

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.3 + phase: 0
(326 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
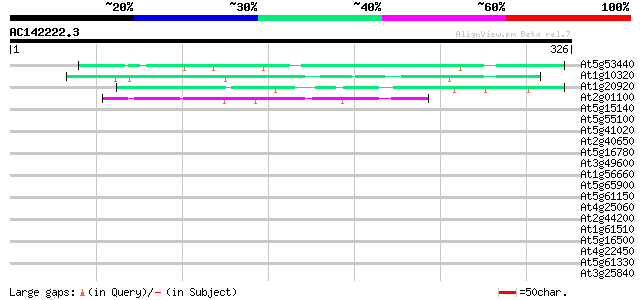
Score E
Sequences producing significant alignments: (bits) Value
At5g53440 putative protein 48 6e-06
At1g10320 unknown protein 47 1e-05
At1g20920 putative RNA helicase 45 4e-05
At2g01100 unknown protein 44 2e-04
At5g15140 putative aldose 1-epimerase - like protein 39 0.004
At5g55100 unknown protein 39 0.005
At5g41020 putative protein 39 0.005
At2g40650 unknown protein 38 0.007
At5g16780 putative protein 37 0.011
At3g49600 putative protein 37 0.011
At1g56660 hypothetical protein 37 0.015
At5g65900 ATP-dependent RNA helicase-like 37 0.019
At5g61150 unknown protein 36 0.032
At4g25060 hypothetical protein 35 0.042
At2g44200 putative protein 35 0.042
At1g61510 hypothetical protein 35 0.042
At5g16500 protein kinase-like protein 35 0.072
At4g22450 hypothetical protein 35 0.072
At5g61330 unknown protein 34 0.094
At3g25840 protein kinase like protein 34 0.094
>At5g53440 putative protein
Length = 1181
Score = 48.1 bits (113), Expect = 6e-06
Identities = 68/302 (22%), Positives = 114/302 (37%), Gaps = 36/302 (11%)
Query: 41 YD-DDHSLDEFLADRRRKKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDS 99
YD DDH + +D+ K R P ++Y+ +RR++ H+D+ D
Sbjct: 159 YDGDDHHKSKAGSDKTESKAQDHARSPG-TENYTEKR---SRRKRDDHGTGDKHHDNSDD 214
Query: 100 L---------DSLLPKQHQAERGHTRY---DDEDSLDEVLPKQRQAKRGRSHYDDDDSL- 146
+ D + +H+ E+ +Y +E+ + + KQR + + H D+ L
Sbjct: 215 VGDRVLTSGDDYIKDGKHKGEKSRDKYREDKEEEDIKQKGDKQRDDRPTKEHLRSDEKLT 274
Query: 147 -DEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKR 205
DE K H H PD L ++R+ YD + D D + R
Sbjct: 275 RDESKKKSKFQDNDHGHE------PDSELDGYHERERNRDYDRESDRNERDRERTRDRDR 328
Query: 206 GRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGD-SLHEDLPKNHQAERGQPRYDD--DD 262
D D + + + ++D D D D ++H+ +R R D D
Sbjct: 329 DYERDRDRDRDRDRERDRDRRDYEHDRYHDRDWDRDRSRDRDRDHERDRTHDREKDRSRD 388
Query: 263 DLPDGVLPKQRQAKKLHPHYDNDED-SLHEDLPKNHQAKR-GRPLYDDTDSLDEVLPARR 320
DG K + + DND D S +D ++ +R GR D D D + +R
Sbjct: 389 YYHDGKRSKSDRER------DNDRDVSRLDDQSGRYKDRRDGRRSPDYQDYQDVITGSRS 442
Query: 321 RR 322
R
Sbjct: 443 SR 444
Score = 36.6 bits (83), Expect = 0.019
Identities = 48/264 (18%), Positives = 94/264 (35%), Gaps = 25/264 (9%)
Query: 68 YDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVL 127
Y ++Y+ + + ++ ++ D+D +S + +E+ R + + +
Sbjct: 64 YYEEYTSSSSKRRKGKSGESGSDRWNGKDDDKGESSKKTKVSSEKSRKRDEGDGEETKKS 123
Query: 128 PKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDL-----PDGVLPKQQAKKR 182
+ K S + +D+ +++ + YD DD D K Q R
Sbjct: 124 SGKSDGKHRESSRRESKDVDKEKDRKYKEGKSDKFYDGDDHHKSKAGSDKTESKAQDHAR 183
Query: 183 HPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLH 242
P +N + +H G H+D D++ D VL GD
Sbjct: 184 SPGTENYTEKRSRRKRDDHGT--GDKHHDNSDDVGDRVL--------------TSGDDYI 227
Query: 243 EDLPKNHQAERGQPRYDDDDDLPD--GVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQAK 300
+D H+ E+ + +Y +D + D KQR + H +DE ++ K + +
Sbjct: 228 KD--GKHKGEKSRDKYREDKEEEDIKQKGDKQRDDRPTKEHLRSDEKLTRDESKKKSKFQ 285
Query: 301 RGRPLYDDTDSLDEVLPARRRRHY 324
++ LD R R Y
Sbjct: 286 DNDHGHEPDSELDGYHERERNRDY 309
>At1g10320 unknown protein
Length = 757
Score = 47.4 bits (111), Expect = 1e-05
Identities = 64/300 (21%), Positives = 106/300 (35%), Gaps = 49/300 (16%)
Query: 34 ARRGHPRYDDDHSLDEFLADRRRKKHL---------ALRRHPHY----DDDYSLDEIFPN 80
+RR R D ++ + R RK H LRRH DD S D
Sbjct: 463 SRRSRSRDHDHANVGSTPSYRSRKYHGDTQDSTREDKLRRHAENCHDGDDSPSRDGSLER 522
Query: 81 RRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLD------EVLPKQRQAK 134
K + + + + D + + +R H RY D+DS D +R+ +
Sbjct: 523 EMYKERRYAKDTLHRDSRWSEHSPGHRVGRKRIHGRYSDDDSADGDDYGRRGTGHKRKPR 582
Query: 135 RGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLH 194
RG + +E K H + R H D +G H D H
