

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142142.4 - phase: 0
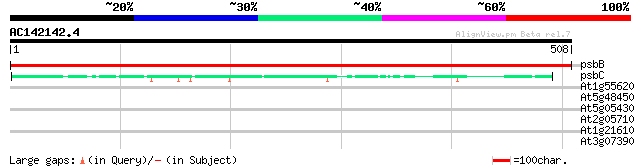
(508 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
psbB -chloroplast genome- PSII 47KDa protein 1042 0.0
psbC -chloroplast genome- PSII 43 KDa protein 95 8e-20
At1g55620 CLC-f chloride channel protein 30 3.1
At5g48450 putative protein 29 5.4
At5g05430 putative protein 29 5.4
At2g05710 cytoplasmic aconitate hydratase 29 7.0
At1g21610 unknown protein 29 7.0
At3g07390 auxin-induced protein (AIR12) 28 9.1
>psbB -chloroplast genome- PSII 47KDa protein
Length = 508
Score = 1042 bits (2694), Expect = 0.0
Identities = 494/508 (97%), Positives = 505/508 (99%)
Query: 1 MGLPWYRVHTVVLNDPGRLLSVHIMHTALVAGWAGSMALYELAVFDPSDPVLDPMWRQGM 60
MGLPWYRVHTVVLNDPGRLL+VHIMHTALVAGWAGSMALYELAVFDPSDPVLDPMWRQGM
Sbjct: 1 MGLPWYRVHTVVLNDPGRLLAVHIMHTALVAGWAGSMALYELAVFDPSDPVLDPMWRQGM 60
Query: 61 FVIPFMTRLGITNSWGGWNITGGTITNPGIWSYEGVAAAHIVFSGLCFLAAIWHWVYWDL 120
FVIPFMTRLGITNSWGGWNITGGTITNPG+WSYEGVA AHIVFSGLCFLAAIWHWVYWDL
Sbjct: 61 FVIPFMTRLGITNSWGGWNITGGTITNPGLWSYEGVAGAHIVFSGLCFLAAIWHWVYWDL 120
Query: 121 EIFCDERTGKPSLDLPKIFGIHLFLAGVACFGFGAFHVTGLFGPGIWVSDPYGLTGRVQS 180
EIFCDERTGKPSLDLPKIFGIHLFL+GVACFGFGAFHVTGL+GPGIWVSDPYGLTG+VQ
Sbjct: 121 EIFCDERTGKPSLDLPKIFGIHLFLSGVACFGFGAFHVTGLYGPGIWVSDPYGLTGKVQP 180
Query: 181 VNPAWGVDGFDPFVPGGIASHHIAAGTLGILAGLFHLSVRPPQRLYKGLRMGNIETVLSS 240
VNPAWGV+GFDPFVPGGIASHHIAAGTLGILAGLFHLSVRPPQRLYKGLRMGNIETVLSS
Sbjct: 181 VNPAWGVEGFDPFVPGGIASHHIAAGTLGILAGLFHLSVRPPQRLYKGLRMGNIETVLSS 240
Query: 241 SIAAVFFAAFVVAGTMWYGSATTPIELFGPTRYQWDQGYFQQEIYRRVGAGLAENQSLSE 300
SIAAVFFAAFVVAGTMWYGSATTPIELFGPTRYQWDQGYFQQEIYRRV AGLAENQSLSE
Sbjct: 241 SIAAVFFAAFVVAGTMWYGSATTPIELFGPTRYQWDQGYFQQEIYRRVSAGLAENQSLSE 300
Query: 301 AWSKIPEKLAFYDYIGNNPAKGGLFRAGSMDNGDGIAVGWLGHPIFRDKEGRELFVRRMP 360
AW+KIPEKLAFYDYIGNNPAKGGLFRAGSMDNGDGIAVGWLGHP+FR+KEGRELFVRRMP
Sbjct: 301 AWAKIPEKLAFYDYIGNNPAKGGLFRAGSMDNGDGIAVGWLGHPVFRNKEGRELFVRRMP 360
Query: 361 TFFETFPVVLVDGDGIVRADVPFRRAESKYSVEQVGVTVEFFGGELNGVSYSDPATVKKY 420
TFFETFPVVLVDGDGIVRADVPFRRAESKYSVEQVGVTVEF+GGELNGVSYSDPATVKKY
Sbjct: 361 TFFETFPVVLVDGDGIVRADVPFRRAESKYSVEQVGVTVEFYGGELNGVSYSDPATVKKY 420
Query: 421 ARRAQLGEIFELDRATLKSDGVFRSSPRGWFTFGHASFALLFFFGHIWHGARTLFRDVFA 480
ARRAQLGEIFELDRATLKSDGVFRSSPRGWFTFGHASFALLFFFGHIWHGARTLFRDVFA
