESVQRSIG 376
Query: 344 SISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDEN 403
+ G RL + A+ G++F+F + + ++ S+ +P E L VNF QL+++ DE+
Sbjct: 377 GLRIIGLRLEQLAEDNGVSFKFKAMPSKTSIVSPSTLGCKPGETLIVNFAFQLHHMPDES 436
Query: 404 TNSV---EKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMA 460
+V ++ L + KSLNPK+VT+ E + + T F RF A+ Y++A FESL+ +
Sbjct: 437 VTTVNQRDELLHMVKSLNPKLVTVVEQDVNTNTS-PFFPRFIEAYEYYSAVFESLDMTLP 495
Query: 461 LDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFESVGLSHYAISQ 515
+S ER VE L R I ++ ER E +W+ M GF +S +
Sbjct: 496 RESQERMNVERQCLARDIVNIVACEGEERIERYEAAGKWRARMMMAGFNPKPMSAKVTNN 555
Query: 516 AKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+ L+ Y + Y L E + L W++ L+ S+WR
Sbjct: 556 IQNLI-KQQYCNKYKLKE-EMGELHFCWEEKSLIVASAWR 593
>At1g63100 transcription factor SCARECROW, putative
Length = 658
Score = 157 bits (398), Expect = 1e-38
Identities = 120/422 (28%), Positives = 205/422 (48%), Gaps = 47/422 (11%)
Query: 168 PQRKNDNTTTETTSSFSSSTNPL-LKTLTEIASLIETQKPNQAIETLTHLNKSISQNGN- 225
P T TT + +++ N L+ E+ +L+ + +N I++ G+
Sbjct: 244 PYSHRGATEERTTGNINNNNNRNDLQRDFELVNLLTGCLDAIRSRNIAAINHFIARTGDL 303
Query: 226 -------PNQRVSFYFSQALTNKITAQ----SSIASSNSSSTTWE-ELTLSYKALNDACP 273
P R+ Y+ +AL ++ IA T E E + + LN P
Sbjct: 304 ASPRGRTPMTRLIAYYIEALALRVARMWPHIFHIAPPREFDRTVEDESGNALRFLNQVTP 363
Query: 274 YSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIP 333
KF H TAN+ +L A EG +HI+DF I QG+QW + Q+ A+R + P+ VRI+GI
Sbjct: 364 IPKFIHFTANEMLLRAFEGKERVHIIDFDIKQGLQWPSFFQSLASRIN-PPHHVRITGIG 422
Query: 334 AMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQPDEALAVNFM 393
L ++ TG+RL FA+ + L FEF P++ +E + ++ E++AVN +
Sbjct: 423 ESKL------ELNETGDRLHGFAEAMNLQFEFHPVVDRLEDVRLWMLHVKEGESVAVNCV 476
Query: 394 LQLYNLLDENTN-SVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFET----AFNYF 448
+Q++ L + T ++ L L +S NP + L E EA + E+ ET + Y+
Sbjct: 477 MQMHKTLYDGTGAAIRDFLGLIRSTNPIALVLAEQEAEHNS-----EQLETRVCNSLKYY 531
Query: 449 AAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV-----RERMEDKEQWKVLMENCGF 503
+A F+++ N+A DS R +VE +L GR I ++ +ER W+ ++E GF
Sbjct: 532 SAMFDAIHTNLATDSLMRVKVEEMLFGREIRNIVACEGSHRQERHVGFRHWRRMLEQLGF 591
Query: 504 ESVGLSHYAISQAKILLWNYSYSS--LYSLVES---------QPAFLSLAWKDVPLLTVS 552
S+G+S + Q+K+LL Y + +++ S + ++L W + PL T+S
Sbjct: 592 RSLGVSEREVLQSKMLLRMYGSDNEGFFNVERSDEDNGGEGGRGGGVTLRWSEQPLYTIS 651
Query: 553 SW 554
+W
Sbjct: 652 AW 653
>At4g37650 SHORT-ROOT (SHR)
Length = 531
Score = 156 bits (395), Expect = 2e-38
Identities = 140/507 (27%), Positives = 225/507 (43%), Gaps = 49/507 (9%)