Sbjct: 583 RGTDSGVQEQMDNEKDRKTHRSSRKHSREGSSADKEEG--------HEHDRVHTVSDKSH 634
Query: 195 EDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERG 254
+ K H+ +R S Y E++ + + H H ++D E P +Q+++
Sbjct: 635 RERSK-HRHERSSSRYSHEEDSTE---------SRHHQHKESDKKRSVETSPVGYQSDKD 684
Query: 255 ------QPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQAKRGRPLYDD 308
+ RY DD D +RQ++ +N + H++ H+ +R DD
Sbjct: 685 RDRSKQRQRYKSDDPESDQSRKGKRQSE------ENSDRETHKERRHRHRKRRRTQNSDD 738
Score = 34.7 bits (78), Expect = 0.072
Identities = 32/156 (20%), Positives = 66/156 (41%), Gaps = 13/156 (8%)
Query: 30 SVSQARRGHPRYDDDHSLDEFLADRRRKKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHK 89
S + GH +D H++ + + R R KH RH YS +E R H
Sbjct: 613 SSADKEEGH-EHDRVHTVSD-KSHRERSKH----RHERSSSRYSHEEDSTESR-----HH 661
Query: 90 RHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEV 149
+H D + S+++ P +Q+++ R P+ Q+++G+ +++ +
Sbjct: 662 QHKESDKKRSVETS-PVGYQSDKDRDRSKQRQRYKSDDPESDQSRKGKRQSEENSDRETH 720
Query: 150 LPKQH-HAKRGHPHYDDDDDLPDGVLPKQQAKKRHP 184
++H H KR DD + + +++ ++ P
Sbjct: 721 KERRHRHRKRRRTQNSDDQNPKESEEVEEEIERWRP 756
>At1g20920 putative RNA helicase
Length = 1166
Score = 45.4 bits (106), Expect = 4e-05
Identities = 60/283 (21%), Positives = 106/283 (37%), Gaps = 46/283 (16%)
Query: 63 RRHPHYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDS 122
RR ++ D+ RR+K + KR D ED D ++ + + R
Sbjct: 26 RRDRDRSNERKKDKGSEKRREKDRRKKRVKSSDSEDDYDRDDDEEREKRKEKERERRRRD 85
Query: 123 LDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQ--HHAKRGHPHYDDDDDLPDGVLPKQQAK 180
D V K+R +R S +DD ++ K+ + +RGH ++ D + K
Sbjct: 86 KDRV--KRRSERRKSSDSEDDVEEEDERDKRRVNEKERGHREHERD-----------RGK 132
Query: 181 KRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDS 240
R + +E +D + + R R + E+ ++ + K+R DG+
Sbjct: 133 DRKRDREREE---RKDKEREREKDRERREREREER-------EKERVKERERREREDGER 182
Query: 241 LHEDLPKNHQAERGQPR--------YDDDDDLPDGVLPKQRQA---KKLHPHYDNDEDSL 289
+ K + R + R + DDD+ + ++++ K+ S
Sbjct: 183 DRREREKERGSRRNRERERSREVGNEESDDDVKRDLKRRRKEGGERKEKEREKSVGRSSR 242
Query: 290 HEDLPKNHQAK----------RGRPLYDDTDSLDEVLPARRRR 322
HED PK + R L D+ LDE + RRRR
Sbjct: 243 HEDSPKRKSVEDNGEKKEKKTREEELEDEQKKLDEEVEKRRRR 285
>At2g01100 unknown protein
Length = 247
Score = 43.5 bits (101), Expect = 2e-04
Identities = 49/204 (24%), Positives = 83/204 (40%), Gaps = 28/204 (13%)
Query: 55 RRKKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGH 114
+RK+ A D S E+ R+ H H+RH H D +DS D K+ + ++
Sbjct: 52 KRKQRAASESSSESD---SSSEVRRKSRRSHNKHRRHAHSDSDDS-DRRKEKKSRRQKRR 107
Query: 115 TRYDDEDSL--------DEVLPKQRQAKRGRSHYD----DDDSLDEVLPKQ-HHAKRGHP 161
+ +DS DE+ K + +R + H DDDS ++V K H +R
Sbjct: 108 SLSPSDDSTGDYESGSEDELRMKIKHHRRHKWHSSRKTCDDDSTEDVRRKHLKHHRRSEV 167
Query: 162 HYDDDDDLPDGVLPKQQAKKRHPHYDNDEDS--LHEDLPKNHQAKRGRSHYDVEDNLLDG 219
D + G +++ K H H S ED K+ + ++ + H E +
Sbjct: 168 VTSSDSEEESG---RRRRGKYHRHNRGSASSSGSEEDSGKSMKRRQHKRHRLAESS---- 220
Query: 220 VLPKQPQAKKRHPHYDNDGDSLHE 243
++ A +R H+ + DS E
Sbjct: 221 --SEEDGAMRRTRHHKHGRDSASE 242
Score = 35.4 bits (80), Expect = 0.042
Identities = 40/172 (23%), Positives = 65/172 (37%), Gaps = 12/172 (6%)
Query: 54 RRRKKHLALRRHPHYD-DDYSLDEIFPNRRQKHQA------HKRHYHYDDEDSLDSLLPK 106
+ R+ H RRH H D DD + +RRQK ++ Y ED L + K
Sbjct: 74 KSRRSHNKHRRHAHSDSDDSDRRKEKKSRRQKRRSLSPSDDSTGDYESGSEDELRMKI-K 132
Query: 107 QHQAERGHT--RYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYD 164
H+ + H+ + D+DS ++V K + R DS +E ++ H
Sbjct: 133 HHRRHKWHSSRKTCDDDSTEDVRRKHLKHHRRSEVVTSSDSEEESGRRRRGKYHRHNRGS 192
Query: 165 DDDDLPDGVLPKQQAKKRHPHYDNDEDSLHED--LPKNHQAKRGRSHYDVED 214
+ K +++H + E S ED + + K GR D
Sbjct: 193 ASSSGSEEDSGKSMKRRQHKRHRLAESSSEEDGAMRRTRHHKHGRDSASESD 244
Score = 33.9 bits (76), Expect = 0.12
Identities = 47/200 (23%), Positives = 75/200 (37%), Gaps = 23/200 (11%)
Query: 128 PKQRQAKRGRSHYDDDDSLDEVLPK---QHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHP 184
