

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142095.2 + phase: 0
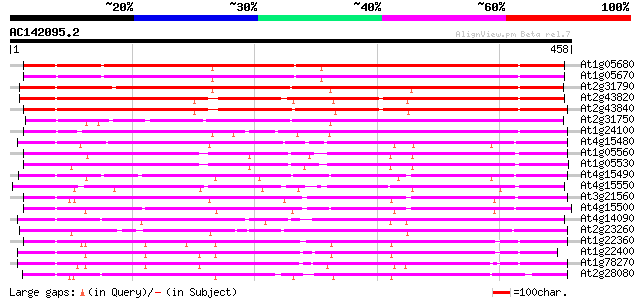
(458 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g05680 indole-3-acetate beta-glucosyltransferase like protein 351 5e-97
At1g05670 UDP glycosyltransferase UGT74E1 350 1e-96
At2g31790 putative glucosyltransferase 339 2e-93
At2g43820 putative glucosyltransferase 332 3e-91
At2g43840 putative glucosyltransferase 328 4e-90
At2g31750 putative glucosyltransferase 324 7e-89
At1g24100 putative indole-3-acetate beta-glucosyltransferase 299 2e-81
At4g15480 indole-3-acetate beta-glucosyltransferase like protein 271 4e-73
At1g05560 UDP-Glucose Transferase (UGT75B2) 253 2e-67
At1g05530 hypothetical protein 249 3e-66
At4g15490 indole-3-acetate beta-glucosyltransferase like protein 248 4e-66
At4g15550 UDP-glucose:indole-3-acetate beta-D-glucosyltransferas... 231 5e-61
At3g21560 UDP-glucose:indole-3-acetate beta-D-glucosyltransferas... 228 7e-60
At4g15500 indole-3-acetate beta-glucosyltransferase like protein 227 1e-59
At4g14090 glucosyltransferase like protein 225 4e-59
At2g23260 putative glucosyltransferase 218 5e-57
At1g22360 unknown protein 216 2e-56
At1g22400 Putative UDP-glucose glucosyltransferase 214 8e-56
At1g78270 similar to glucoronosyl transferase-like protein; simi... 206 2e-53
At2g28080 putative glucosyltransferase 203 1e-52
>At1g05680 indole-3-acetate beta-glucosyltransferase like protein
Length = 453
Score = 351 bits (900), Expect = 5e-97
Identities = 186/449 (41%), Positives = 273/449 (60%), Gaps = 11/449 (2%)
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIETISDGF 71
H +VLPFP GH PM +F K L + G+K+TLV K ++SIT+ IS+GF
Sbjct: 6 HLIVLPFPGQGHITPMSQFCKRLASK-GLKLTLVLVSDKPSPPYKTEHDSITVFPISNGF 64
Query: 72 DKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAKEFG 131
+G +D Y+ + +L L+ ++ + ++YDS MPW LDVA +G
Sbjct: 65 QEGE-EPLQDLDDYMERVETSIKNTLPKLVEDMKLSGNPPRAIVYDSTMPWLLDVAHSYG 123
Query: 132 IVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQE----ITLPALPQLQPRDMPSFYFTYEQ 187
+ GA F TQ ++ +IYYHV G P + + P+ P L D+PSF
Sbjct: 124 LSGAVFFTQPWLVTAIYYHVFKGSFSVPSTKYGHSTLASFPSFPMLTANDLPSFLCESSS 183
Query: 188 DPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLPYTFLDKRV 247
P L I V Q SNI + D +LCN+F +LE+++ W +W +GP +P +LDKR+
Sbjct: 184 YPNILRIVVDQLSNIDRVDIVLCNTFDKLEEKLLKWVQSLWPVLN-IGPTVPSMYLDKRL 242
Query: 248 KDDEDH--SIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCGSYF 305
+D+++ S+ K E +EWLN+K S VY+SFGS+ L E+Q+ E+A LK G +F
Sbjct: 243 SEDKNYGFSLFNAKVAECMEWLNSKEPNSVVYLSFGSLVILKEDQMLELAAGLKQSGRFF 302
Query: 306 LWVVKTSEETKLPKDF-EKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIG 364
LWVV+ +E KLP+++ E+ E GL+V+W PQL+VLAH++IGCF+THCGWNSTLE LS+G
Sbjct: 303 LWVVRETETHKLPRNYVEEIGEKGLIVSWSPQLDVLAHKSIGCFLTHCGWNSTLEGLSLG 362
Query: 365 VPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIM 424
VP++ +P ++DQ +AKF+ D+WKVG+R + VR++ + + E+M EKGKEI
Sbjct: 363 VPMIGMPHWTDQPTNAKFMQDVWKVGVRVKAEGDGFVRREEIMRSVEEVME-GEKGKEIR 421
Query: 425 NNVMQWKTLATRAVGKDGSSHKNMIEFVN 453
N +WK LA AV + GSS K++ EFV+
Sbjct: 422 KNAEKWKVLAQEAVSEGGSSDKSINEFVS 450
>At1g05670 UDP glycosyltransferase UGT74E1
Length = 453
Score = 350 bits (897), Expect = 1e-96
Identities = 188/449 (41%), Positives = 272/449 (59%), Gaps = 11/449 (2%)
