

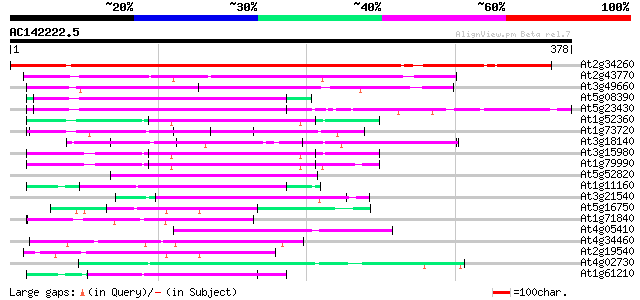
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.5 + phase: 0
(378 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g34260 unknown protein 481 e-136
At2g43770 putative splicing factor 79 3e-15
At3g49660 putative WD-40 repeat - protein 67 2e-11
At5g08390 katanin p80 subunit - like protein 57 1e-08
At5g23430 unknown protein 56 3e-08
At1g52360 coatomer complex subunit, putative 55 8e-08
At1g73720 unknown protein 54 1e-07
At3g18140 unknown protein 54 2e-07
At3g15980 putative coatomer complex subunit 54 2e-07
At1g79990 coatomer protein like complex, subunit beta 2 (beta pr... 53 2e-07
At5g52820 Notchless protein homolog 53 3e-07
At1g11160 hypothetical protein 52 5e-07
At3g21540 WD repeat like protein 52 7e-07
At5g16750 WD40-repeat protein 51 9e-07
At1g71840 unknown protein (At1g71840) 51 9e-07
At4g05410 U3 snoRNP-associated -like protein 50 2e-06
At4g34460 GTP binding protein beta subunit 50 3e-06
At2g19540 putative WD-40 repeat protein 50 3e-06
At4g02730 putative WD-repeat protein 49 3e-06
At1g61210 49 5e-06
>At2g34260 unknown protein
Length = 353
Score = 481 bits (1238), Expect = e-136
Identities = 241/365 (66%), Positives = 291/365 (79%), Gaps = 12/365 (3%)
Query: 1 MEINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAA 60
MEI+LG AF IDFHPS NLVA GLIDG LHLYRY SD S VR ++ AH ESCRA
Sbjct: 1 MEIDLGANAFGIDFHPSTNLVAAGLIDGHLHLYRYDSD---SSLVRERKVRAHKESCRAV 57
Query: 61 RFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCI 120
RFI+ G+ ++T S D SILATDVETG+ +A L+NAHE AVN L+N+TE+T+ASGDD GC+
Sbjct: 58 RFIDDGQRIVTASADCSILATDVETGAQVAHLENAHEDAVNTLINVTETTIASGDDKGCV 117
Query: 121 KVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSE 180
K+WDTR+RSC + F AHEDYIS +TFASD+MKL+ TSGDGTLSVCNLR +KVQ+QSEFSE
Sbjct: 118 KIWDTRQRSCSHEFNAHEDYISGMTFASDSMKLVVTSGDGTLSVCNLRTSKVQSQSEFSE 177
Query: 181 DELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIIT 240
DELLSVV+MKNGRKV+CG+Q G +LLYSWG FKDCSDRFVDL+ NS+D +LKLDEDR+IT
Sbjct: 178 DELLSVVIMKNGRKVICGTQNGTLLLYSWGFFKDCSDRFVDLAPNSVDALLKLDEDRLIT 237
Query: 241 GSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWD 300
G +NG+I+LVGILPNR+IQP+ H +YP+E L A SHD+KFLGS AHD MLKLW+
Sbjct: 238 GCDNGIISLVGILPNRIIQPIGSH-DYPIEDL------ALSHDKKFLGSTAHDSMLKLWN 290
Query: 301 LDNILQGSRNTQRNENGGVDNDVDSDNEDEMDVDNNTSKFTKGNKRKNASKGHAAGDSNN 360
L+ IL+GS N +G + D DSDN D MD+DN+ SK +KG+KRK SK + +NN
Sbjct: 291 LEEILEGSNVNSGNASGAAE-DSDSDN-DGMDLDNDPSKSSKGSKRKTKSKANTLNATNN 348
Query: 361 FFADL 365
FFADL
Sbjct: 349 FFADL 353
>At2g43770 putative splicing factor
Length = 343
Score = 79.3 bits (194), Expect = 3e-15
Identities = 64/301 (21%), Positives = 126/301 (41%), Gaps = 23/301 (7%)
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+ + F+P+ L+A+G D ++ L+R D N + + H + + + G +
Sbjct: 57 YTMKFNPAGTLIASGSHDREIFLWRVHGDCKN-----FMVLKGHKNAILDLHWTSDGSQI 111
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTES--TVASGDDDGCIKVWDTRE 127
++ SPD ++ A DVETG I ++ H + VN + SG DDG K+WD R+
Sbjct: 112 VSASPDKTVRAWDVETGKQIKKMAE-HSSFVNSCCPTRRGPPLIISGSDDGTAKLWDMRQ 170
Query: 128 RSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVV 187
R +F + I+ ++F+ A K+ D + V +LR+ + E +D + +
Sbjct: 171 RGAIQTF-PDKYQITAVSFSDAADKIFTGGVDNDVKVWDLRKGEATMTLEGHQDTITGMS 229
Query: 188 LMKNGRKVVCGSQTGIMLLYSW-------GCFKDCSDRFVDLSSNSIDTMLKLDEDRIIT 240
L +G ++ + ++ C K + N + D ++
Sbjct: 230 LSPDGSYLLTNGMDNKLCVWDMRPYAPQNRCVKIFEGHQHNFEKNLLKCSWSPDGTKVTA 289
Query: 241 GSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWD 300
GS + M+++ R I + H+ G+ F +GS + D+ + L +
Sbjct: 290 GSSDRMVHIWDTTSRRTIYKLPGHT-------GSVNECVFHPTEPIIGSCSSDKNIYLGE 342
Query: 301 L 301
+
Sbjct: 343 I 343
>At3g49660 putative WD-40 repeat - protein
Length = 317
Score = 67.0 bits (162), Expect = 2e-11
Identities = 61/298 (20%), Positives = 122/298 (40%), Gaps = 25/298 (8%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVR--LLEIHAHTESCRAARFINGGRAL 69
+ F L+A+ D + Y NT +DP+ + E H F + R +
Sbjct: 30 VKFSSDGRLLASASADKTIRTYTI---NTINDPIAEPVQEFTGHENGISDVAFSSDARFI 86
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERS 129
++ S D ++ DVETGS I L A N + + SG D +++WD
Sbjct: 87 VSASDDKTLKLWDVETGSLIKTLIGHTNYAFCVNFNPQSNMIVSGSFDETVRIWDVTTGK 146
Query: 130 CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVV-L 188
C AH D ++ + F D ++++S DG + + E+ +S V
Sbjct: 147 CLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRIWDSGTGHCVKTLIDDENPPVSFVRF 206
Query: 189 MKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDE-------DRIITG 241
NG+ ++ G+ + L++ S +F+ + ++ + RI++G
Sbjct: 207 SPNGKFILVGTLDNTLRLWNIS-----SAKFLKTYTGHVNAQYCISSAFSVTNGKRIVSG 261
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLW 299
SE+ +++ + +++Q + H+E + VA + S + D+ +++W
Sbjct: 262 SEDNCVHMWELNSKKLLQKLEGHTETVMN-------VACHPTENLIASGSLDKTVRIW 312
Score = 47.0 bits (110), Expect = 2e-05
Identities = 25/117 (21%), Positives = 57/117 (48%), Gaps = 5/117 (4%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARF-INGGRALL 70
