

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.6 - phase: 0
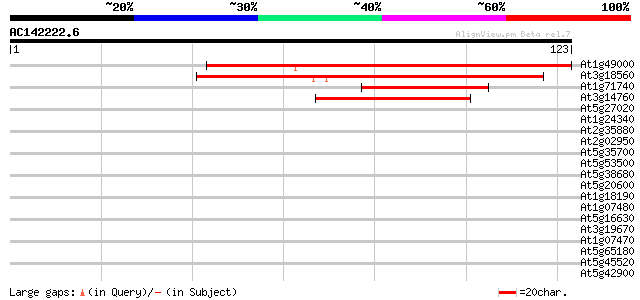
(123 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g49000 hypothetical protein; similar to ESTs gb|R64892.1, gb|... 85 8e-18
At3g18560 unknown protein 83 3e-17
At1g71740 hypothetical protein 45 1e-05
At3g14760 unknown protein 39 6e-04
At5g27020 putative protein 32 0.059
At1g24340 unknown protein 28 1.5
At2g35880 unknown protein 27 2.5
At2g02950 phytochrome kinase substrate 1 (PKS1) 27 3.3
At5g35700 fimbrin 26 4.3
At5g53500 unknown protein 26 5.6
At5g38680 putative protein 26 5.6
At5g20600 unknown protein 26 5.6
At1g18190 hypothetical protein 26 5.6
At1g07480 transcription factor IIA large subunit 26 5.6
At5g16630 unknown protein 25 7.3
At3g19670 unknown protein 25 7.3
At1g07470 putative transcription factor IIA large subunit protein 25 7.3
At5g65180 unknown protein 25 9.5
At5g45520 putative protein 25 9.5
At5g42900 unknown protein 25 9.5
>At1g49000 hypothetical protein; similar to ESTs gb|R64892.1,
gb|T04316.1
Length = 156
Score = 85.1 bits (209), Expect = 8e-18
Identities = 43/81 (53%), Positives = 53/81 (65%), Gaps = 1/81 (1%)
Query: 44 KTKKLLSNMSNKALSQFG-KMKKTKKQRDNELNDGVWQKEILMGGKCEPLDFSGVIHYDI 102
+ K+L + +SNKA++ K +K E G+WQ+EILMGGKCEPLDFSGVI+YD
Sbjct: 72 RPKELFTTLSNKAMTMVRRKNPPEEKATAMEEEHGLWQREILMGGKCEPLDFSGVIYYDS 131
Query: 103 NGRQTGEVPLRSPRAIPFSRY 123
NGR EVP RSPR P Y
Sbjct: 132 NGRLLNEVPPRSPRGTPLPSY 152
>At3g18560 unknown protein
Length = 195
Score = 83.2 bits (204), Expect = 3e-17
Identities = 43/88 (48%), Positives = 58/88 (65%), Gaps = 12/88 (13%)
Query: 42 KAKTKKLLSNMSNKALSQFGKMKK------TKK------QRDNELNDGVWQKEILMGGKC 89
+ + K+L + +SNKA++ G+ K TKK ++E GVWQ+EILMGGKC
Sbjct: 85 RVRPKELYTTLSNKAMTMVGRKNKAYDGGPTKKTAVEMVMEEDEEEYGVWQREILMGGKC 144
Query: 90 EPLDFSGVIHYDINGRQTGEVPLRSPRA 117
EPLD+SGVI+YD +G Q +VP RSPRA
Sbjct: 145 EPLDYSGVIYYDCSGHQLKQVPPRSPRA 172
>At1g71740 hypothetical protein
Length = 130
Score = 44.7 bits (104), Expect = 1e-05
Identities = 19/28 (67%), Positives = 23/28 (81%)
Query: 78 VWQKEILMGGKCEPLDFSGVIHYDINGR 105
+WQK ILMGGKC+ DFSGVI YD +G+
Sbjct: 88 LWQKNILMGGKCQLPDFSGVILYDADGQ 115
>At3g14760 unknown protein
Length = 168
Score = 38.9 bits (89), Expect = 6e-04
Identities = 19/34 (55%), Positives = 22/34 (63%)
Query: 68 KQRDNELNDGVWQKEILMGGKCEPLDFSGVIHYD 101
K+R D VWQ+ ILMG KCE FSG+I YD
Sbjct: 106 KERSLNAVDPVWQRPILMGEKCELPRFSGLILYD 139
>At5g27020 putative protein
Length = 164
Score = 32.3 bits (72), Expect = 0.059
Identities = 17/49 (34%), Positives = 31/49 (62%), Gaps = 7/49 (14%)
Query: 61 GKMKKTKKQRDNELNDGVWQKE-----ILMGGKCEPLDFSGVIHYDING 104
G+ K ++ R L++G+ ++E ILMG +C+P+ +GV+ YD +G
Sbjct: 112 GQRLKRRRNRRRSLSEGMEEEELCKKRILMGERCKPM--NGVLQYDGDG 158
>At1g24340 unknown protein
