

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142142.2 + phase: 0
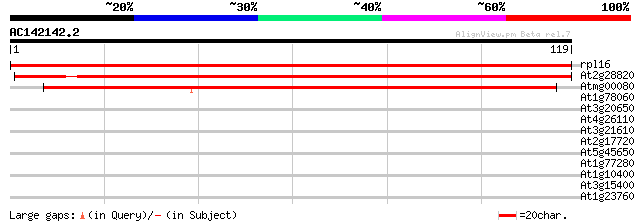
(119 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
rpl16 -chloroplast genome- ribosomal protein L16 220 1e-58
At2g28820 unknown protein 164 9e-42
Atmg00080 -mitochondrial genome- ribosomal protein L16 (RPL16) 98 7e-22
At1g78060 putative protein 27 1.2
At3g20650 unknown protein 27 2.1
At4g26110 nucleosome assembly protein I-like protein 26 3.6
At3g21610 unknown protein 26 3.6
At2g17720 similar to prolyl 4-hydroxylase alpha subunit 25 4.7
At5g45650 subtilisin-like protease 25 6.2
At1g77280 hypothetical protein 25 6.2
At1g10400 glucosyl transferase - like protein 25 6.2
At3g15400 anther development protein, ATA20 25 8.1
At1g23760 putative polygalacuronase isoenzyme 1 beta subunit 25 8.1
>rpl16 -chloroplast genome- ribosomal protein L16
Length = 135
Score = 220 bits (560), Expect = 1e-58
Identities = 107/119 (89%), Positives = 113/119 (94%)
Query: 1 MKGKAYRGNTISFGKYALQALEPAWITSRQIEAGRRAMSRNVRRGGKIWVRIFPDKPVTV 60
+KG + RGN I FG+YALQ LEPAWITSRQIEAGRRAM+RNVRRGGKIWVRIFPDKPVTV
Sbjct: 17 LKGISSRGNRICFGRYALQTLEPAWITSRQIEAGRRAMTRNVRRGGKIWVRIFPDKPVTV 76
Query: 61 RPTETRMGSGKGSPEYWVAVVKPGKILYEMGGVPENIARKAISIAASKMPIRTQFILSE 119
RP ETRMGSGKGSPEYWVAVVKPGKILYEMGGVPENIARKAISIAASKMPI+TQFI+SE
Sbjct: 77 RPAETRMGSGKGSPEYWVAVVKPGKILYEMGGVPENIARKAISIAASKMPIKTQFIISE 135
>At2g28820 unknown protein
Length = 291
Score = 164 bits (414), Expect = 9e-42
Identities = 82/118 (69%), Positives = 98/118 (82%), Gaps = 2/118 (1%)
Query: 2 KGKAYRGNTISFGKYALQALEPAWITSRQIEAGRRAMSRNVRRGGKIWVRIFPDKPVTVR 61
KG + +G S +YALQ LEPAWITSRQIEAGRRAM+RN+ RG + V IF DKPVTVR
Sbjct: 176 KGVSSQGYICS--RYALQTLEPAWITSRQIEAGRRAMTRNIGRGLTVRVHIFADKPVTVR 233
Query: 62 PTETRMGSGKGSPEYWVAVVKPGKILYEMGGVPENIARKAISIAASKMPIRTQFILSE 119
P ETRMG GKG+P +WVAVVKPGKI+YEMGGV E +AR+AISIAASK+P +T+FI+S+
Sbjct: 234 PPETRMGRGKGAPAFWVAVVKPGKIIYEMGGVSEKVAREAISIAASKLPAKTKFIISK 291
>Atmg00080 -mitochondrial genome- ribosomal protein L16 (RPL16)
Length = 179
Score = 97.8 bits (242), Expect = 7e-22
Identities = 52/117 (44%), Positives = 71/117 (60%), Gaps = 8/117 (6%)
Query: 8 GNTISFGKYALQALEPAWITSRQIEAGRRA--------MSRNVRRGGKIWVRIFPDKPVT 59
G + FG+Y ++ + ++ R IEA RRA MS RR GKIWVR+F D P+T
Sbjct: 60 GTKLGFGRYGTKSCKAGRLSYRAIEAARRAIIGHFHRAMSGQFRRNGKIWVRVFADLPIT 119
Query: 60 VRPTETRMGSGKGSPEYWVAVVKPGKILYEMGGVPENIARKAISIAASKMPIRTQFI 116
+PTE RMG GKG+P W+A V G+I +EM GV AR+A ++AA K T+F+
Sbjct: 120 GKPTEVRMGRGKGNPTGWIARVSTGQIPFEMDGVSLANARQAATLAAHKPCSSTKFV 176
>At1g78060 putative protein
Length = 767
Score = 27.3 bits (59), Expect = 1.2
Identities = 21/64 (32%), Positives = 30/64 (46%), Gaps = 5/64 (7%)
Query: 32 EAGRRAMSRNV----RRGGKIWVRIFPDKPVTVRPTETRMGSGKGSPEYWVAVVKPGKIL 87
EAG A+S + GG++ V +P V ++ T+ RM S G P K K+
Sbjct: 558 EAGGIAISEIIFGDHNPGGRLPVTWYPQSFVNIQMTDMRMRSATGYPGRTYKFYKGPKV- 616
Query: 88 YEMG 91
YE G
Sbjct: 617 YEFG 620
>At3g20650 unknown protein
Length = 370
Score = 26.6 bits (57), Expect = 2.1
Identities = 14/59 (23%), Positives = 26/59 (43%), Gaps = 1/59 (1%)
Query: 20 ALEPAWITSRQIEAGRRAMSRNVRRGGKIWVRIFPDKPVTVRPTETRMGSGKGSPEYWV 78