Query: 87 SGPENTDPAFNFPSPLDPHSTTFRFSDFDSDDWMDTLMSAADSHSDFH--------LNPF 138
+G T +NFP D F F D +D +S++ SH + H +PF
Sbjct: 34 TGSPQTAYHYNFPQN-DVVEECFNFF-MDEED-----LSSSSSHHNHHNHNNPNTYYSPF 86
Query: 139 SSCPTRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTTT----ETTSSFSSSTNPLL--K 192
++ PT+ P +TA P + +++ + +T SF S N
Sbjct: 87 TT-PTQYHPATSSTPSSTAAAAALASPYSSSGHHNDPSAFSIPQTPPSFDFSANAKWADS 145
Query: 193 TLTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQSS----- 247
L E A + +A + L LN+ S G+ Q+++ YF QAL N++T
Sbjct: 146 VLLEAARAFSDKDTARAQQILWTLNELSSPYGDTEQKLASYFLQALFNRMTGSGERCYRT 205
Query: 248 --IASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQ 305
A++ + ++E + + P++ F H+ AN AILEA +G IHIVD
Sbjct: 206 MVTAAATEKTCSFESTRKTVLKFQEVSPWATFGHVAANGAILEAVDGEAKIHIVDISSTF 265
Query: 306 GIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSS---ISATGNRLSEFAKLLGLN 362
QW LL+A ATRS P+ + + A +S + GNR+ +FA+L+G+
Sbjct: 266 CTQWPTLLEALATRSDDTPHLRLTTVVVANKFVNDQTASHRMMKEIGNRMEKFARLMGVP 325
Query: 363 FEFTPI--LTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPK 420
F+F I + + D + ++PDE LA+N + ++ + + + + + L P+
Sbjct: 326 FKFNIIHHVGDLSEFDLNELDVKPDEVLAINCVGAMHGIASRGSPR-DAVISSFRRLRPR 384
Query: 421 IVTLGEYEASLT--TRVGFVERFETAF----NYFAAFFESLEPNMALDSPERFQVESLLL 474
IVT+ E EA L GF + F F +F FES E + S ER +E
Sbjct: 385 IVTVVEEEADLVGEEEGGFDDEFLRGFGECLRWFRVCFESWEESFPRTSNERLMLER-AA 443
Query: 475 GRRIDGVIGVR-----ERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLY 529
GR I ++ ER E +W M N GF +VG S + LL Y ++
Sbjct: 444 GRAIVDLVACEPSDSTERRETARKWSRRMRNSGFGAVGYSDEVADDVRALLRRYK-EGVW 502
Query: 530 SLVE-SQPAFLSLAWKDVPLLTVSSWR 555
S+V+ A + L W+D P++ S+WR
Sbjct: 503 SMVQCPDAAGIFLCWRDQPVVWASAWR 529
Score = 28.9 bits (63), Expect = 7.8
Identities = 29/110 (26%), Positives = 46/110 (41%), Gaps = 15/110 (13%)
Query: 24 QQEQHEQQQHQHHHHHQNILGITNPLSVSLHPWHNNNIPVSSSTSLPPLSFPPTDFTDPF 83
+++ H +HH+H N +P + +H P +SST P + P+
Sbjct: 61 EEDLSSSSSHHNHHNHNNPNTYYSPFTTPTQ-YH----PATSST--PSSTAAAAALASPY 113
Query: 84 QVGSGPENTDPAFNFPSPLDPHSTTFRFSDFDSDDWMDT-LMSAADSHSD 132
SG N AF+ P +F FS + W D+ L+ AA + SD
Sbjct: 114 S-SSGHHNDPSAFSIPQT----PPSFDFS--ANAKWADSVLLEAARAFSD 156
>At4g08250 putative protein
Length = 483
Score = 156 bits (395), Expect = 2e-38
Identities = 134/499 (26%), Positives = 219/499 (43%), Gaps = 39/499 (7%)
Query: 72 LSFPPTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTTFRFSDFDSD---DWMDTLMSAAD 128
+++P DF D F +TDP S S + +D D D D+ D + S
Sbjct: 1 MNYPYEDFLDLFF----STHTDPLATAAST---SSNGYSLNDLDIDWDCDFRDVIESIMG 53