Length = 709
Score = 27.7 bits (60), Expect = 1.5
Identities = 15/45 (33%), Positives = 23/45 (50%)
Query: 49 LSNMSNKALSQFGKMKKTKKQRDNELNDGVWQKEILMGGKCEPLD 93
L+N+ K L G + K+ D D V ++ILMG +C +D
Sbjct: 160 LTNLLLKRLEDLGFHVRGSKESDGLEADSVVARQILMGHECVGID 204
>At2g35880 unknown protein
Length = 432
Score = 26.9 bits (58), Expect = 2.5
Identities = 21/89 (23%), Positives = 40/89 (44%), Gaps = 7/89 (7%)
Query: 6 SSPESLSPKRQSNSSFFITISAILAMLSRQARRLKGKAKTKKLLSNMS------NKALSQ 59
++P PK S+S+ I+ L Q + +K K K KK+ + KA +
Sbjct: 344 AAPRVTKPKDSSSSTVKKPITKSQPKLETQEKSVKAKEKKKKVKKEEAEKRGEEEKATAV 403
Query: 60 FGKMKKTKKQRDN-ELNDGVWQKEILMGG 87
K ++ K +N ++ + E+++GG
Sbjct: 404 AAKPEEQKPNSNNIQVKAEIMASEVVVGG 432
>At2g02950 phytochrome kinase substrate 1 (PKS1)
Length = 439
Score = 26.6 bits (57), Expect = 3.3
Identities = 27/106 (25%), Positives = 42/106 (39%), Gaps = 9/106 (8%)
Query: 1 MANLTSSPESLSPK--------RQSNSSFFITISAILAMLSRQARRLKGKAKTKKLLSNM 52
M LT S S +PK S+SS +++ S+ L+ K + K L+
Sbjct: 1 MVTLTPSSAS-TPKTSFDFMKNNNSHSSLYVSSSSYLSSKEDALVTTKKLMEPSKTLNMS 59
Query: 53 SNKALSQFGKMKKTKKQRDNELNDGVWQKEILMGGKCEPLDFSGVI 98
N +FG KK K+ + GV+ E G + S V+
Sbjct: 60 INPKQEEFGDEKKMVKKAPEDPEIGVFGAEKYFNGDMDSDQGSSVL 105
>At5g35700 fimbrin
Length = 687
Score = 26.2 bits (56), Expect = 4.3
Identities = 14/57 (24%), Positives = 28/57 (48%)
Query: 28 ILAMLSRQARRLKGKAKTKKLLSNMSNKALSQFGKMKKTKKQRDNELNDGVWQKEIL 84
+L +L +GK T + N +N+ + + G+ + RD L+ G++ E+L
Sbjct: 498 MLQLLRNLRSHSQGKEITDADILNWANRKVKRGGRTSQADSFRDKNLSSGMFFLELL 554
>At5g53500 unknown protein
Length = 654
Score = 25.8 bits (55), Expect = 5.6
Identities = 18/62 (29%), Positives = 36/62 (58%), Gaps = 2/62 (3%)
Query: 10 SLSPKRQSNSSFFITISAILAMLSRQARRLKGKAKTKKLLSNMSNKALSQFGKMKKTKKQ 69
S S S S + +S L++ R R + G T++ S+M++ + S++ ++K+T+KQ
Sbjct: 91 SSSSSEVSLSGSSVEVSEELSL--RVDRNVGGCDVTRRQSSSMASCSSSRYCQVKETEKQ 148
Query: 70 RD 71
R+
Sbjct: 149 RN 150
>At5g38680 putative protein
Length = 357
Score = 25.8 bits (55), Expect = 5.6
Identities = 12/32 (37%), Positives = 21/32 (65%)
Query: 6 SSPESLSPKRQSNSSFFITISAILAMLSRQAR 37
SSPE SP +SNS+ + + I++ ++R +R
Sbjct: 2 SSPEKFSPAPESNSNPSLPDALIISCIARVSR 33
>At5g20600 unknown protein
Length = 274
Score = 25.8 bits (55), Expect = 5.6
Identities = 16/51 (31%), Positives = 25/51 (48%), Gaps = 2/51 (3%)
Query: 43 AKTKKLLSNMSNKALSQFGKMKKTKKQRDNELNDGVWQKEILMGGKCEPLD 93
+K+KK+++N S+ K KTKK D+ V + +G K E D
Sbjct: 73 SKSKKIVANGSSSKKKGLKKCSKTKK--DSVKKGAVMDSVVDLGNKDESFD 121
>At1g18190 hypothetical protein
Length = 649
Score = 25.8 bits (55), Expect = 5.6
Identities = 16/58 (27%), Positives = 32/58 (54%)
Query: 25 ISAILAMLSRQARRLKGKAKTKKLLSNMSNKALSQFGKMKKTKKQRDNELNDGVWQKE 82
+ A +++L+ + RL+ KA + M K L + +++ K+R N+L D + QK+
Sbjct: 453 LEAEISLLTDKIGRLQDKATKLEADIEMMRKELEEPTEVEIELKRRLNQLTDHLIQKQ 510