PK++Q S + DS EV K H+ R H H D DD ++ K+R
Sbjct: 51 PKRKQRAASESS-SESDSSSEVRRKSRRSHNKHRRHAHSDSDDSDRRKEKKSRRQKRRSL 109
Query: 185 HYDND---------EDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRH---P 232
+D ED L + + + K S +D+ + V K + +R
Sbjct: 110 SPSDDSTGDYESGSEDELRMKIKHHRRHKWHSSRKTCDDDSTEDVRRKHLKHHRRSEVVT 169
Query: 233 HYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHED 292
D++ +S K H+ RG ++ G K+RQ H + E S ED
Sbjct: 170 SSDSEEESGRRRRGKYHRHNRGSASSSGSEE-DSGKSMKRRQ----HKRHRLAESSSEED 224
Query: 293 --LPKNHQAKRGRPLYDDTD 310
+ + K GR ++D
Sbjct: 225 GAMRRTRHHKHGRDSASESD 244
Score = 31.2 bits (69), Expect = 0.80
Identities = 30/113 (26%), Positives = 45/113 (39%), Gaps = 7/113 (6%)
Query: 36 RGHPRYDDDHSLDEFLADRRRKKHLAL-RRHPHYDDDYSLDEIFPNRRQKHQAHKR---H 91
R H + + D+ + R+KHL RR S +E RR K+ H R
Sbjct: 135 RRHKWHSSRKTCDDDSTEDVRRKHLKHHRRSEVVTSSDSEEESGRRRRGKYHRHNRGSAS 194
Query: 92 YHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDD 144
+EDS S+ +QH+ R +ED + + R K GR + D
Sbjct: 195 SSGSEEDSGKSMKRRQHKRHRLAESSSEEDG---AMRRTRHHKHGRDSASESD 244
Score = 29.6 bits (65), Expect = 2.3
Identities = 25/105 (23%), Positives = 41/105 (38%), Gaps = 8/105 (7%)
Query: 229 KRHPHYDNDGDSLHEDLPKNHQAERG-QPRYDDDDDLPDG------VLPKQRQAKKLHPH 281
+RH H D+D ++ Q R P D D G + K + K H
Sbjct: 83 RRHAHSDSDDSDRRKEKKSRRQKRRSLSPSDDSTGDYESGSEDELRMKIKHHRRHKWHSS 142
Query: 282 YDNDEDSLHEDLPKNH-QAKRGRPLYDDTDSLDEVLPARRRRHYR 325
+D ED+ + H + R + +DS +E RR +++R
Sbjct: 143 RKTCDDDSTEDVRRKHLKHHRRSEVVTSSDSEEESGRRRRGKYHR 187
>At5g15140 putative aldose 1-epimerase - like protein
Length = 490
Score = 38.9 bits (89), Expect = 0.004
Identities = 29/112 (25%), Positives = 46/112 (40%), Gaps = 14/112 (12%)
Query: 95 DDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQH 154
DDED+ K+H ++ +++++D DE ++ G DDD+ K+H
Sbjct: 64 DDEDNDKKEKKKEHDVQKKDKQHENKDKDDEKKHVDKKKSGGHDKDDDDE-------KKH 116
Query: 155 HAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRG 206
K+ H DDDD D D+D+D D N + K G
Sbjct: 117 KDKKKDGHNDDDDSDDD-------TDDDDDDDDDDDDDDEVDGDDNEKEKIG 161
Score = 37.4 bits (85), Expect = 0.011
Identities = 24/92 (26%), Positives = 41/92 (44%), Gaps = 12/92 (13%)
Query: 81 RRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHY 140
++++H K+ ++++D D + GH + DD++ K+ + K+ H
Sbjct: 73 KKKEHDVQKKDKQHENKDKDDEKKHVDKKKSGGHDKDDDDE-------KKHKDKKKDGHN 125
Query: 141 DDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDG 172
DDDDS D+ DDDDD DG
Sbjct: 126 DDDDSDDDT-----DDDDDDDDDDDDDDEVDG 152
Score = 32.0 bits (71), Expect = 0.47
Identities = 28/104 (26%), Positives = 44/104 (41%), Gaps = 16/104 (15%)
Query: 164 DDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPK 223
D+D+D K++ KK H D+ HE+ K+ + K H D + + G K
Sbjct: 65 DEDND-------KKEKKKEHDVQKKDKQ--HENKDKDDEKK----HVDKKKS---GGHDK 108
Query: 224 QPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDG 267
+K+H DG + +D + + DDDDD DG
Sbjct: 109 DDDDEKKHKDKKKDGHNDDDDSDDDTDDDDDDDDDDDDDDEVDG 152
>At5g55100 unknown protein
Length = 844
Score = 38.5 bits (88), Expect = 0.005
Identities = 43/197 (21%), Positives = 78/197 (38%), Gaps = 22/197 (11%)
Query: 42 DDDHSLDEFLADRRR-KKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSL 100
+++ S+DE + + KKH R+ + YS + +H K + +D
Sbjct: 636 EEESSMDEVTEETKTDKKHSCSRKRHKHKTRYS-------SKDRHSRDKHKHESSSDDEY 688
Query: 101 DSLLPKQHQAERGHTRYDDEDSLD-EVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRG 159
S +H+ + R++ DS D E + R +K + D D S D+ HH R
Sbjct: 689 HSRSRHRHRHSKSSDRHELYDSSDNEGEHRHRSSKHSK---DVDYSKDK--RSHHHRSRK 743
Query: 160 HPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDG 219
H + D D + ++++H EDS D+ H+ K + E + +
Sbjct: 744 HEKHRDSSDDEHHHHRHRSSRRKH------EDS--SDVEHGHRHKSSKRIKKDEKTVEEE 795
Query: 220 VLPKQPQAKKRHPHYDN 236
+ K Q+ + DN
Sbjct: 796 TVSKSDQSDLKASPGDN 812
>At5g41020 putative protein
Length = 588
Score = 38.5 bits (88), Expect = 0.005