Query: 129 SHSDFHLNPFSSCPTRLSPQPDLLLPATAPLQIPT-----QPPTPQRKNDNTTTETTSSF 183
+ P S L Q L A+ L + +P T + K ++
Sbjct: 54 DEGAM-MEPESEAVPMLHDQEGLCNSASTGLSVADGVSFGEPKTDESKGLRLVHLLVAAA 112
Query: 184 SSSTNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPN-QRVSFYFSQALTNKI 242
+ST E+ +I L L +S N +R++ +F+ L+ +
Sbjct: 113 DASTGA--NKSRELTRVI-----------LARLKDLVSPGDRTNMERLAAHFTNGLSKLL 159
Query: 243 TAQSSIASSNSSSTTWEELTL--SYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVD 300
S + +++ + +++ L + PY F +LTA QAILEA + IHIVD
Sbjct: 160 ERDSVLCPQQHRDDVYDQADVISAFELLQNMSPYVNFGYLTATQAILEAVKYERRIHIVD 219
Query: 301 FGIVQGIQWAALLQAFATRSSG-KPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLL 359
+ I +G+QWA+L+QA +R++G +RI+ + G V+++ TG RL+ FA +
Sbjct: 220 YDINEGVQWASLMQALVSRNTGPSAQHLRITALSRATNGKKSVAAVQETGRRLTAFADSI 279
Query: 360 GLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNP 419
G F + SS + EA+ +N ML L + +SV L AK+LNP
Sbjct: 280 GQPFSYQHCKLDTNAFSTSSLKLVRGEAVVINCMLHLPRFSHQTPSSVISFLSEAKTLNP 339
Query: 420 KIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLG---- 475
K+VTL E L GF+ RF + F+A F+SLE +++ +P R VE + +G
Sbjct: 340 KLVTLVHEEVGLMGNQGFLYRFMDLLHQFSAIFDSLEAGLSIANPARGFVERVFIGPWVA 399
Query: 476 RRIDGVIGVRERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQ 535
+ + +E W +E GF+ + +S QAK+LL ++ + + E
Sbjct: 400 NWLTRITANDAEVESFASWPQWLETNGFKPLEVSFTNRCQAKLLL--SLFNDGFRVEELG 457
Query: 536 PAFLSLAWKDVPLLTVSSW 554
L L WK L++ S W
Sbjct: 458 QNGLVLGWKSRRLVSASFW 476
>At2g01570 putative RGA1, giberellin repsonse modulation protein
Length = 587
Score = 154 bits (389), Expect = 1e-37
Identities = 146/524 (27%), Positives = 240/524 (44%), Gaps = 59/524 (11%)
Query: 53 LHPWHNNNIPVSSSTSLPPLSFPPTDFTDPFQVGSGPENTD-PAFNFPSPLDPHSTTFRF 111
L+ W +N + S + PPL ++ DP V PE PA ++ + P + ++F
Sbjct: 96 LYSWLDNML---SELNPPPLP-ASSNGLDP--VLPSPEICGFPASDYDLKVIPGNAIYQF 149
Query: 112 SDFDSDDWMDTLMSAADSHSDFHLNPFSSCPTRLSPQPDLLLPATAPLQIPTQPPTPQRK 171
DS S++ ++ + L SS PD ++ +T+ TQ
Sbjct: 150 PAIDS--------SSSSNNQNKRLKSCSS--------PDSMVTSTST---GTQIGGVIGT 190
Query: 172 NDNTTTETTSSFSSSTNP------------LLKTLTEIASLIETQKPNQAIETLTHLNKS 219
TTT TT++ ST L+ L A I+ A + +
Sbjct: 191 TVTTTTTTTTAAGESTRSVILVDSQENGVRLVHALMACAEAIQQNNLTLAEALVKQIGCL 250
Query: 220 ISQNGNPNQRVSFYFSQALTNKITAQSSIASSNSSSTTWEELTLSYKALNDACPYSKFAH 279
++V+ YF++AL +I S + + + L + + + CPY KFAH
Sbjct: 251 AVSQAGAMRKVATYFAEALARRIYRLSPPQNQIDHCLS-DTLQMHFY---ETCPYLKFAH 306
Query: 280 LTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGT 339
TANQAILEA EG +H++DF + QG+QW AL+QA A R G P + R++GI A