+ F P+ + G +D L L+ SS ++ H + + C ++ F + G+ ++
Sbjct: 204 VRFSPNGKFILVGTLDNTLRLWNISSAKF----LKTYTGHVNAQYCISSAFSVTNGKRIV 259
Query: 71 TGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRE 127
+GS D + ++ + + +L+ E +N + TE+ +ASG D +++W ++
Sbjct: 260 SGSEDNCVHMWELNSKKLLQKLEGHTETVMNVACHPTENLIASGSLDKTVRIWTQKK 316
Score = 40.8 bits (94), Expect = 0.001
Identities = 49/257 (19%), Positives = 99/257 (38%), Gaps = 54/257 (21%)
Query: 52 AHTESCRAARFINGGRALLTGSPDFSILATDVET-GSTIA---RLDNAHEAAVNRLVNLT 107
+H + + +F + GR L + S D +I + T IA + HE ++ + +
Sbjct: 22 SHNRAVSSVKFSSDGRLLASASADKTIRTYTINTINDPIAEPVQEFTGHENGISDVAFSS 81
Query: 108 EST-VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
++ + S DD +K+WD S + H +Y + F + +++ S D T+ + +
Sbjct: 82 DARFIVSASDDKTLKLWDVETGSLIKTLIGHTNYAFCVNFNPQSNMIVSGSFDETVRIWD 141
Query: 167 LRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNS 226
+ K D + +V ++G +V S G+ ++ G + +
Sbjct: 142 VTTGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRIWDSG------------TGHC 189
Query: 227 IDTMLKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKF 286
+ T++ DE+ PV+ FV FS + KF
Sbjct: 190 VKTLID-DEN----------------------PPVS--------------FVRFSPNGKF 212
Query: 287 LGSIAHDQMLKLWDLDN 303
+ D L+LW++ +
Sbjct: 213 ILVGTLDNTLRLWNISS 229
Score = 38.9 bits (89), Expect = 0.005
Identities = 36/195 (18%), Positives = 81/195 (41%), Gaps = 10/195 (5%)
Query: 9 AFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRA 68
AF ++F+P N++ +G D + ++ D T +++L AH++ A F G
Sbjct: 116 AFCVNFNPQSNMIVSGSFDETVRIW----DVTTGKCLKVLP--AHSDPVTAVDFNRDGSL 169
Query: 69 LLTGSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRE 127
+++ S D D TG + L + V+ + + + G D +++W+
Sbjct: 170 IVSSSYDGLCRIWDSGTGHCVKTLIDDENPPVSFVRFSPNGKFILVGTLDNTLRLWNISS 229
Query: 128 RSCCNSFEAH---EDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL 184
++ H + IS ++ ++++ S D + + L K+ + E + ++
Sbjct: 230 AKFLKTYTGHVNAQYCISSAFSVTNGKRIVSGSEDNCVHMWELNSKKLLQKLEGHTETVM 289
Query: 185 SVVLMKNGRKVVCGS 199
+V + GS
Sbjct: 290 NVACHPTENLIASGS 304
>At5g08390 katanin p80 subunit - like protein
Length = 823
Score = 57.4 bits (137), Expect = 1e-08
Identities = 47/193 (24%), Positives = 76/193 (39%), Gaps = 8/193 (4%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ F S+ LVA G G + L+ D + VR L H +C + F G +
Sbjct: 158 VTFDASEGLVAAGAASGTIKLW----DLEEAKVVRTLT--GHRSNCVSVNFHPFGEFFAS 211
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-VASGDDDGCIKVWDTRERSC 130
GS D ++ D+ I H VN L + + SG +D +KVWD
Sbjct: 212 GSLDTNLKIWDIRKKGCIHTY-KGHTRGVNVLRFTPDGRWIVSGGEDNVVKVWDLTAGKL 270
Query: 131 CNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMK 190
+ F++HE I + F L S D T+ +L ++ + +
Sbjct: 271 LHEFKSHEGKIQSLDFHPHEFLLATGSADKTVKFWDLETFELIGSGGTETTGVRCLTFNP 330
Query: 191 NGRKVVCGSQTGI 203
+G+ V+CG Q +
Sbjct: 331 DGKSVLCGLQESL 343
Score = 48.5 bits (114), Expect = 6e-06
Identities = 33/170 (19%), Positives = 72/170 (41%), Gaps = 6/170 (3%)
Query: 17 SDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDF 76
S ++ TG D ++L+ N +L ++ H+ + F + G+
Sbjct: 121 SSRVLVTGGEDHKVNLWAIGKPNA------ILSLYGHSSGIDSVTFDASEGLVAAGAASG 174
Query: 77 SILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFEA 136
+I D+E + L V+ + ASG D +K+WD R++ C ++++
Sbjct: 175 TIKLWDLEEAKVVRTLTGHRSNCVSVNFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYKG 234
Query: 137 HEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSV 186
H ++ + F D +++ D + V +L K+ + + E ++ S+
Sbjct: 235 HTRGVNVLRFTPDGRWIVSGGEDNVVKVWDLTAGKLLHEFKSHEGKIQSL 284
>At5g23430 unknown protein
Length = 837
Score = 56.2 bits (134), Expect = 3e-08
Identities = 83/376 (22%), Positives = 154/376 (40%), Gaps = 31/376 (8%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ F S+ LVA G G + L+ D + VR L H +C + F G +
Sbjct: 65 VTFDASEVLVAAGAASGTIKLW----DLEEAKIVRTLT--GHRSNCISVDFHPFGEFFAS 118
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-VASGDDDGCIKVWDTRERSC 130
GS D ++ D+ I H VN L + V SG +D +KVWD
Sbjct: 119 GSLDTNLKIWDIRKKGCIHTY-KGHTRGVNVLRFTPDGRWVVSGGEDNIVKVWDLTAGKL 177
Query: 131 CNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMK 190
F++HE I + F L S D T+ +L ++ + +
Sbjct: 178 LTEFKSHEGQIQSLDFHPHEFLLATGSADRTVKFWDLETFELIGSGGPETAGVRCLSFNP 237
Query: 191 NGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLV 250
+G+ V+CG Q + ++SW + C D VD+ + + M + E +++ S N V
Sbjct: 238 DGKTVLCGLQESLK-IFSWEPIR-CHDG-VDVGWSRLSDM-NVHEGKLLGCSYNQSCVGV 293
Query: 251 GILPNRVIQP-----VAEHSEYPVERLGN*RFVAFSHD---RKFLGSIAHDQMLKLWDLD 302
++ +P A+ + +P +R + R +D + LG ++ Q ++D
Sbjct: 294 WVVDLSRTEPCMAGDTAQSNGHPEKRSCSGRDPVVLNDNNSKTVLGKLSVSQ-----NVD 348
Query: 303 NILQGSRNTQRNENGGVDNDVDSDNEDEMDVDNNTSKFTKGNKRKNASKGHAAGDSNNFF 362
+L+ +++ R N S E + ++TS+ ++ + +++ G + N+
Sbjct: 349 PLLKETKSLGRLSVS--QNSDPSTKETKSIGRSSTSQNSESSMKESKPLGRLSVSQNSDV 406
Query: 363 ADL*EIMAATFSTTSS 378
+ + TFS+T S
Sbjct: 407 SK----ESRTFSSTGS 418