Identities = 55/315 (17%), Positives = 132/315 (41%), Gaps = 40/315 (12%)
Query: 32 SQARRGHPRYDDDHSLDEFLAD--------RRRKKHLALRRHPHYDDDYSL--------- 74
++ ++ + D + + +EF+++ ++RKK+ + +D +
Sbjct: 7 NKKKKSDAKVDSEETGEEFISEHSSMKDKEKKRKKNKRENKDGFTGEDMEITGRESEKLG 66
Query: 75 DEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAK 134
DE+F +++K K+ D E ++D++ K + + T+ D E D V K ++
Sbjct: 67 DEVFIVKKKKKS--KKPIRIDSE-AVDAVKKKSKKRSK-ETKADSEAEDDGVEKKSKE-- 120
Query: 135 RGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYD-NDEDSL 193
+ + D ++ D V K+ +K+ ++ V K++ K++ D E+++
Sbjct: 121 KSKETKVDSEAHDGVKRKKKKSKKESGGDVIENTESSKVSDKKKGKRKRDDTDLGAEENI 180
Query: 194 HEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDND----GDSLHEDLPKNH 249
+++ + + K+ DVED LD + + KK+ D++ G + +D K
Sbjct: 181 DKEVKRKNNKKKPSVDSDVEDINLDSTNDGKKKRKKKKQSEDSETEENGLNSTKDAKKRR 240
Query: 250 ------------QAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNH 297
+AE + D+D P + + + ++ +D++ +D +
Sbjct: 241 KKKKKKKQSEVSEAEEKSDKSDEDLTTPSTSSKRVKFSDQVEFFPSDDDEGTEDDDEEEV 300
Query: 298 QAKRGRPLYDDTDSL 312
+ RG+ + D +
Sbjct: 301 KVVRGKRFTKEEDEM 315
>At2g40650 unknown protein
Length = 355
Score = 38.1 bits (87), Expect = 0.007
Identities = 33/164 (20%), Positives = 69/164 (41%), Gaps = 10/164 (6%)
Query: 42 DDDHSLDEFLADRRRKKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLD 101
+++ +E +AD + R+ P + + D + R + + + R Y D + D
Sbjct: 191 EEEKEENEGIADGSEDEMDQRRKSPERERERDRDRRRDSHRHRDRDYDRDYDMDRDHDRD 250
Query: 102 SLLPKQHQAERGHTR-YDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGH 160
++ ERGH R D E D + R +RGR + D D + R
Sbjct: 251 ------YERERGHGRDRDRERDRDHYRERDRDRERGRDR--ERDRRDRARRRSRSRSRDR 302
Query: 161 PHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAK 204
+ + DD+ D PK++ +K+ ++ D + ++ + ++ +
Sbjct: 303 KRH-ETDDVRDREEPKKKKEKKEKMKEDGTDHPNPEIAEMNRLR 345
>At5g16780 putative protein
Length = 820
Score = 37.4 bits (85), Expect = 0.011
Identities = 32/160 (20%), Positives = 64/160 (40%), Gaps = 11/160 (6%)
Query: 53 DRRRKKHLALRRHPHYDDDYSLDEIFPNRRQK-HQAHKRHYHYDDEDSLDSLLPKQHQAE 111
++ + +H Y+ + R++K H++ + YD E D + + E
Sbjct: 4 EKSKSRHEIREERADYEGSPVREHRDGRRKEKDHRSKDKEKDYDREKIRDKDHRRDKEKE 63
Query: 112 RGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPD 171
R R DED+ +++ RGR + D + + ++ K + H D +++ +
Sbjct: 64 RDRKRSRDEDT-------EKEISRGRDKEREKDKSRDRVKEKDKEKERNRHKDRENERDN 116
Query: 172 GVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYD 211
K++ K R + HED + H+A H D
Sbjct: 117 ---EKEKDKDRARVKERASKKSHEDDDETHKAAERYEHSD 153
Score = 32.3 bits (72), Expect = 0.36
Identities = 34/134 (25%), Positives = 55/134 (40%), Gaps = 28/134 (20%)
Query: 42 DDDHSLD-EFLADRRR------KKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHKRHYHY 94
D DH D E DR+R +K ++ R + D S D + ++K + +R+ H
Sbjct: 53 DKDHRRDKEKERDRKRSRDEDTEKEISRGRDKEREKDKSRDRV----KEKDKEKERNRHK 108
Query: 95 DDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQH 154
D E+ D+ + E D K+R +K+ SH DDD++ +H
Sbjct: 109 DRENERDN---------------EKEKDKDRARVKERASKK--SHEDDDETHKAAERYEH 151
Query: 155 HAKRGHPHYDDDDD 168
RG D+ D
Sbjct: 152 SDNRGLNEGGDNVD 165
>At3g49600 putative protein
Length = 1672
Score = 37.4 bits (85), Expect = 0.011
Identities = 58/280 (20%), Positives = 103/280 (36%), Gaps = 44/280 (15%)
Query: 70 DDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPK 129
DD + E F R ++ + H + D DS G + D++ +DE K
Sbjct: 152 DDEPMREGFEGRLKERREHS----FLDRDS-------------GRKKVDED--VDEKDAK 192
Query: 130 QRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDD---------LPDGVLPKQQAK 180
+++K+ R DDD + + K+ KR H + D+ KQ+++
Sbjct: 193 VKESKKQRGGDDDDVDVVKRHKKKESKKRRHDDSSESDEHGRDRRRRSKKKAKGRKQESE 252
Query: 181 KRHPHYDNDEDSLHED-------LPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPH 233
D++ DS +D P KR R V + + +K H
Sbjct: 253 SDSSSSDSESDSDSDDGKKRGRKKPTKTTKKRSRRKRSVSSESEEVESDDSKKLRKSHKK 312