Sbjct: 307 FTANQAILEAFEGKKRVHVIDFSMNQGLQWPALMQALALREGGPP-TFRLTGIGPPAPDN 365
Query: 340 SPVSSISATGNRLSEFAKLLGLNFEFTP-ILTPIELLDESSFCIQPD--EALAVNFMLQL 396
S + G +L++ A+ + + FE+ + + LD S ++P EA+AVN + +L
Sbjct: 366 S--DHLHEVGCKLAQLAEAIHVEFEYRGFVANSLADLDASMLELRPSDTEAVAVNSVFEL 423
Query: 397 YNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLE 456
+ LL +EK L + K + P I T+ E E++ V F++RF + +Y++ F+SLE
Sbjct: 424 HKLLG-RPGGIEKVLGVVKQIKPVIFTVVEQESNHNGPV-FLDRFTESLHYYSTLFDSLE 481
Query: 457 PNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFESVGLSHY 511
+ + + + + LG++I ++ ER E QW + G L
Sbjct: 482 ---GVPNSQDKVMSEVYLGKQICNLVACEGPDRVERHETLSQWGNRFGSSGLAPAHLGSN 538
Query: 512 AISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
A QA +LL ++ Y + ES L L W PL+T S+W+
Sbjct: 539 AFKQASMLLSVFNSGQGYRVEESN-GCLMLGWHTRPLITTSAWK 581
>At1g14920 signal response protein (GAI)
Length = 533
Score = 150 bits (380), Expect = 1e-36
Identities = 118/406 (29%), Positives = 200/406 (49%), Gaps = 35/406 (8%)
Query: 171 KNDNTTTETTSSFSSSTNP------------LLKTLTEIASLIETQKPNQAIETLTHLN- 217
K N ETT++ + ST L+ L A ++ + A + +
Sbjct: 138 KCSNGVVETTTATAESTRHVVLVDSQENGVRLVHALLACAEAVQKENLTVAEALVKQIGF 197
Query: 218 KSISQNGNPNQRVSFYFSQALTNKITAQSSIASSNSSSTTWEELTLSYKALNDACPYSKF 277
++SQ G ++V+ YF++AL +I S S S + + L + + + CPY KF
Sbjct: 198 LAVSQIG-AMRKVATYFAEALARRIYRLSPSQSPIDHSLS-DTLQMHFY---ETCPYLKF 252
Query: 278 AHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMAL 337
AH TANQAILEA +G +H++DF + QG+QW AL+QA A R G P R++GI A
Sbjct: 253 AHFTANQAILEAFQGKKRVHVIDFSMSQGLQWPALMQALALRPGGPP-VFRLTGIGPPA- 310
Query: 338 GTSPVSSISATGNRLSEFAKLLGLNFEFTP-ILTPIELLDESSFCIQPD--EALAVNFML 394
+ G +L+ A+ + + FE+ + + LD S ++P E++AVN +
Sbjct: 311 -PDNFDYLHEVGCKLAHLAEAIHVEFEYRGFVANTLADLDASMLELRPSEIESVAVNSVF 369
Query: 395 QLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFES 454
+L+ LL +++K L + + P+I T+ E E++ + + F++RF + +Y++ F+S
Sbjct: 370 ELHKLLG-RPGAIDKVLGVVNQIKPEIFTVVEQESNHNSPI-FLDRFTESLHYYSTLFDS 427
Query: 455 LEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFESVGLS 509
LE + S + + + LG++I V+ ER E QW+ + GF + +
Sbjct: 428 LE---GVPSGQDKVMSEVYLGKQICNVVACDGPDRVERHETLSQWRNRFGSAGFAAAHIG 484
Query: 510 HYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
A QA +LL ++ Y VE L L W PL+ S+W+
Sbjct: 485 SNAFKQASMLLALFNGGEGYR-VEESDGCLMLGWHTRPLIATSAWK 529
>At1g07530 transcription factor scarecrow-like 14, putative
Length = 769
Score = 147 bits (371), Expect = 2e-35
Identities = 105/399 (26%), Positives = 175/399 (43%), Gaps = 21/399 (5%)