Sbjct: 421 ARRAQLGEIFELDRATLKSDGVFRSSPRGWFTFGHASFALLFFFGHIWHGARTLFRDVFA 480
Query: 481 GIDPDLDAQVEFGAFQKLGDPTTKKQAV 508
GIDPDLDAQVEFGAFQKLGDPTTK+QAV
Sbjct: 481 GIDPDLDAQVEFGAFQKLGDPTTKRQAV 508
>psbC -chloroplast genome- PSII 43 KDa protein
Length = 473
Score = 95.1 bits (235), Expect = 8e-20
Identities = 122/516 (23%), Positives = 195/516 (37%), Gaps = 109/516 (21%)
Query: 2 GLPWYRVHTVVLNDPGRLLSVHIMHTALVAGWAGSMALYELAVFDPSDPVLDPMWRQGMF 61
G W+ + ++N G+LL H+ H L+ WAG+M L+E+A F P PM+ QG+
Sbjct: 32 GFAWWAGNARLINLSGKLLGAHVAHAGLIVFWAGAMNLFEVAHFVPE----KPMYEQGLI 87
Query: 62 VIPFMTRLGITNSWGGWNITGGTITNPGIWSYEGVAAAHIVFSGLCFLAAIWHWVYWDLE 121
++P + LG WG GG + + + GV H++ S + I+H +
Sbjct: 88 LLPHLATLG----WGVG--PGGEVIDTFPYFVSGV--LHLISSAVLGFGGIYHALLGPET 139
Query: 122 IFCDER------TGKPSLDLPKIFGIHLFLAGVACF--GFGAFHVTGLF---GPGIWVSD 170
+ +E K + I GIHL L GV F F A + G++ PG D
Sbjct: 140 L--EESFPFFGYVWKDRNKMTTILGIHLILLGVGAFLLVFKALYFGGVYDTWAPG--GGD 195
Query: 171 PYGLTGRVQSVNPAWGVDGFDPFVPGG----------IASHHIAAGTLGILAGLFHLSVR 220
+T S + +G PF G I H+ G++ I G++H+ +
Sbjct: 196 VRKITNLTLSPSVIFGYLLKSPFGGEGWIVSVDDLEDIIGGHVWLGSICIFGGIWHILTK 255
Query: 221 PPQRLYKGLRMGNIETVLSSSIAAVFFAAFVVAGTMWYGSATTPIELFGPTRYQWDQGYF 280
P + L + + E LS S+AA+ F+ +W+ + P E +GPT + Q
Sbjct: 256 PFAWARRAL-VWSGEAYLSYSLAALSVCGFIACCFVWFNNTAYPSEFYGPTGPEASQAQA 314
Query: 281 QQEIYR--RVGAGLAENQSLSEAWSKIPEKLAFYDYIGNNPAKGGLFRAGSMDNGDGIAV 338
+ R R+GA + Q P L Y+ +P +F +M D +
Sbjct: 315 FTFLVRDQRLGANVGSAQG--------PTGLG--KYLMRSPTGEVIFGGETMRFWD-LRA 363
Query: 339 GWLGHPIFRDKEGRELFVRRMPTFFETFPVVLVDGDGIVRADVPFRRAESKYSVEQVGVT 398
WL P+ R G +L R+ + + R ++Y +
Sbjct: 364 PWL-EPL-RGPNGLDL--SRLKKDIQPWQ----------------ERRSAEYMTHAPLGS 403
Query: 399 VEFFGG---ELNGVSYSDPATVKKYARRAQLGEIFELDRATLKSDGVFRSSPRGWFTFGH 455
+ GG E+N V+Y P R W + H
Sbjct: 404 LNSVGGVATEINAVNYVSP---------------------------------RSWLSTSH 430
Query: 456 ASFALLFFFGHIWHGARTLFRDVFAGIDPDLDAQVE 491
F GH+WH R R AG + +D E
Sbjct: 431 FVLGFFLFVGHLWHAGRA--RAAAAGFEKGIDRDFE 464
>At1g55620 CLC-f chloride channel protein
Length = 781
Score = 30.0 bits (66), Expect = 3.1
Identities = 15/38 (39%), Positives = 21/38 (54%), Gaps = 5/38 (13%)
Query: 75 WGGWNI-----TGGTITNPGIWSYEGVAAAHIVFSGLC 107
WG N+ TG + + PGIW +AAA +V + LC
Sbjct: 416 WGFTNVEEILHTGKSASAPGIWLLAQLAAAKVVATALC 453
>At5g48450 putative protein
Length = 544
Score = 29.3 bits (64), Expect = 5.4