Query: 234 YDNDGDSLHEDLPKNH--QAERGQPRYDDDDDLPDGVLPKQRQAKKLHPH-----YDNDE 286
S ++L H Q+ G+ R+D D P+ KQ KK + D+
Sbjct: 313 SLPSNRSGSKELRDKHDEQSRAGRKRHDSDVSEPESEDNKQPLRKKEEAYRGGQKQKRDD 372
Query: 287 DSLHEDLPKNHQAKRGRPLYDDTDSLDEVLPARRRRHYRS 326
+ + D K+ + + D+D D + + ++ RS
Sbjct: 373 EDVEADHLKDRYTRDDKKAARDSD--DSEIEYQNKKQLRS 410
Score = 32.3 bits (72), Expect = 0.36
Identities = 37/148 (25%), Positives = 58/148 (39%), Gaps = 20/148 (13%)
Query: 32 SQARRGHPRYDDDHSLDEFLADRRRKKHLALRRHPHY-DDDYSLDEIFPNRRQKHQAHKR 90
S +R D D S E+ ++ K R H ++D D +R + A KR
Sbjct: 439 SDSRGKEVARDSDDSEAEYENRKKLKNESYQRGRKHKREEDEDNDNHGRDRYRGDDAVKR 498
Query: 91 H--YHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHY-------- 140
+ DD+ + ++ +RG R E D+ + + K GR Y
Sbjct: 499 YGTIKEDDDRYRGRAIEEEGDDDRGRYRPRRESVKDD----EEEYKHGRDRYRGDGRRAT 554
Query: 141 ---DDDDSLDEVLPKQHHAKRGHPHYDD 165
DDDD D V ++ ++ RG YDD
Sbjct: 555 GKEDDDD--DRVSREREYSSRGRSRYDD 580
Score = 32.0 bits (71), Expect = 0.47
Identities = 54/262 (20%), Positives = 97/262 (36%), Gaps = 41/262 (15%)
Query: 40 RYDDDHSLDEFLADRRR--KKHLALRRHPHYDDDYSLDEIF--------------PNRRQ 83
R+DD DE DRRR KK R+ D S D P +
Sbjct: 222 RHDDSSESDEHGRDRRRRSKKKAKGRKQESESDSSSSDSESDSDSDDGKKRGRKKPTKTT 281
Query: 84 KHQAHKRHYHYDDEDSLDS----LLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSH 139
K ++ ++ + + ++S L K H+ R ++ D+ Q++ GR
Sbjct: 282 KKRSRRKRSVSSESEEVESDDSKKLRKSHKKSLPSNRSGSKELRDK---HDEQSRAGRKR 338
Query: 140 YDDDDSLDE------VLPKQHHAKRG--HPHYDDDDDLPDGVLPKQQAKKRHPHYDNDED 191
+D D S E L K+ A RG DD+D D + + + D+D+
Sbjct: 339 HDSDVSEPESEDNKQPLRKKEEAYRGGQKQKRDDEDVEADHLKDRYTRDDKKAARDSDDS 398
Query: 192 SLHEDLPKN-------HQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYD-NDGDSLHE 243
+ K + A + + ED G + ++ + D +D ++ +E
Sbjct: 399 EIEYQNKKQLRSKVEVYSAGMSQKRKEEEDVTKHGKDKYRSDSRGKEVARDSDDSEAEYE 458
Query: 244 DLP--KNHQAERGQPRYDDDDD 263
+ KN +RG+ ++D+
Sbjct: 459 NRKKLKNESYQRGRKHKREEDE 480
Score = 27.7 bits (60), Expect = 8.8
Identities = 26/113 (23%), Positives = 43/113 (38%), Gaps = 23/113 (20%)
Query: 63 RRHPHYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDS 122
RR DD+ E + + R +++ R ++D D + ++ + RG +RYDD S
Sbjct: 528 RRESVKDDE----EEYKHGRDRYRGDGRRATGKEDDDDDRVSREREYSSRGRSRYDDSRS 583
Query: 123 L-------------------DEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHA 156
D + + K R D DS ++L K H A
Sbjct: 584 SGKRSSHGFSIVTGEISGGGDTMSRPNTRNKNKRQRPDAVDSSSQILRKIHEA 636
>At1g56660 hypothetical protein
Length = 522
Score = 37.0 bits (84), Expect = 0.015
Identities = 52/288 (18%), Positives = 115/288 (39%), Gaps = 43/288 (14%)
Query: 17 QARRARQSAGAFTSVSQARRGHPRYDDDHSLDEFLADRRRKKHLALRRHPHYDDDYSLDE 76
+A++ +S+G + ++G + D ++ D+++ + ++H D + E
Sbjct: 46 KAKKDEESSGKSKKDKEKKKG--KNVDSEVKEDKDDDKKKDGKMVSKKHEEGHGDLEVKE 103
Query: 77 IFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRG 136
+ + HK+ E+ + E+ + ++ DE P+++ K
Sbjct: 104 SDVKVEEHEKEHKKGKEKKHEEL---------EEEKEGKKKKNKKEKDESGPEEKNKKAD 154
Query: 137 RSHYDDDDSLD-EVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHE 195
+ +D S + E L ++ K D+ PK++ K++ +E +E
Sbjct: 155 KEKKHEDVSQEKEELEEEDGKKNKKKEKDESGTEEKKKKPKKEKKQK------EESKSNE 208
Query: 196 DLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQ 255
D + ++G G L K+ + KK+ H + D + +D KN + E+ +
Sbjct: 209 DKKVKGKKEKGEK----------GDLEKEDEEKKKE-HDETDQEMKEKDSKKNKKKEKDE 257
Query: 256 PRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQAKRGR 303
++ PD K+++ K DE + ED K + K+G+
Sbjct: 258 SCAEEKKKKPD----KEKKEK--------DESTEKED--KKLKGKKGK 291
>At5g65900 ATP-dependent RNA helicase-like
Length = 633
Score = 36.6 bits (83), Expect = 0.019
Identities = 27/136 (19%), Positives = 61/136 (44%), Gaps = 14/136 (10%)
Query: 80 NRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSH 139
N K + HK+ + + + ++ E G + + ++L + PK+++ K+ +
Sbjct: 13 NEEIKKKKHKKRARDEAKKLKQPAMEEEPDHEDGDAK--ENNALIDEEPKKKKKKKNKKR 70