Query: 170 RKNDNTTTETTSSFSSSTNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQR 229
R +T+T ++ T L L A + A E L + + S GN ++R
Sbjct: 373 RGKKSTSTSHSNDSKKETADLRTLLVLCAQAVSVDDRRTANEMLRQIREHSSPLGNGSER 432
Query: 230 VSFYFSQALTNKI--TAQSSIASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAIL 287
++ YF+ +L ++ T + +S T+ ++ +Y+ CP+ K A + AN +++
Sbjct: 433 LAHYFANSLEARLAGTGTQIYTALSSKKTSAADMLKAYQTYMSVCPFKKAAIIFANHSMM 492
Query: 288 EATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISA 347
T +N IHI+DFGI G QW AL+ + G +RI+GI G P +
Sbjct: 493 RFTANANTIHIIDFGISYGFQWPALIHRLSLSRPGGSPKLRITGIELPQRGFRPAEGVQE 552
Query: 348 TGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDEN--TN 405
TG+RL+ + + + FE+ I E + ++ E + VN + + NLLDE N
Sbjct: 553 TGHRLARYCQRHNVPFEYNAIAQKWETIQVEDLKLRQGEYVVVNSLFRFRNLLDETVLVN 612
Query: 406 SVEKA-LRLAKSLNPKI----VTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMA 460
S A L+L + +NP + + G Y A FV RF A +++A F+ + +A
Sbjct: 613 SPRDAVLKLIRKINPNVFIPAILSGNYNAPF-----FVTRFREALFHYSAVFDMCDSKLA 667
Query: 461 LDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFESVGLSHYAISQ 515
+ R E GR I V+ ER E +QW+ + GF + L +
Sbjct: 668 REDEMRLMYEKEFYGREIVNVVACEGTERVERPETYKQWQARLIRAGFRQLPLEKELMQN 727
Query: 516 AKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 554
K+ + N Y + V+ +L WK + S W
Sbjct: 728 LKLKIEN-GYDKNFD-VDQNGNWLLQGWKGRIVYASSLW 764
>At1g07520 transcription factor scarecrow-like 14, putative
Length = 695
Score = 145 bits (366), Expect = 6e-35
Identities = 107/377 (28%), Positives = 171/377 (44%), Gaps = 19/377 (5%)
Query: 194 LTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITA------QSS 247
LT A + A + L + K S G+ +QR++ +F+ AL ++ QS
Sbjct: 319 LTLCAQSVSAGDKITADDLLRQIRKQCSPVGDASQRLAHFFANALEARLEGSTGTMIQSY 378
Query: 248 IASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGI 307
S +S T ++ SY A P+ + +N+ IL+A + ++ +HIVDFGI+ G
Sbjct: 379 YDSISSKKRTAAQILKSYSVFLSASPFMTLIYFFSNKMILDAAKDASVLHIVDFGILYGF 438
Query: 308 QWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTP 367
QW +Q + + G +RI+GI G P I TG RL+E+ K G+ FE+
Sbjct: 439 QWPMFIQHLSKSNPGL-RKLRITGIEIPQHGLRPTERIQDTGRRLTEYCKRFGVPFEYNA 497
Query: 368 ILTP-IELLDESSFCIQPDEALAVNFMLQLYNLLD----ENTNSVEKALRLAKSLNPKIV 422
I + E + F I+P+E LAVN +L+ NL D E + L+L + +NP +
Sbjct: 498 IASKNWETIKMEEFKIRPNEVLAVNAVLRFKNLRDVIPGEEDCPRDGFLKLIRDMNPNVF 557
Query: 423 TLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVI 482
S F RF+ A +++A F+ ++ ++PER E GR + VI
Sbjct: 558 LSSTVNGSFNAPF-FTTRFKEALFHYSALFDLFGATLSKENPERIHFEGEFYGREVMNVI 616