Identities = 12/28 (42%), Positives = 19/28 (67%)
Query: 406 LNGVSYSDPATVKKYARRAQLGEIFELD 433
LNG+SY PAT K A++ + +++LD
Sbjct: 373 LNGISYLPPATPLKLAQQYNISGVYKLD 400
>At5g05430 putative protein
Length = 158
Score = 29.3 bits (64), Expect = 5.4
Identities = 19/79 (24%), Positives = 39/79 (49%), Gaps = 13/79 (16%)
Query: 14 NDPGRL--------LSVHIMHTALVAGWAGSMALYELAV---FDPSDPVLDPMWRQGMFV 62
ND GR+ L + ++TAL ++ ++E+ + FD ++ +L+ W +
Sbjct: 20 NDVGRIKVTGYDTGLPLDDLYTALAEHFSSCGEIWEIYIPLKFDETNTILNRFW--WLLP 77
Query: 63 IPFMTRLGITNSWGGWNIT 81
PF++ + GGWN++
Sbjct: 78 PPFLSNSSAQVNVGGWNVS 96
>At2g05710 cytoplasmic aconitate hydratase
Length = 990
Score = 28.9 bits (63), Expect = 7.0
Identities = 17/46 (36%), Positives = 27/46 (57%), Gaps = 3/46 (6%)
Query: 395 VGVTVEFFGGELNGVSYSDPATVKKYARR--AQLGEIFELDRATLK 438
VG VEF+G ++G+S +D AT+ + A +G F +D TL+
Sbjct: 370 VGKFVEFYGNGMSGLSLADRATIANMSPEYGATMG-FFPVDHVTLQ 414
>At1g21610 unknown protein
Length = 684
Score = 28.9 bits (63), Expect = 7.0
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query: 403 GGELNGVSYSDPA-TVKKYARRAQLGEIFELDRATLKSDGVF 443
G EL+G D T + A+L E FE+D +T+K DG +
Sbjct: 116 GEELDGAPDDDEYDTEDSFIDDAELDEYFEVDNSTVKHDGFY 157
>At3g07390 auxin-induced protein (AIR12)
Length = 273
Score = 28.5 bits (62), Expect = 9.1
Identities = 26/114 (22%), Positives = 39/114 (33%), Gaps = 21/114 (18%)
Query: 115 WVYWDL----------EIFCDERTGKPSLDLPKIFGIHLF---LAGVACFGFGAFHVTGL 161
WV W + + F R+G + + K + I + + G F F L
Sbjct: 100 WVAWAINPTGTKMAGSQAFLAYRSGGGAAPVVKTYNISSYSSLVEGKLAFDFWNLRAESL 159
Query: 162 FGPGIWVSDPYGLTGRVQSVNPAW--------GVDGFDPFVPGGIASHHIAAGT 207
G I + + SVN W G G PF P + SH + + T
Sbjct: 160 SGGRIAIFTTVKVPAGADSVNQVWQIGGNVTNGRPGVHPFGPDNLGSHRVLSFT 213
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.143 0.469
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,318,763
Number of Sequences: 26719
Number of extensions: 560269
Number of successful extensions: 1344
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1332
Number of HSP's gapped (non-prelim): 12
length of query: 508
length of database: 11,318,596
effective HSP length: 104
effective length of query: 404
effective length of database: 8,539,820
effective search space: 3450087280
effective search space used: 3450087280
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC142142.4