Query: 140 YDDDDSLDEVLPKQHHAK----------RGHPHYDDDDDLPDGVLPKQQAKKRHPHYDND 189
D DD DE + ++ K RG + ++D+ + + PK++ KK+ D +
Sbjct: 71 GDTDDGEDEAVAEEEPKKKKKKNKKLQQRGDTNDEEDEVIAEEEEPKKKKKKQRK--DTE 128
Query: 190 EDSLHEDLPKNHQAKR 205
S E++ + K+
Sbjct: 129 AKSEEEEVEDKEEEKK 144
Score = 36.6 bits (83), Expect = 0.019
Identities = 37/166 (22%), Positives = 72/166 (43%), Gaps = 33/166 (19%)
Query: 141 DDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHP-HYDND---EDSLHED 196
+ S +E + K+ H KR D+ L KQ A + P H D D ++L ++
Sbjct: 7 EQHSSENEEIKKKKHKKRAR---DEAKKL------KQPAMEEEPDHEDGDAKENNALIDE 57
Query: 197 LPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQ-AERGQ 255
PK + K+ + D +D + V ++P+ KK+ KN + +RG
Sbjct: 58 EPKKKKKKKNKKRGDTDDGEDEAVAEEEPKKKKK----------------KNKKLQQRGD 101
Query: 256 PRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQAKR 301
++D+ + + PK+++ K+ D + S E++ + K+
Sbjct: 102 TNDEEDEVIAEEEEPKKKKKKQ---RKDTEAKSEEEEVEDKEEEKK 144
>At5g61150 unknown protein
Length = 625
Score = 35.8 bits (81), Expect = 0.032
Identities = 44/203 (21%), Positives = 80/203 (38%), Gaps = 17/203 (8%)
Query: 23 QSAGAFTSVSQAR----RGHPRYDDDHSLDE-FLADRRRKKHLALRRHPHYDDDYSLDEI 77
Q+ A T +SQAR R +P + L +L D + R + Y++D L+
Sbjct: 427 QNLKASTKLSQAREKIKRKYPLPVERRQLSTGYLEDALDEDDEDYRSNRGYEED--LEAE 484
Query: 78 FPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGR 137
R+ A K H S+ S P + Q E + ++ + E +++ RGR
Sbjct: 485 AQRERRILNAKKSHKGIPGRSSMTSARPSRRQMEYSESEREESEYETEEEEEEKSPARGR 544
Query: 138 SHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAK------KRHPHYDNDED 191
D +D +E + + Y D+D+ + V + K ++ ++DE+
Sbjct: 545 GK-DSEDEYEEDAEEDEEERGKSNRYSDEDEEEEEVAGGRAEKDHRGSGRKRKGIESDEE 603
Query: 192 SLHEDLPKNHQAKRGRSHYDVED 214
E P+ R ++ D D
Sbjct: 604 ---ESPPRKAPTHRRKAVIDDSD 623
>At4g25060 hypothetical protein
Length = 412
Score = 35.4 bits (80), Expect = 0.042
Identities = 53/233 (22%), Positives = 86/233 (36%), Gaps = 48/233 (20%)
Query: 92 YHYDDEDSLDSL----LPKQHQAERGHTRYDDEDSLDEV-LPKQRQAKRGRSHYDDDDSL 146
Y YDDED + L LP Q+ R + + S++E P ++ +DD+
Sbjct: 84 YDYDDEDGGEDLYQVHLPPLTQSRRRDDKRIGKGSVNEKKAPAKKPIILPAPKFDDESED 143
Query: 147 DE----------VLPKQHHAKRGH-----------PHYDDDDDLPDGVLPKQQAKKRH-- 183
DE ++PK++ + P + + +P + + +
Sbjct: 144 DETRANFRPNPVIIPKKNAREPRKAPATKPVVLPPPKFSNVRRIPVEISGRNARGSKEAS 203
Query: 184 --------PHYDN--DEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPH 233
P +D+ D D + ED+P+N RG V + V P K
Sbjct: 204 TVKQVVSTPKFDDEYDSDEITEDVPRNPVEIRG----PVRGKVAPAVKPVVVLPTKFDDE 259
Query: 234 YDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDE 286
YD+D + ED+P+N RG R + V P K YD+DE
Sbjct: 260 YDSD--EITEDVPRNPVEIRGTVR----GKVAPAVKPVVVLPTKFDDEYDSDE 306
Score = 33.1 bits (74), Expect = 0.21
Identities = 56/222 (25%), Positives = 89/222 (39%), Gaps = 37/222 (16%)
Query: 103 LLPKQHQAERGHTRYDDEDSLDEV----LPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKR 158
LL K + YDDED +++ LP Q++R S++E K+ AK+
Sbjct: 72 LLAKVMEKREYDYDYDDEDGGEDLYQVHLPPLTQSRRRDDKRIGKGSVNE---KKAPAKK 128
Query: 159 G----HPHYDDDDDLPDG---------VLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKR 205
P +DD+ + + ++PK+ A R P + PK +R
Sbjct: 129 PIILPAPKFDDESEDDETRANFRPNPVIIPKKNA--REPRKAPATKPVVLPPPKFSNVRR 186
Query: 206 GRSHYDVEDNLLDGVLPKQPQAKKR---HPHYDN--DGDSLHEDLPKNHQAERGQPRYDD 260
VE + + K+ K+ P +D+ D D + ED+P+N RG R
Sbjct: 187 ----IPVEISGRNARGSKEASTVKQVVSTPKFDDEYDSDEITEDVPRNPVEIRGPVR--- 239
Query: 261 DDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQAKRG 302
+ V P K YD+DE + ED+P+N RG
Sbjct: 240 -GKVAPAVKPVVVLPTKFDDEYDSDE--ITEDVPRNPVEIRG 278
>At2g44200 putative protein
Length = 493
Score = 35.4 bits (80), Expect = 0.042
Identities = 36/180 (20%), Positives = 73/180 (40%), Gaps = 17/180 (9%)
Query: 129 KQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQ--AKKRHPHY 186
+++ + R RS ++D + K HH K H D + P L + + R HY
Sbjct: 192 QKQSSSRQRSDSEEDSGEENNGRKSHHQKTSGTH-DRHYERPRSDLEDESKGRESRDRHY 250