Score = 47.4 bits (111), Expect = 1e-05
Identities = 32/170 (18%), Positives = 72/170 (41%), Gaps = 6/170 (3%)
Query: 17 SDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDF 76
S ++ TG D ++L+ N +L ++ H+ + F + G+
Sbjct: 28 SSRVLVTGGEDHKVNLWAIGKPNA------ILSLYGHSSGIDSVTFDASEVLVAAGAASG 81
Query: 77 SILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFEA 136
+I D+E + L ++ + ASG D +K+WD R++ C ++++
Sbjct: 82 TIKLWDLEEAKIVRTLTGHRSNCISVDFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYKG 141
Query: 137 HEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSV 186
H ++ + F D +++ D + V +L K+ + + E ++ S+
Sbjct: 142 HTRGVNVLRFTPDGRWVVSGGEDNIVKVWDLTAGKLLTEFKSHEGQIQSL 191
>At1g52360 coatomer complex subunit, putative
Length = 926
Score = 54.7 bits (130), Expect = 8e-08
Identities = 55/243 (22%), Positives = 97/243 (39%), Gaps = 13/243 (5%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+D HP++ + L G L ++ Y + V R+A+F+ + ++
Sbjct: 21 VDLHPTEPWILASLYSGTLCIWNYQTQ------VMAKSFEVTELPVRSAKFVARKQWVVA 74
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRER-S 129
G+ D I + T + ++ AH + + V+ T V S DD IK+WD + +
Sbjct: 75 GADDMYIRVYNYNTMDKV-KVFEAHSDYIRCVAVHPTLPYVLSSSDDMLIKLWDWEKGWA 133
Query: 130 CCNSFEAHEDYISDITF-ASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVL 188
C FE H Y+ +TF D + S D T+ + NL + + + V
Sbjct: 134 CTQIFEGHSHYVMQVTFNPKDTNTFASASLDRTIKIWNLGSPDPNFTLDAHQKGVNCVDY 193
Query: 189 MKNGRK--VVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGM 246
G K ++ GS ++ + K C + N + IITGSE+G
Sbjct: 194 FTGGDKPYLITGSDDHTAKVWDYQT-KSCVQTLEGHTHNVSAVCFHPELPIIITGSEDGT 252
Query: 247 INL 249
+ +
Sbjct: 253 VRI 255
Score = 53.5 bits (127), Expect = 2e-07
Identities = 28/116 (24%), Positives = 59/116 (50%), Gaps = 3/116 (2%)
Query: 94 NAHEAAVNRLVNLT---ESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDA 150
+AH+ VN + T + + +G DD KVWD + +SC + E H +S + F +
Sbjct: 182 DAHQKGVNCVDYFTGGDKPYLITGSDDHTAKVWDYQTKSCVQTLEGHTHNVSAVCFHPEL 241
Query: 151 MKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLL 206
++ S DGT+ + + +++ + + + ++ +K+ R+VV G G +++
Sbjct: 242 PIIITGSEDGTVRIWHATTYRLENTLNYGLERVWAIGYIKSSRRVVIGYDEGTIMV 297
>At1g73720 unknown protein
Length = 511
Score = 53.9 bits (128), Expect = 1e-07
Identities = 35/137 (25%), Positives = 60/137 (43%), Gaps = 9/137 (6%)
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRN 170
+ASG DG IK+W R C F+AH ++ ++F+ D +LL+TS D T + L+
Sbjct: 278 LASGSQDGKIKIWRIRTGVCIRRFDAHSQGVTSLSFSRDGSQLLSTSFDQTARIHGLKSG 337
Query: 171 KVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRF--------VDL 222
K+ + + + +G +++ S + ++ DC F D
Sbjct: 338 KLLKEFRGHTSYVNHAIFTSDGSRIITASSDCTVKVWD-SKTTDCLQTFKPPPPLRGTDA 396
Query: 223 SSNSIDTMLKLDEDRII 239
S NSI K E ++
Sbjct: 397 SVNSIHLFPKNTEHIVV 413
Score = 48.1 bits (113), Expect = 8e-06
Identities = 35/157 (22%), Positives = 58/157 (36%), Gaps = 10/157 (6%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHA------HTESCRAARFINGGR 67
F P +A+ +DG + ++ Y S D L+ A H + F
Sbjct: 221 FSPDGQFLASSSVDGFIEVWDYISGKLKKD----LQYQADESFMMHDDPVLCIDFSRDSE 276
Query: 68 ALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRE 127
L +GS D I + TG I R D + + + S + S D ++ +
Sbjct: 277 MLASGSQDGKIKIWRIRTGVCIRRFDAHSQGVTSLSFSRDGSQLLSTSFDQTARIHGLKS 336
Query: 128 RSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
F H Y++ F SD +++ S D T+ V
Sbjct: 337 GKLLKEFRGHTSYVNHAIFTSDGSRIITASSDCTVKV 373
Score = 44.3 bits (103), Expect = 1e-04
Identities = 29/125 (23%), Positives = 57/125 (45%), Gaps = 8/125 (6%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
IDF ++A+G DG + ++R + V + AH++ + F G LL+
Sbjct: 269 IDFSRDSEMLASGSQDGKIKIWRIRTG------VCIRRFDAHSQGVTSLSFSRDGSQLLS 322
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTE-STVASGDDDGCIKVWDTRERSC 130
S D + +++G + H + VN + ++ S + + D +KVWD++ C
Sbjct: 323 TSFDQTARIHGLKSGKLLKEF-RGHTSYVNHAIFTSDGSRIITASSDCTVKVWDSKTTDC 381
Query: 131 CNSFE 135
+F+
Sbjct: 382 LQTFK 386
Score = 33.5 bits (75), Expect = 0.20
Identities = 23/97 (23%), Positives = 43/97 (43%), Gaps = 8/97 (8%)
Query: 111 VASGDDDGCIKVWD--------TRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTL 162
+AS DG I+VWD + SF H+D + I F+ D+ L + S DG +
Sbjct: 228 LASSSVDGFIEVWDYISGKLKKDLQYQADESFMMHDDPVLCIDFSRDSEMLASGSQDGKI 287
Query: 163 SVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGS 199
+ +R + + + S+ ++G +++ S
Sbjct: 288 KIWRIRTGVCIRRFDAHSQGVTSLSFSRDGSQLLSTS 324
>At3g18140 unknown protein
Length = 305
Score = 53.5 bits (127), Expect = 2e-07
Identities = 46/161 (28%), Positives = 75/161 (46%), Gaps = 9/161 (5%)
Query: 39 NTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEA 98
N+NS P ++ +HT + A F + + +GS D ++ D+ ++ A
Sbjct: 64 NSNS-PQPVMTYDSHTNNVMAVGFQCDAKWMYSGSEDGTVKIWDLRAPGCQKEYESV--A 120
Query: 99 AVNRLV-NLTESTVASGDDDGCIKVWDTRERSC-CNSFEAHEDYISDITFASDAMKLLAT 156
AVN +V + ++ + SGD +G I+VWD R SC C + + +T D ++A