>At1g07480 transcription factor IIA large subunit
Length = 375
Score = 25.8 bits (55), Expect = 5.6
Identities = 15/47 (31%), Positives = 22/47 (45%), Gaps = 3/47 (6%)
Query: 51 NMSNKALSQFGKMKKTKKQRDNELNDGVW---QKEILMGGKCEPLDF 94
N + L+QF K+ +TK + L DG+ K+IL DF
Sbjct: 329 NTQHLVLAQFDKVTRTKSRWKCSLKDGIMHINDKDILFNKAAGEFDF 375
>At5g16630 unknown protein
Length = 865
Score = 25.4 bits (54), Expect = 7.3
Identities = 17/72 (23%), Positives = 33/72 (45%), Gaps = 2/72 (2%)
Query: 50 SNMSNKALSQFGKMKKTK--KQRDNELNDGVWQKEILMGGKCEPLDFSGVIHYDINGRQT 107
+ + +K L + K KK K + DN + G E+ + EPL ++ + +
Sbjct: 612 NEVPSKILKRNSKFKKVKDFEDGDNNIKGGSSCMELYGKWQMEPLCLPPAVNGIVPKNER 671
Query: 108 GEVPLRSPRAIP 119
G+V + S + +P
Sbjct: 672 GQVDVWSEKCLP 683
>At3g19670 unknown protein
Length = 960
Score = 25.4 bits (54), Expect = 7.3
Identities = 10/45 (22%), Positives = 25/45 (55%)
Query: 29 LAMLSRQARRLKGKAKTKKLLSNMSNKALSQFGKMKKTKKQRDNE 73
+++L Q+ R+K K + + +K ++G+ K ++RD++
Sbjct: 813 VSLLKEQSNRIKQNKKVPEDVREEHDKGRDKYGREKDRVRERDSD 857
>At1g07470 putative transcription factor IIA large subunit protein
Length = 375
Score = 25.4 bits (54), Expect = 7.3
Identities = 15/47 (31%), Positives = 22/47 (45%), Gaps = 3/47 (6%)
Query: 51 NMSNKALSQFGKMKKTKKQRDNELNDGVW---QKEILMGGKCEPLDF 94
N + L+QF K+ +TK + L DG+ K+IL DF
Sbjct: 329 NTQHLVLAQFDKVTRTKSRWKCSLKDGIMHINDKDILFNKATGEFDF 375
>At5g65180 unknown protein
Length = 439
Score = 25.0 bits (53), Expect = 9.5
Identities = 12/43 (27%), Positives = 20/43 (45%)
Query: 33 SRQARRLKGKAKTKKLLSNMSNKALSQFGKMKKTKKQRDNELN 75
S+ A+R KTK +S K +S F ++ + E+N
Sbjct: 148 SKSAKRDSKSTKTKLSSGGVSEKIVSAFNLVRAENSNEETEMN 190
>At5g45520 putative protein
Length = 1167
Score = 25.0 bits (53), Expect = 9.5
Identities = 11/40 (27%), Positives = 21/40 (52%)
Query: 49 LSNMSNKALSQFGKMKKTKKQRDNELNDGVWQKEILMGGK 88
+ N + + G +++T KQ D + DG+ ++ L GK
Sbjct: 590 VQNYGQTSKEEKGNVEETGKQEDGDQGDGINEEANLEDGK 629
>At5g42900 unknown protein
Length = 246
Score = 25.0 bits (53), Expect = 9.5
Identities = 9/24 (37%), Positives = 16/24 (66%)
Query: 58 SQFGKMKKTKKQRDNELNDGVWQK 81
++FG +K +++ L+DG WQK
Sbjct: 85 TRFGSGRKPSQEQFKVLHDGFWQK 108
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.130 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,608,082
Number of Sequences: 26719
Number of extensions: 97324
Number of successful extensions: 328
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 313
Number of HSP's gapped (non-prelim): 25
length of query: 123
length of database: 11,318,596
effective HSP length: 87
effective length of query: 36
effective length of database: 8,994,043
effective search space: 323785548
effective search space used: 323785548
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 53 (25.0 bits)
Medicago: description of AC142222.6