Query: 187 DNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLP 246
+ L + + + +R +HY+ + +D + + + R HY+ L ++
Sbjct: 251 EKRRSELDDG---HKRRERHDTHYERRRSEMD---DESKRRESRDNHYERRRSDLDDESK 304
Query: 247 K--NHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQAKRGRP 304
+ +H + R D DD+ K+R+++ D D++ D N + + P
Sbjct: 305 RRESHDKHFERQRSDLDDEY------KRRESQDKRRRSDIDDEPKRRDARPNEKYRNRSP 358
Score = 31.6 bits (70), Expect = 0.61
Identities = 30/143 (20%), Positives = 55/143 (37%), Gaps = 13/143 (9%)
Query: 6 RQSAGAFTSVSQARRARQSAGAFTSVSQARRGHPRYDDDHSLDEFLADRRRKKHLAL--R 63
R+S TS + R + S+ R R+ + + +RR++H R
Sbjct: 214 RKSHHQKTSGTHDRHYERPRSDLEDESKGRESRDRHYEKRRSELDDGHKRRERHDTHYER 273
Query: 64 RHPHYDDDY----SLDEIFPNRR-------QKHQAHKRHYHYDDEDSLDSLLPKQHQAER 112
R DD+ S D + RR ++ ++H +H+ D D ++ Q +R
Sbjct: 274 RRSEMDDESKRRESRDNHYERRRSDLDDESKRRESHDKHFERQRSDLDDEYKRRESQDKR 333
Query: 113 GHTRYDDEDSLDEVLPKQRQAKR 135
+ DDE + P ++ R
Sbjct: 334 RRSDIDDEPKRRDARPNEKYRNR 356
>At1g61510 hypothetical protein
Length = 608
Score = 35.4 bits (80), Expect = 0.042
Identities = 24/99 (24%), Positives = 39/99 (39%)
Query: 218 DGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKK 277
+G PKQ + K H D+DGDS + ++ E Q + DDD +LP Q
Sbjct: 14 EGWAPKQGEEKLDSMHQDDDGDSTQIPITQDDDGESRQIPLNLDDDEAAHILPTQSNVGN 73
Query: 278 LHPHYDNDEDSLHEDLPKNHQAKRGRPLYDDTDSLDEVL 316
+ + S + + D +S D++L
Sbjct: 74 GPSYSSKSKSSKKRSRSVQDEQGVAEVIKDSIESRDKIL 112
Score = 28.5 bits (62), Expect = 5.2
Identities = 28/117 (23%), Positives = 45/117 (37%), Gaps = 19/117 (16%)
Query: 128 PKQRQAKRGRSHYDDDDSLDEV-LPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHY 186
PKQ + K H DDD ++ + + + + DDD +LP Q P Y
Sbjct: 18 PKQGEEKLDSMHQDDDGDSTQIPITQDDDGESRQIPLNLDDDEAAHILPTQSNVGNGPSY 77
Query: 187 DNDEDSLHEDLPKNHQAKRGRSHYD-------VEDNL--LDGVLPKQPQAKKRHPHY 234
+ S KR RS D ++D++ D +L + Q + HP +
Sbjct: 78 SSKSKS---------SKKRSRSVQDEQGVAEVIKDSIESRDKILSHKNQLIEMHPEF 125
>At5g16500 protein kinase-like protein
Length = 636
Score = 34.7 bits (78), Expect = 0.072
Identities = 46/212 (21%), Positives = 81/212 (37%), Gaps = 36/212 (16%)
Query: 97 EDSLDSLLPKQHQAERGHTRYDDE-DSLDEVLPKQRQAK----RGRSHYDDDD-----SL 146
E +S K+ + E T DDE DS E ++ Q++ R S D S+
Sbjct: 418 EKEEESTSKKRQEQEETATDSDDESDSNSEKDQEEEQSQLEKARESSSSSSDSGSERRSI 477
Query: 147 DEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHP----HYDNDEDSLHEDLPKNHQ 202
DE K + +Y +++ + + K K YD+ D H+D +N
Sbjct: 478 DETNATAQSLKISYSNYSSEEEDNEKLSSKSSCKSNEESTFSRYDSGRD--HDDSSRNTS 535
Query: 203 AK-RGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDD 261
+ +H D E++ +++ S H+D P+N +DDD
Sbjct: 536 MRINSLAHDDKEED--------------EEENHETRSYSDHDDSPRNTSMRINSLSHDDD 581
Query: 262 DDLPDGVLPKQRQAKKLHPHYDNDED-SLHED 292
++ + + Q + H H ED S++ D
Sbjct: 582 EEEEE----ENHQTRLEHIHSSKSEDQSVYSD 609
Score = 29.6 bits (65), Expect = 2.3
Identities = 45/227 (19%), Positives = 75/227 (32%), Gaps = 38/227 (16%)
Query: 113 GHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDG 172
G+ DS DE K+++A++ +++S + +Q D DD D
Sbjct: 397 GNKSSSSSDSEDEEEEKEQKAEK------EEESTSKKRQEQEETAT------DSDDESDS 444
Query: 173 VLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHP 232
+ + D++ L K ++ S E +D K +
Sbjct: 445 ------------NSEKDQEEEQSQLEKARESSSSSSDSGSERRSIDETNATAQSLKISYS 492
Query: 233 HY---DNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDED-- 287
+Y + D + L E RYD D D + L H D +ED
Sbjct: 493 NYSSEEEDNEKLSSKSSCKSNEESTFSRYDSGRDHDDSSRNTSMRINSL-AHDDKEEDEE 551
Query: 288 --------SLHEDLPKNHQAKRGRPLYDDTDSLDEVLPARRRRHYRS 326
S H+D P+N + +DD + +E R H S
Sbjct: 552 ENHETRSYSDHDDSPRNTSMRINSLSHDDDEEEEEENHQTRLEHIHS 598
>At4g22450 hypothetical protein
Length = 457
Score = 34.7 bits (78), Expect = 0.072
Identities = 19/55 (34%), Positives = 27/55 (48%)
Query: 218 DGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQ 272
+G PKQ + K H D+DGDS + ++ E Q + DDD +LP Q