Sbjct: 121 AVNTVVLHPNQTELISGDQNGNIRVWDLRANSCSCELVPEVDTAVRSLTVMWDGTMVVAA 180
Query: 157 SGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVC 197
+ GT V L R K Q +EF E L + NG + C
Sbjct: 181 NNRGTCYVWRLLRGK-QTMTEF---EPLHKLQAHNGHILKC 217
Score = 47.4 bits (111), Expect = 1e-05
Identities = 50/237 (21%), Positives = 102/237 (42%), Gaps = 6/237 (2%)
Query: 69 LLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRER 128
L T S D +I + ETG + ++ VNRL + + + I+++D
Sbjct: 8 LATASYDHTIRFWEAETGRCYRTIQYP-DSHVNRLEITPDKHYLAAACNPHIRLFDVNSN 66
Query: 129 SC--CNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSV 186
S ++++H + + + F DA + + S DGT+ + +LR Q + E S + +V
Sbjct: 67 SPQPVMTYDSHTNNVMAVGFQCDAKWMYSGSEDGTVKIWDLRAPGCQKEYE-SVAAVNTV 125
Query: 187 VLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKL-DEDRIITGSENG 245
VL N +++ G Q G + ++ CS V ++ ++ + D ++ + G
Sbjct: 126 VLHPNQTELISGDQNGNIRVWDLRA-NSCSCELVPEVDTAVRSLTVMWDGTMVVAANNRG 184
Query: 246 MINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLD 302
+ +L + E G+ S K+L + + D+ +K+W++D
Sbjct: 185 TCYVWRLLRGKQTMTEFEPLHKLQAHNGHILKCLLSPANKYLATASSDKTVKIWNVD 241
Score = 43.1 bits (100), Expect = 2e-04
Identities = 38/196 (19%), Positives = 81/196 (40%), Gaps = 15/196 (7%)
Query: 113 SGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKV 172
SG +DG +K+WD R C +E+ ++ + + +L++ +G + V +LR N
Sbjct: 95 SGSEDGTVKIWDLRAPGCQKEYES-VAAVNTVVLHPNQTELISGDQNGNIRVWDLRANSC 153
Query: 173 QAQSEFSEDELL-SVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDL------SSN 225
+ D + S+ +M +G VV + G ++ K F L + +
Sbjct: 154 SCELVPEVDTAVRSLTVMWDGTMVVAANNRGTCYVWRLLRGKQTMTEFEPLHKLQAHNGH 213
Query: 226 SIDTMLKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRK 285
+ +L + T S + + + + ++ + + H + + FS D +
Sbjct: 214 ILKCLLSPANKYLATASSDKTVKIWNVDGFKLEKVLTGHQRWVWD-------CVFSVDGE 266
Query: 286 FLGSIAHDQMLKLWDL 301
FL + + D +LW +
Sbjct: 267 FLVTASSDMTARLWSM 282
>At3g15980 putative coatomer complex subunit
Length = 921
Score = 53.5 bits (127), Expect = 2e-07
Identities = 57/244 (23%), Positives = 99/244 (40%), Gaps = 15/244 (6%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDN-TNSDPVRLLEIHAHTESCRAARFINGGRALL 70
+D HP++ + L G + ++ Y + T S V L + R+A+FI + ++
Sbjct: 24 VDLHPTEPWILASLYSGTVCIWNYQTQTITKSFEVTELPV-------RSAKFIPRKQWVV 76
Query: 71 TGSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRER- 128
G+ D I + T + ++ AH + + V+ T V S DD IK+WD
Sbjct: 77 AGADDMYIRVYNYNTMDKV-KVFEAHSDYIRCVAVHPTLPYVLSSSDDMLIKLWDWENGW 135
Query: 129 SCCNSFEAHEDYISDITF-ASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVV 187
+C FE H Y+ + F D + S D T+ + NL + + + V
Sbjct: 136 ACTQIFEGHSHYVMQVVFNPKDTNTFASASLDRTIKIWNLGSPDPNFTLDAHQKGVNCVD 195
Query: 188 LMKNGRK--VVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENG 245
G K ++ GS ++ + K C + N + IITGSE+G
Sbjct: 196 YFTGGDKPYLITGSDDHTAKVWDYQT-KSCVQTLDGHTHNVSAVCFHPELPIIITGSEDG 254
Query: 246 MINL 249
+ +
Sbjct: 255 TVRI 258
Score = 52.4 bits (124), Expect = 4e-07
Identities = 27/116 (23%), Positives = 59/116 (50%), Gaps = 3/116 (2%)
Query: 94 NAHEAAVNRLVNLT---ESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDA 150
+AH+ VN + T + + +G DD KVWD + +SC + + H +S + F +
Sbjct: 185 DAHQKGVNCVDYFTGGDKPYLITGSDDHTAKVWDYQTKSCVQTLDGHTHNVSAVCFHPEL 244
Query: 151 MKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLL 206
++ S DGT+ + + +++ + + + ++ +K+ R+VV G G +++
Sbjct: 245 PIIITGSEDGTVRIWHATTYRLENTLNYGLERVWAIGYIKSSRRVVIGYDEGTIMV 300
>At1g79990 coatomer protein like complex, subunit beta 2 (beta
prime)
Length = 920
Score = 53.1 bits (126), Expect = 2e-07
Identities = 60/247 (24%), Positives = 102/247 (41%), Gaps = 21/247 (8%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDN-TNSDPVRLLEIHAHTESCRAARFINGGRALL 70
+D HP++ + L G L ++ Y + S V L + R+A+FI + ++
Sbjct: 21 VDLHPTEPWILASLYSGTLCIWNYQTQTMVKSFDVTELPV-------RSAKFIARKQWVV 73
Query: 71 TGSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRERS 129
G+ D I + T I ++ AH + + V+ T V S DD IK+WD +
Sbjct: 74 AGADDMFIRVYNYNTMDKI-KVFEAHADYIRCVAVHPTLPYVLSSSDDMLIKLWDWEKGW 132
Query: 130 CCNS-FEAHEDYISDITF-ASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVV 187
C FE H Y+ +TF D + S D T+ + NL + + V
Sbjct: 133 LCTQIFEGHSHYVMQVTFNPKDTNTFASASLDRTIKIWNLGSPDPNFTLDAHLKGVNCVD 192
Query: 188 LMKNGRK--VVCGSQTGIMLLYSW---GCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGS 242
G K ++ GS ++ + C + ++S+ S L + IITGS
Sbjct: 193 YFTGGDKPYLITGSDDHTAKVWDYQTKSCVQTLEGHTHNVSAVSFHPELPI----IITGS 248
Query: 243 ENGMINL 249
E+G + +
Sbjct: 249 EDGTVRI 255
Score = 50.8 bits (120), Expect = 1e-06
Identities = 28/116 (24%), Positives = 58/116 (49%), Gaps = 3/116 (2%)
Query: 94 NAHEAAVNRLVNLT---ESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDA 150
+AH VN + T + + +G DD KVWD + +SC + E H +S ++F +
Sbjct: 182 DAHLKGVNCVDYFTGGDKPYLITGSDDHTAKVWDYQTKSCVQTLEGHTHNVSAVSFHPEL 241
Query: 151 MKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLL 206
++ S DGT+ + + +++ + + + ++ +K R+VV G G +++