A+ +W T + +S +R GG +++ PD V ++ G G+ YW+
Sbjct: 174 AMHYSWTTEARARRALANVSALLRPGG-VFIGTMPDANVIIKKLREAEGLEIGNSVYWI 231
>At4g26110 nucleosome assembly protein I-like protein
Length = 372
Score = 25.8 bits (55), Expect = 3.6
Identities = 11/44 (25%), Positives = 19/44 (43%)
Query: 46 GKIWVRIFPDKPVTVRPTETRMGSGKGSPEYWVAVVKPGKILYE 89
G V + P+ V E + KG P +W+ +K ++ E
Sbjct: 100 GTTEVELAPEDDTKVDQGEEKTAEEKGVPSFWLTALKNNDVISE 143
>At3g21610 unknown protein
Length = 194
Score = 25.8 bits (55), Expect = 3.6
Identities = 19/80 (23%), Positives = 30/80 (36%), Gaps = 14/80 (17%)
Query: 22 EPAWITSRQIEAGRRAMSRNVRRGGKIWVRIFPDKPVTVRPTETRMGSGKGSPEYWVAVV 81
E W + R I +G S + VT G G+P + +AVV
Sbjct: 58 EKRWDSKRMISSGGMPSSHSAT--------------VTALAVAIGFEEGAGAPAFAIAVV 103
Query: 82 KPGKILYEMGGVPENIARKA 101
++Y+ GV + R+A
Sbjct: 104 LACVVMYDASGVRLHAGRQA 123
>At2g17720 similar to prolyl 4-hydroxylase alpha subunit
Length = 291
Score = 25.4 bits (54), Expect = 4.7
Identities = 12/31 (38%), Positives = 19/31 (60%), Gaps = 2/31 (6%)
Query: 60 VRPTETRMGSGKGSPEYWVAVV--KPGKILY 88
VR +ET G +G+ E WV V+ +P ++Y
Sbjct: 62 VRKSETSSGDEEGNGERWVEVISWEPRAVVY 92
>At5g45650 subtilisin-like protease
Length = 791
Score = 25.0 bits (53), Expect = 6.2
Identities = 12/31 (38%), Positives = 17/31 (54%)
Query: 56 KPVTVRPTETRMGSGKGSPEYWVAVVKPGKI 86
K VTV+ T T +G+G + Y +V P I
Sbjct: 698 KTVTVKRTVTNVGTGNSTSTYLFSVKPPSGI 728
>At1g77280 hypothetical protein
Length = 781
Score = 25.0 bits (53), Expect = 6.2
Identities = 11/26 (42%), Positives = 15/26 (57%)
Query: 71 KGSPEYWVAVVKPGKILYEMGGVPEN 96
K S + WV V GKIL++ G P +
Sbjct: 172 KLSKDCWVIAVNNGKILFQKEGSPSS 197
>At1g10400 glucosyl transferase - like protein
Length = 467
Score = 25.0 bits (53), Expect = 6.2
Identities = 13/29 (44%), Positives = 18/29 (61%)
Query: 2 KGKAYRGNTISFGKYALQALEPAWITSRQ 30
KGK R N ++GK A +ALE +SR+
Sbjct: 425 KGKELRRNVEAYGKMAKKALEEGIGSSRK 453
>At3g15400 anther development protein, ATA20
Length = 416
Score = 24.6 bits (52), Expect = 8.1
Identities = 20/55 (36%), Positives = 25/55 (45%), Gaps = 5/55 (9%)
Query: 50 VRIFPDKPV----TVRPTETRMGSGKGSPEYWVAVVKPGKILYEMGGVPENIARK 100
V + PD V TV+P T + S G E+ VKPG L M G E+ K
Sbjct: 50 VMLQPDHEVQPEHTVKPGHT-LQSMSGVSEHPEHTVKPGHTLQSMSGALEHPEHK 103
>At1g23760 putative polygalacuronase isoenzyme 1 beta subunit
Length = 622
Score = 24.6 bits (52), Expect = 8.1
Identities = 13/53 (24%), Positives = 22/53 (40%)
Query: 4 KAYRGNTISFGKYALQALEPAWITSRQIEAGRRAMSRNVRRGGKIWVRIFPDK 56
K Y + F KY +L ++ +E G+ ++ G IW+ DK
Sbjct: 375 KDYTKTGVEFAKYNRSSLGGGKTVNKWVEPGKFFRESMLKEGTLIWMPDIKDK 427
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,614,543
Number of Sequences: 26719
Number of extensions: 101187
Number of successful extensions: 165
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 153
Number of HSP's gapped (non-prelim): 15
length of query: 119
length of database: 11,318,596
effective HSP length: 95
effective length of query: 24
effective length of database: 8,780,291
effective search space: 210726984
effective search space used: 210726984
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC142142.2