Query: 483 GVR-----ERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPA 537
ER E +QW+V M GF+ + + + + + Y + L E
Sbjct: 617 ACEGVDRVERPETYKQWQVRMIRAGFKQKPVEAELVQLFREKMKKWGYHKDFVLDEDSNW 676
Query: 538 FLSLAWKDVPLLTVSSW 554
FL WK L + S W
Sbjct: 677 FLQ-GWKGRILFSSSCW 692
>At5g59450 putative SCARECROW transcriptional regulatory protein
(F2O15.13)
Length = 610
Score = 145 bits (365), Expect = 7e-35
Identities = 107/407 (26%), Positives = 182/407 (44%), Gaps = 18/407 (4%)
Query: 163 TQPPTPQRKND-NTTTETTSSFSSSTNPLLKTLTEIASLIETQKPNQAIETLTHLNKSIS 221
T+ TP R ++ + + ++T L LT+ A + + +A + L + S
Sbjct: 196 TKQSTPNRAGRAKGSSNKSKTHKTNTVDLRSLLTQCAQAVASFDQRRATDKLKEIRAHSS 255
Query: 222 QNGNPNQRVSFYFSQALTNKITAQSSIASSN---SSSTTWEELTLSYKALNDACPYSKFA 278
NG+ QR++FYF++AL +IT S SN SS+T+ ++ +YK CP
Sbjct: 256 SNGDGTQRLAFYFAEALEARITGNISPPVSNPFPSSTTSMVDILKAYKLFVHTCPIYVTD 315
Query: 279 HLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALG 338
+ AN++I E + +HIVDFG++ G QW LL+A + R G P +R++GI G
Sbjct: 316 YFAANKSIYELAMKATKLHIVDFGVLYGFQWPCLLRALSKRPGGPP-MLRVTGIELPQAG 374
Query: 339 TSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYN 398
P + TG RL F + FEF I E + I P E VN + +L
Sbjct: 375 FRPSDRVEETGRRLKRFCDQFNVPFEFNFIAKKWETITLDELMINPGETTVVNCIHRLQY 434
Query: 399 LLDENT---NSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESL 455
DE + + L+L + +NP + E + F+ RF A ++++ F+
Sbjct: 435 TPDETVSLDSPRDTVLKLFRDINPDLFVFAEINGMYNSPF-FMTRFREALFHYSSLFDMF 493
Query: 456 EPNMALDS--PERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFESVGL 508
+ + + R +E LL R VI R E +QW+V + GF+ +
Sbjct: 494 DTTIHAEDEYKNRSLLERELLVRDAMSVISCEGAERFARPETYKQWRVRILRAGFKPATI 553
Query: 509 SHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
S + +AK ++ + +++S ++ WK + S W+
Sbjct: 554 SKQIMKEAKEIVRKRYHRDF--VIDSDNNWMLQGWKGRVIYAFSCWK 598
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.129 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,523,479
Number of Sequences: 26719
Number of extensions: 564568
Number of successful extensions: 4315
Number of sequences better than 10.0: 145
Number of HSP's better than 10.0 without gapping: 68
Number of HSP's successfully gapped in prelim test: 78
Number of HSP's that attempted gapping in prelim test: 3583
Number of HSP's gapped (non-prelim): 385
length of query: 555
length of database: 11,318,596
effective HSP length: 104
effective length of query: 451
effective length of database: 8,539,820
effective search space: 3851458820
effective search space used: 3851458820
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC142222.7