Sbjct: 242 PIIITGSEDGTVRIWHATTYRLENTLNYGLERVWAIGHIKGSRRVVIGYDEGSIMV 297
>At5g52820 Notchless protein homolog
Length = 473
Score = 52.8 bits (125), Expect = 3e-07
Identities = 32/140 (22%), Positives = 65/140 (45%), Gaps = 1/140 (0%)
Query: 69 LLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-VASGDDDGCIKVWDTRE 127
L++GS DF++ + + H+ VN + + +AS D +++W+
Sbjct: 332 LVSGSDDFTMFLWEPSVSKQPKKRLTGHQQLVNHVYFSPDGKWIASASFDKSVRLWNGIT 391
Query: 128 RSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVV 187
F H + +++++D+ LL+ S D TL + +R K++ DE+ +V
Sbjct: 392 GQFVTVFRGHVGPVYQVSWSADSRLLLSGSKDSTLKIWEIRTKKLKQDLPGHADEVFAVD 451
Query: 188 LMKNGRKVVCGSQTGIMLLY 207
+G KVV G + ++ L+
Sbjct: 452 WSPDGEKVVSGGKDRVLKLW 471
Score = 33.5 bits (75), Expect = 0.20
Identities = 32/165 (19%), Positives = 74/165 (44%), Gaps = 12/165 (7%)
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRN 170
+ASG D +++WD + + + H++++ + ++ D L++ S G + N ++
Sbjct: 124 LASGSGDTTVRLWDLYTETPLFTCKGHKNWVLTVAWSPDGKHLVSGSKSGEICCWNPKKG 183
Query: 171 KVQAQSEFSEDELLS------VVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSS 224
+++ + ++ V L R+ V S+ G ++ K + LS
Sbjct: 184 ELEGSPLTGHKKWITGISWEPVHLSSPCRRFVTSSKDGDARIWDITLKKS----IICLSG 239
Query: 225 NSID-TMLKLDEDRII-TGSENGMINLVGILPNRVIQPVAEHSEY 267
+++ T +K D II TGS++ I + ++I+ + H +
Sbjct: 240 HTLAVTCVKWGGDGIIYTGSQDCTIKMWETTQGKLIRELKGHGHW 284
Score = 28.5 bits (62), Expect = 6.3
Identities = 18/47 (38%), Positives = 25/47 (52%), Gaps = 7/47 (14%)
Query: 255 NRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
NR Q +A H+E + V+FS D K L S + D ++LWDL
Sbjct: 99 NRCSQTIAGHAEAVL-------CVSFSPDGKQLASGSGDTTVRLWDL 138
>At1g11160 hypothetical protein
Length = 974
Score = 52.0 bits (123), Expect = 5e-07
Identities = 47/199 (23%), Positives = 77/199 (38%), Gaps = 9/199 (4%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ F+ + LV G G + L+ D S VR H +C A F G L +
Sbjct: 13 VAFNSEEVLVLAGASSGVIKLW----DLEESKMVRAFT--GHRSNCSAVEFHPFGEFLAS 66
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-VASGDDDGCIKVWDTRERSC 130
GS D ++ D I H ++ + + V SG D +KVWD
Sbjct: 67 GSSDTNLRVWDTRKKGCIQTY-KGHTRGISTIEFSPDGRWVVSGGLDNVVKVWDLTAGKL 125
Query: 131 CNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMK 190
+ F+ HE I + F L S D T+ +L ++ + + ++
Sbjct: 126 LHEFKCHEGPIRSLDFHPLEFLLATGSADRTVKFWDLETFELIGTTRPEATGVRAIAFHP 185
Query: 191 NGRKVVCGSQTGIMLLYSW 209
+G+ + CG G+ +YSW
Sbjct: 186 DGQTLFCGLDDGLK-VYSW 203
Score = 45.1 bits (105), Expect = 7e-05
Identities = 33/140 (23%), Positives = 64/140 (45%), Gaps = 2/140 (1%)
Query: 48 LEIHAHTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNL 106
+ + HT + F + +L G+ I D+E S + R H + + + +
Sbjct: 1 MSLCGHTSPVDSVAFNSEEVLVLAGASSGVIKLWDLEE-SKMVRAFTGHRSNCSAVEFHP 59
Query: 107 TESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
+ASG D ++VWDTR++ C +++ H IS I F+ D +++ D + V +
Sbjct: 60 FGEFLASGSSDTNLRVWDTRKKGCIQTYKGHTRGISTIEFSPDGRWVVSGGLDNVVKVWD 119
Query: 167 LRRNKVQAQSEFSEDELLSV 186
L K+ + + E + S+
Sbjct: 120 LTAGKLLHEFKCHEGPIRSL 139
>At3g21540 WD repeat like protein
Length = 949
Score = 51.6 bits (122), Expect = 7e-07
Identities = 45/157 (28%), Positives = 63/157 (39%), Gaps = 2/157 (1%)
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRERSC 130
G D SI D E G+ N+H+ AV L N S +ASG D I +WD S
Sbjct: 82 GYADGSIRIWDTEKGTCEVNF-NSHKGAVTALRYNKVGSMLASGSKDNDIILWDVVGESG 140
Query: 131 CNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMK 190
H D ++D+ F KL+++S D L V +L E+ SV
Sbjct: 141 LFRLRGHRDQVTDLVFLDGGKKLVSSSKDKFLRVWDLETQHCMQIVSGHHSEVWSVDTDP 200
Query: 191 NGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSI 227
R VV GS + Y+ + D ++N I
Sbjct: 201 EERYVVTGSADQELRFYAVKEYSSNGSLVSDSNANEI 237
Score = 45.8 bits (107), Expect = 4e-05
Identities = 32/147 (21%), Positives = 68/147 (45%), Gaps = 7/147 (4%)
Query: 99 AVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSG 158
AV + + S VA G DG I++WDT + +C +F +H+ ++ + + L + S
Sbjct: 67 AVTSIASSASSLVAVGYADGSIRIWDTEKGTCEVNFNSHKGAVTALRYNKVGSMLASGSK 126
Query: 159 DGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLY---SWGCFKDC 215
D + + ++ + D++ +V + G+K+V S+ + ++ + C +
Sbjct: 127 DNDIILWDVVGESGLFRLRGHRDQVTDLVFLDGGKKLVSSSKDKFLRVWDLETQHCMQIV 186
Query: 216 SDRFVDLSSNSIDTMLKLDEDRIITGS 242
S ++ S D +E ++TGS
Sbjct: 187 SGHHSEVWSVDTDP----EERYVVTGS 209
Score = 38.1 bits (87), Expect = 0.008
Identities = 51/294 (17%), Positives = 111/294 (37%), Gaps = 21/294 (7%)
Query: 16 PSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPD 75
P ++L L + L YS ++ + +E H R+ ++ LL +
Sbjct: 368 PKESLGTLALSLNNNSLEFYSLKSSENAKTVTIEHQGHRSDVRSVT-LSEDNTLLMSTSH 426
Query: 76 FSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFE 135
+ + TGS + +D+ + + L+ G G +++ D + +
Sbjct: 427 SEVKIWNPSTGSCLRTIDSGY--GLCSLIVPQNKYGIVGTKSGVLEIIDIGSATKVEEVK 484
Query: 136 AHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSE----DELLSVVLMKN 191
AH I IT + + S D + + K S D++L+V + +