Sbjct: 139 EGWAPKQGEEKLDSMHQDDDGDSTQIPITQDDDGESRQIPLNLDDDEATHILPTQ 193
Score = 28.5 bits (62), Expect = 5.2
Identities = 28/117 (23%), Positives = 45/117 (37%), Gaps = 19/117 (16%)
Query: 128 PKQRQAKRGRSHYDDDDSLDEV-LPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHY 186
PKQ + K H DDD ++ + + + + DDD +LP Q P Y
Sbjct: 143 PKQGEEKLDSMHQDDDGDSTQIPITQDDDGESRQIPLNLDDDEATHILPTQSNVGNGPSY 202
Query: 187 DNDEDSLHEDLPKNHQAKRGRSHYD-------VEDNL--LDGVLPKQPQAKKRHPHY 234
+ S KR RS D ++D++ D +L + Q + HP +
Sbjct: 203 SSKSKS---------SKKRSRSVQDEQGVAEVIKDSIKSRDKILSHKNQLIEMHPKF 250
>At5g61330 unknown protein
Length = 436
Score = 34.3 bits (77), Expect = 0.094
Identities = 40/189 (21%), Positives = 79/189 (41%), Gaps = 31/189 (16%)
Query: 131 RQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDE 190
+++KR R + +D D+ K ++DD LPDG+ D++
Sbjct: 6 KRSKRARLDSESEDISDQENLKAESD-------NEDDQLPDGI------------EDDEV 46
Query: 191 DSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQ 250
DS+ +D ++ + G + D E G + + + + D DG+S + + +
Sbjct: 47 DSMEDDEGESEEDDEGDTEEDDE-----GDSEEDDEGENKE---DEDGESEDFEDGNDKE 98
Query: 251 AERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQAKRGRPLYDDTD 310
+E G DD+ D L +++ K+L + +L D K A +G+ + +
Sbjct: 99 SESGDEGNDDNKDAQMEEL--EKEVKELRSQEQDILKNLKRD--KGEDAVKGQAVKNQKA 154
Query: 311 SLDEVLPAR 319
D++L R
Sbjct: 155 LWDKILEFR 163
>At3g25840 protein kinase like protein
Length = 935
Score = 34.3 bits (77), Expect = 0.094
Identities = 53/240 (22%), Positives = 86/240 (35%), Gaps = 32/240 (13%)
Query: 64 RHPHYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSL 123
RH DY D + R ++ + Y +D +L + ER D E S+
Sbjct: 213 RHHDSSRDYHHDRVDSVRTEERYHRRGRYEENDRQYSREVLERDRSKERD---MDREGSI 269
Query: 124 DEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRH 183
+ R ++ + D D E + KR D + +++ K+R
Sbjct: 270 -----RDRDSEGSKRRDKDSDRRRE----REREKRREIESDRE---------RRKEKERE 311
Query: 184 PHYDNDEDSLHE-DLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKK-----RHPHYDND 237
D D E D ++ +RGRS N + K+ + +K R D +
Sbjct: 312 RSIDRDRRKEREGDYLRDRANERGRSRDRTRYNSRERKREKEREGEKDWERGRETQKDRE 371
Query: 238 G-DSLHEDLPKNHQAERGQPRYDDDDDL----PDGVLPKQRQAKKLHPHYDNDEDSLHED 292
S D + + RYD DDL P+ + + KL + NDE S +ED
Sbjct: 372 KYISSDVDYGEMRHKQSRNTRYDAVDDLEMRKPNSLKAHNSKGDKLEETWANDERSRNED 431
Score = 28.9 bits (63), Expect = 4.0
Identities = 50/270 (18%), Positives = 105/270 (38%), Gaps = 51/270 (18%)
Query: 63 RRHPHYDDDYSLDEIFPNRRQKHQAHKRHYH----------------YDDEDSLDSLLP- 105
R+H H S D ++R KH+ HK H+H DE + + P
Sbjct: 12 RKH-HRSSSSSDDTEKASKRHKHRHHKHHHHRHRHHRDKKRENEIPSAGDETEILDVTPA 70
Query: 106 ---------KQHQAERGHTRYDDEDSLDEVLPK--QRQAKRGRSHYDDDDSLDEVLPKQH 154
++ E G +ED + +VL K + G ++D++ + P
Sbjct: 71 APIVVSNGCEEEDVEEGEIL--EEDGIGDVLMKTADSDGESGEIKFEDNN----LPPLGE 124
Query: 155 HAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVED 214
++G ++ +GVL + ++++ +D + E + K GRS + +
Sbjct: 125 KGRQG------EEKSSNGVLTR-ESEREDKRWDKEAGGPSERVSK-LSYDNGRSSFSPSN 176
Query: 215 NLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDD---DLPDGVLPK 271
+ + +A+ R +D + ++ ++ ++RG+ D D D V +
Sbjct: 177 SRQS----NEGRARSRSKSHDRERETSRSRFAEDEFSDRGRHHDSSRDYHHDRVDSVRTE 232
Query: 272 QRQAKKLHPHYDNDEDSLHEDLPKNHQAKR 301
+R ++ + +ND E L ++ +R
Sbjct: 233 ERYHRR-GRYEENDRQYSREVLERDRSKER 261
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,238,918
Number of Sequences: 26719
Number of extensions: 506076
Number of successful extensions: 1677
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 78
Number of HSP's that attempted gapping in prelim test: 1419
Number of HSP's gapped (non-prelim): 246
length of query: 326
length of database: 11,318,596
effective HSP length: 100
effective length of query: 226
effective length of database: 8,646,696
effective search space: 1954153296
effective search space used: 1954153296
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC142222.3