Sbjct: 485 AHGGTIWSITPIPNDSGFVTVSADHEVKFWEYQATKKLTVSNVKSMKMNDDVLAVAISPD 544
Query: 192 GRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTM---LKLDEDRIITGSENGMIN 248
+ + + ++ + D ++ L + + M + D + I+TGS++ +
Sbjct: 545 AKHIAVALLDSTVKVF----YMDSLKFYLSLYGHKLPVMCIDISSDGELIVTGSQDKNLK 600
Query: 249 LVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLD 302
+ G+ + + H + + V F + +L SI D+++K WD D
Sbjct: 601 IWGLDFGDCHKSIFAHGDSVMG-------VKFVRNTHYLFSIGKDRLVKYWDAD 647
Score = 31.6 bits (70), Expect = 0.75
Identities = 23/111 (20%), Positives = 49/111 (43%), Gaps = 6/111 (5%)
Query: 19 NLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDFSI 78
+++A+G D D+ L+ ++ L + H + F++GG+ L++ S D +
Sbjct: 119 SMLASGSKDNDIILWDVVGESG------LFRLRGHRDQVTDLVFLDGGKKLVSSSKDKFL 172
Query: 79 LATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERS 129
D+ET + + H + + E V +G D ++ + +E S
Sbjct: 173 RVWDLETQHCMQIVSGHHSEVWSVDTDPEERYVVTGSADQELRFYAVKEYS 223
Score = 28.1 bits (61), Expect = 8.3
Identities = 18/89 (20%), Positives = 34/89 (37%)
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRN 170
+ +G D +K+W C S AH D + + F + L + D + + +
Sbjct: 590 IVTGSQDKNLKIWGLDFGDCHKSIFAHGDSVMGVKFVRNTHYLFSIGKDRLVKYWDADKF 649
Query: 171 KVQAQSEFSEDELLSVVLMKNGRKVVCGS 199
+ E E+ + + G +V GS
Sbjct: 650 EHLLTLEGHHAEIWCLAISNRGDFLVTGS 678
>At5g16750 WD40-repeat protein
Length = 876
Score = 51.2 bits (121), Expect = 9e-07
Identities = 43/222 (19%), Positives = 85/222 (37%), Gaps = 18/222 (8%)
Query: 28 GDLHLYRYSSDNTNSD---PVRLLE---IHAHTESCRAARFINGGRALLTGSPDFSILAT 81
GD L +S D + D P+ L + AH + + + TGS D +
Sbjct: 467 GDRTLKVWSLDGISEDSEEPINLKTRSVVAAHDKDINSVAVARNDSLVCTGSEDRTASIW 526
Query: 82 DVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYI 141
+ + L + + + V + D +K+W + SC +FE H +
Sbjct: 527 RLPDLVHVVTLKGHKRRIFSVEFSTVDQCVMTASGDKTVKIWAISDGSCLKTFEGHTSSV 586
Query: 142 SDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQT 201
+F +D + ++ DG L + N+ ++ A + ED++ ++ + K + G
Sbjct: 587 LRASFITDGTQFVSCGADGLLKLWNVNTSECIATYDQHEDKVWALAVGKKTEMIATGGGD 646
Query: 202 GIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSE 243
++ L+ D D F + +E+ I+ G E
Sbjct: 647 AVINLWHDSTASDKEDDF------------RKEEEAILRGQE 676
Score = 42.0 bits (97), Expect = 6e-04
Identities = 26/109 (23%), Positives = 48/109 (43%), Gaps = 8/109 (7%)
Query: 66 GRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLV---NLTESTVASGDDDGCIKV 122
G L T D +L DV+ G H+ V+ ++ + ++ + SG DD ++V
Sbjct: 114 GGLLATAGADRKVLVWDVD-GGFCTHYFRGHKGVVSSILFHPDSNKNILISGSDDATVRV 172
Query: 123 WDTR----ERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNL 167
WD E+ C E H ++ I + D + L + D +++ +L
Sbjct: 173 WDLNAKNTEKKCLAIMEKHFSAVTSIALSEDGLTLFSAGRDKVVNLWDL 221
Score = 37.0 bits (84), Expect = 0.018
Identities = 41/177 (23%), Positives = 73/177 (41%), Gaps = 10/177 (5%)
Query: 38 DNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHE 97
D+T+S +E +++ A + L + I D+ET I R HE
Sbjct: 46 DSTDSSVKSTIE--GESDTLTALALSPDDKLLFSAGHSRQIRVWDLETLKCI-RSWKGHE 102
Query: 98 AAVNRLV-NLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMK--LL 154
V + + + +A+ D + VWD C + F H+ +S I F D+ K L+
Sbjct: 103 GPVMGMACHASGGLLATAGADRKVLVWDVDGGFCTHYFRGHKGVVSSILFHPDSNKNILI 162
Query: 155 ATSGDGTLSVCNLR----RNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLY 207
+ S D T+ V +L K A E + S+ L ++G + + ++ L+
Sbjct: 163 SGSDDATVRVWDLNAKNTEKKCLAIMEKHFSAVTSIALSEDGLTLFSAGRDKVVNLW 219
Score = 33.1 bits (74), Expect = 0.26
Identities = 23/101 (22%), Positives = 42/101 (40%), Gaps = 11/101 (10%)
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLAT-SGDGTLSVCNLR- 168
+ +G D +++W+ +SC H I + FA + + SGD TL V +L
Sbjct: 419 IVTGSKDKTVRLWNATSKSCIGVGTGHNGDILAVAFAKKSFSFFVSGSGDRTLKVWSLDG 478
Query: 169 ---------RNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQ 200
K ++ + ++ SV + +N V GS+
Sbjct: 479 ISEDSEEPINLKTRSVVAAHDKDINSVAVARNDSLVCTGSE 519
>At1g71840 unknown protein (At1g71840)
Length = 407
Score = 51.2 bits (121), Expect = 9e-07
Identities = 34/157 (21%), Positives = 73/157 (45%), Gaps = 12/157 (7%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL-- 69
+D + + +L +G DG +H+ + S +++HT+S +F +
Sbjct: 248 LDINSNSSLAISGSKDGSVHIVNIVTGKVVSS------LNSHTDSVECVKFSPSSATIPL 301
Query: 70 -LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNL-TESTVASGDDDGCIKVWDTRE 127
TG D ++ D++ + R HE V L + T +A+G +G + +WD+
Sbjct: 302 AATGGMDKKLIIWDLQHSTP--RFICEHEEGVTSLTWIGTSKYLATGCANGTVSIWDSLL 359
Query: 128 RSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
+C +++ H+D + I+ +++ +++ S D T V
Sbjct: 360 GNCVHTYHGHQDAVQAISVSTNTDFIVSVSVDNTARV 396
Score = 43.5 bits (101), Expect = 2e-04
Identities = 36/157 (22%), Positives = 70/157 (43%), Gaps = 11/157 (7%)
Query: 13 DFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHA-HTESCRAARFINGGRALLT 71
DF P L+ TG D L ++ + + + +++ H HTE + ++
Sbjct: 204 DFTPDGKLICTGSDDASLIVWNPKT----CESIHIVKGHPYHTEGLTCLDINSNSSLAIS 259
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRL----VNLTESTVASGDDDGCIKVWDTRE 127
GS D S+ ++ TG ++ L N+H +V + + T A+G D + +WD +
Sbjct: 260 GSKDGSVHIVNIVTGKVVSSL-NSHTDSVECVKFSPSSATIPLAATGGMDKKLIIWDL-Q 317
Query: 128 RSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
S HE+ ++ +T+ + L +GT+S+
Sbjct: 318 HSTPRFICEHEEGVTSLTWIGTSKYLATGCANGTVSI 354
Score = 38.5 bits (88), Expect = 0.006
Identities = 59/289 (20%), Positives = 110/289 (37%), Gaps = 51/289 (17%)
Query: 16 PSD-NLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSP 74
P+D LVATG D L++ + + + E+ H +S F G+ L +G
Sbjct: 80 PTDATLVATGGGDDKAFLWKIGNGDWAA------ELPGHKDSVSCLAFSYDGQLLASGGL 133
Query: 75 DFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSF 134
D + D +G+ LD + V +G +D + +W+ + + N F
Sbjct: 134 DGVVQIFDASSGTLKCVLDGPGAGIEWVRWHPRGHIVLAGSEDCSLWMWNADKEAYLNMF 193
Query: 135 EAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRK 194
H ++ F D + S D +L V N + + S+ ++K
Sbjct: 194 SGHNLNVTCGDFTPDGKLICTGSDDASLIVWNPKTCE-------------SIHIVKGHPY 240
Query: 195 VVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLVGILP 254
G +D++SNS I+GS++G +++V I+
Sbjct: 241 HTEGLTC------------------LDINSNS---------SLAISGSKDGSVHIVNIVT 273
Query: 255 NRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDN 303
+V+ + H++ VE + +F S + D+ L +WDL +
Sbjct: 274 GKVVSSLNSHTD-SVECV---KFSPSSATIPLAATGGMDKKLIIWDLQH 318
>At4g05410 U3 snoRNP-associated -like protein
Length = 504
Score = 50.1 bits (118), Expect = 2e-06
Identities = 29/148 (19%), Positives = 70/148 (46%), Gaps = 4/148 (2%)
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRN 170
+A+G D + +WD R R +F H + +S + F +L + S D T+ V N+
Sbjct: 237 LATGGVDRHVHIWDVRTREHVQAFPGHRNTVSCLCFRYGTSELYSGSFDRTVKVWNVEDK 296
Query: 171 KVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTM 230
++ + E+L++ ++ R + G ++ + + + ++S+++
Sbjct: 297 AFITENHGHQGEILAIDALRKERALTVGRDRTML----YHKVPESTRMIYRAPASSLESC 352
Query: 231 LKLDEDRIITGSENGMINLVGILPNRVI 258
+ ++ ++GS+NG + L G+L + +
Sbjct: 353 CFISDNEYLSGSDNGTVALWGMLKKKPV 380
Score = 32.0 bits (71), Expect = 0.57
Identities = 36/170 (21%), Positives = 56/170 (32%), Gaps = 23/170 (13%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVA 112
H+ A + GR L TG D + DV T + S +
Sbjct: 221 HSRESLALAVSSDGRYLATGGVDRHVHIWDVRTREHVQAFPGHRNTVSCLCFRYGTSELY 280
Query: 113 SGDDDGCIKVWDTRERSCCNSFEAHEDYISDI---------TFASDAMKLLATSGDGTLS 163
SG D +KVW+ +++ H+ I I T D L + T
Sbjct: 281 SGSFDRTVKVWNVEDKAFITENHGHQGEILAIDALRKERALTVGRDRTMLYHKVPESTRM 340
Query: 164 VCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFK 213
+ + +++ S++E LS GS G + L WG K
Sbjct: 341 IYRAPASSLESCCFISDNEYLS------------GSDNGTVAL--WGMLK 376
>At4g34460 GTP binding protein beta subunit
Length = 377
Score = 49.7 bits (117), Expect = 3e-06
Identities = 54/201 (26%), Positives = 89/201 (43%), Gaps = 19/201 (9%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSS--DNTNSDPV-RLLEIHAHTESCRAARFI-NGGRAL 69
F P+ VA G +D ++ SS D + PV R+L H SC +++ N L
Sbjct: 115 FSPNGQSVACGGLDSVCSIFSLSSTADKDGTVPVSRMLTGHRGYVSC--CQYVPNEDAHL 172
Query: 70 LTGSPDFSILATDVETGSTIA----RLDNAHEAAVNRLVNLTEST---VASGDDDGCIKV 122
+T S D + + DV TG + + H A V V+++ S SG D ++
Sbjct: 173 ITSSGDQTCILWDVTTGLKTSVFGGEFQSGHTADVLS-VSISGSNPNWFISGSCDSTARL 231
Query: 123 WDTRERS-CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRR-NKVQAQSEFSE 180
WDTR S +F HE ++ + F D + S DGT + ++R +++Q +
Sbjct: 232 WDTRAASRAVRTFHGHEGDVNTVKFFPDGYRFGTGSDDGTCRLYDIRTGHQLQVYQPHGD 291
Query: 181 DE---LLSVVLMKNGRKVVCG 198
E + S+ +GR + G
Sbjct: 292 GENGPVTSIAFSVSGRLLFAG 312
Score = 33.5 bits (75), Expect = 0.20
Identities = 15/79 (18%), Positives = 35/79 (43%)
Query: 130 CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLM 189
CC + + H + + + + ++++ S DG L V N ++ + +++
Sbjct: 57 CCRTLQGHTGKVYSLDWTPERNRIVSASQDGRLIVWNALTSQKTHAIKLPCAWVMTCAFS 116
Query: 190 KNGRKVVCGSQTGIMLLYS 208
NG+ V CG + ++S
Sbjct: 117 PNGQSVACGGLDSVCSIFS 135
>At2g19540 putative WD-40 repeat protein
Length = 469
Score = 49.7 bits (117), Expect = 3e-06
Identities = 52/183 (28%), Positives = 82/183 (44%), Gaps = 19/183 (10%)
Query: 10 FDIDFHPSDNLVATGLIDGD----LHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFING 65
+ ID+ P+ A L+ GD +HL+ +S + DP+ HT S ++
Sbjct: 227 YAIDWSPA---TAGRLLSGDCKSMIHLWEPASGSWAVDPIPFA---GHTASVEDLQWSPA 280
Query: 66 GRALLTG-SPDFSILATDVETGSTIARLDNAHEAAVNRLV--NLTESTVASGDDDGCIKV 122
+ S D S+ D+ G + A AH A VN + L +ASG DDG +
Sbjct: 281 EENVFASCSVDGSVAVWDIRLGKSPALSFKAHNADVNVISWNRLASCMLASGSDDGTFSI 340
Query: 123 WDTR----ERSCCNSFEAHEDYISDITF-ASDAMKLLATSGDGTLSVCNLRRNK-VQAQS 176
D R + FE H+ I+ I + A +A L TSGD L++ +L K + ++
Sbjct: 341 RDLRLIKGGDAVVAHFEYHKHPITSIEWSAHEASTLAVTSGDNQLTIWDLSLEKDEEEEA 400
Query: 177 EFS 179
EF+
Sbjct: 401 EFN 403
>At4g02730 putative WD-repeat protein
Length = 333
Score = 49.3 bits (116), Expect = 3e-06
Identities = 60/271 (22%), Positives = 108/271 (39%), Gaps = 23/271 (8%)
Query: 47 LLEIHAHTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNL 106
L + HT + +F N G L + S D +++ S I R + H + ++ L
Sbjct: 36 LKTLEGHTAAISCVKFSNDGNLLASASVDKTMILWSATNYSLIHRYE-GHSSGISDLAWS 94
Query: 107 TES-TVASGDDDGCIKVWDTRE-RSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
++S S DD +++WD R C H +++ + F + +++ S D T+ +
Sbjct: 95 SDSHYTCSASDDCTLRIWDARSPYECLKVLRGHTNFVFCVNFNPPSNLIVSGSFDETIRI 154
Query: 165 CNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFK-DCSDRFVDLS 223
++ K + + SV ++G +V S G + W + C +D
Sbjct: 155 WEVKTGKCVRMIKAHSMPISSVHFNRDGSLIVSASHDGSCKI--WDAKEGTCLKTLIDDK 212
Query: 224 SNSIDTMLKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFV----- 278
S ++ S NG LV L + + ++ G+ V
Sbjct: 213 SPAVS---------FAKFSPNGKFILVATLDSTLKLSNYATGKFLKVYTGHTNKVFCITS 263
Query: 279 AFS-HDRKFLGSIAHDQMLKLWDLD--NILQ 306
AFS + K++ S + D + LWDL NILQ
Sbjct: 264 AFSVTNGKYIVSGSEDNCVYLWDLQARNILQ 294
Score = 40.4 bits (93), Expect = 0.002
Identities = 47/261 (18%), Positives = 100/261 (38%), Gaps = 14/261 (5%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ F NL+A+ +D + L+ TN + E H+ + + +
Sbjct: 49 VKFSNDGNLLASASVDKTMILW----SATNYSLIHRYE--GHSSGISDLAWSSDSHYTCS 102
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRERSC 130
S D ++ D + ++ H V + N + + SG D I++W+ + C
Sbjct: 103 ASDDCTLRIWDARSPYECLKVLRGHTNFVFCVNFNPPSNLIVSGSFDETIRIWEVKTGKC 162
Query: 131 CNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMK 190
+AH IS + F D +++ S DG+ + + + +D+ +V K
Sbjct: 163 VRMIKAHSMPISSVHFNRDGSLIVSASHDGSCKIWDAKEG--TCLKTLIDDKSPAVSFAK 220
Query: 191 ---NGRKVVCGSQTGIMLL--YSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENG 245
NG+ ++ + + L Y+ G F + + I++GSE+
Sbjct: 221 FSPNGKFILVATLDSTLKLSNYATGKFLKVYTGHTNKVFCITSAFSVTNGKYIVSGSEDN 280
Query: 246 MINLVGILPNRVIQPVAEHSE 266
+ L + ++Q + H++
Sbjct: 281 CVYLWDLQARNILQRLEGHTD 301
Score = 39.7 bits (91), Expect = 0.003
Identities = 33/181 (18%), Positives = 69/181 (37%), Gaps = 50/181 (27%)
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIH------------------ 51
F ++F+P NL+ +G D + ++ + VR+++ H
Sbjct: 132 FCVNFNPPSNLIVSGSFDETIRIWEVKTGKC----VRMIKAHSMPISSVHFNRDGSLIVS 187
Query: 52 -AHTESCRA----------------------ARFINGGRALLTGSPDFSILATDVETGST 88
+H SC+ A+F G+ +L + D ++ ++ TG
Sbjct: 188 ASHDGSCKIWDAKEGTCLKTLIDDKSPAVSFAKFSPNGKFILVATLDSTLKLSNYATGKF 247
Query: 89 IARLDNAHEAAVNRLVNLTEST----VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDI 144
+ ++ H V + + T + SG +D C+ +WD + R+ E H D + +
Sbjct: 248 L-KVYTGHTNKVFCITSAFSVTNGKYIVSGSEDNCVYLWDLQARNILQRLEGHTDAVISV 306
Query: 145 T 145
+
Sbjct: 307 S 307
Score = 34.7 bits (78), Expect = 0.088
Identities = 21/113 (18%), Positives = 53/113 (46%), Gaps = 7/113 (6%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARF-INGGRALLTG 72
F P+ + +D L L Y++ +++ H + C + F + G+ +++G
Sbjct: 221 FSPNGKFILVATLDSTLKLSNYATGKF----LKVYTGHTNKVFCITSAFSVTNGKYIVSG 276
Query: 73 SPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDD--DGCIKVW 123
S D + D++ + + RL+ +A ++ + ++ ++S + D I++W
Sbjct: 277 SEDNCVYLWDLQARNILQRLEGHTDAVISVSCHPVQNEISSSGNHLDKTIRIW 329
>At1g61210
Length = 282
Score = 48.9 bits (115), Expect = 5e-06
Identities = 33/135 (24%), Positives = 61/135 (44%), Gaps = 2/135 (1%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTV 111
HT + + F + +L G+ I DVE + R H + + + + +
Sbjct: 87 HTSAVDSVAFDSAEVLVLAGASSGVIKLWDVEEAKMV-RAFTGHRSNCSAVEFHPFGEFL 145
Query: 112 ASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNK 171
ASG D +K+WD R++ C +++ H IS I F D +++ D + V +L K
Sbjct: 146 ASGSSDANLKIWDIRKKGCIQTYKGHSRGISTIRFTPDGRWVVSGGLDNVVKVWDLTAGK 205
Query: 172 VQAQSEFSEDELLSV 186
+ + +F E + S+
Sbjct: 206 LLHEFKFHEGPIRSL 220
Score = 41.6 bits (96), Expect = 7e-04
Identities = 39/157 (24%), Positives = 60/157 (37%), Gaps = 8/157 (5%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ F ++ LV G G + L+ D + VR H +C A F G L +
Sbjct: 94 VAFDSAEVLVLAGASSGVIKLW----DVEEAKMVRAFT--GHRSNCSAVEFHPFGEFLAS 147
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-VASGDDDGCIKVWDTRERSC 130
GS D ++ D+ I H ++ + + V SG D +KVWD
Sbjct: 148 GSSDANLKIWDIRKKGCIQTY-KGHSRGISTIRFTPDGRWVVSGGLDNVVKVWDLTAGKL 206
Query: 131 CNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNL 167
+ F+ HE I + F L S D T+ +L
Sbjct: 207 LHEFKFHEGPIRSLDFHPLEFLLATGSADRTVKFWDL 243
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,500,669
Number of Sequences: 26719
Number of extensions: 374526
Number of successful extensions: 2257
Number of sequences better than 10.0: 169
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 86
Number of HSP's that attempted gapping in prelim test: 1706
Number of HSP's gapped (non-prelim): 497
length of query: 378
length of database: 11,318,596
effective HSP length: 101
effective length of query: 277
effective length of database: 8,619,977
effective search space: 2387733629
effective search space used: 2387733629
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC142222.5