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIETISDGF 71
H +VLPFPA GH PM +F K L + +K+TLV K +++IT+ IS+GF
Sbjct: 6 HVIVLPFPAQGHITPMSQFCKRLASKS-LKITLVLVSDKPSPPYKTEHDTITVVPISNGF 64
Query: 72 DKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAKEFG 131
+G ++D Y+ + L LI ++ + L+YDS MPW LDVA +G
Sbjct: 65 QEGQ-ERSEDLDEYMERVESSIKNRLPKLIEDMKLSGNPPRALVYDSTMPWLLDVAHSYG 123
Query: 132 IVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQE----ITLPALPQLQPRDMPSFYFTYEQ 187
+ GA F TQ ++++IYYHV G P + + P+LP L D+PSF
Sbjct: 124 LSGAVFFTQPWLVSAIYYHVFKGSFSVPSTKYGHSTLASFPSLPILNANDLPSFLCESSS 183
Query: 188 DPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLPYTFLDKRV 247
P L + Q SNI + D +LCN+F +LE+++ W +W +GP +P +LDKR+
Sbjct: 184 YPYILRTVIDQLSNIDRVDIVLCNTFDKLEEKLLKWIKSVWPVLN-IGPTVPSMYLDKRL 242
Query: 248 KDDEDH--SIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCGSYF 305
+D+++ S+ K E +EWLN+K S VYVSFGS+ L ++Q+ E+A LK G +F
Sbjct: 243 AEDKNYGFSLFGAKIAECMEWLNSKQPSSVVYVSFGSLVVLKKDQLIELAAGLKQSGHFF 302
Query: 306 LWVVKTSEETKLPKDF-EKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIG 364
LWVV+ +E KLP+++ E+ E GL V+W PQLEVL H++IGCFVTHCGWNSTLE LS+G
Sbjct: 303 LWVVRETERRKLPENYIEEIGEKGLTVSWSPQLEVLTHKSIGCFVTHCGWNSTLEGLSLG 362
Query: 365 VPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSEKGKEIM 424
VP++ +P ++DQ +AKF+ D+WKVG+R D VR++ + E+M +E+GKEI
Sbjct: 363 VPMIGMPHWADQPTNAKFMEDVWKVGVRVKADSDGFVRREEFVRRVEEVME-AEQGKEIR 421
Query: 425 NNVMQWKTLATRAVGKDGSSHKNMIEFVN 453
N +WK LA AV + GSS KN+ EFV+
Sbjct: 422 KNAEKWKVLAQEAVSEGGSSDKNINEFVS 450
>At2g31790 putative glucosyltransferase
Length = 457
Score = 339 bits (869), Expect = 2e-93
Identities = 182/454 (40%), Positives = 273/454 (60%), Gaps = 15/454 (3%)
Query: 9 KSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIETIS 68
K H L P+P GH NPM++ +K L ++ G+ TL+ +++ + SIT+ TI
Sbjct: 5 KKGHVLFFPYPLQGHINPMIQLAKRLSKK-GITSTLIIASKDHREPYTSDDYSITVHTIH 63
Query: 69 DGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAK 128
DGF AK L ++F +SL I++ ++ LIYD FMP+ LD+AK
Sbjct: 64 DGFFPHEHPHAKFVDL--DRFHNSTSRSLTDFISSAKLSDNPPKALIYDPFMPFALDIAK 121
Query: 129 EFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQE----ITLPALPQLQPRDMPSFYFT 184
+ + ++ TQ + + +YYH++ G P E + P P L D+PSF
Sbjct: 122 DLDLYVVAYFTQPWLASLVYYHINEGTYDVPVDRHENPTLASFPGFPLLSQDDLPSFACE 181
Query: 185 YEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLPYTFLD 244
P + V QFSN+ +AD ILCN+F +LE +V W W + +GP +P FLD
Sbjct: 182 KGSYPLLHEFVVRQFSNLLQADCILCNTFDQLEPKVVKWMNDQWP-VKNIGPVVPSKFLD 240
Query: 245 KRVKDDEDHSIAQLKS--DESI-EWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDC 301
R+ +D+D+ + K+ DES+ +WL N+P +S VYV+FG++ +L+E+Q++E+A +
Sbjct: 241 NRLPEDKDYELENSKTEPDESVLKWLGNRPAKSVVYVAFGTLVALSEKQMKEIAMAISQT 300
Query: 302 GSYFLWVVKTSEETKLPKDFEKKSE---NGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTL 358
G +FLW V+ SE +KLP F +++E +GLV W PQLEVLAHE+IGCFV+HCGWNSTL
Sbjct: 301 GYHFLWSVRESERSKLPSGFIEEAEEKDSGLVAKWVPQLEVLAHESIGCFVSHCGWNSTL 360
Query: 359 EALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIMSMSE 418
EAL +GVP+V +P ++DQ +AKF+ D+WK+G+R D + + K+ + CI E+M E
Sbjct: 361 EALCLGVPMVGVPQWTDQPTNAKFIEDVWKIGVRVRTDGEGLSSKEEIARCIVEVME-GE 419
Query: 419 KGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFV 452
+GKEI NV + K LA A+ + GSS K + EFV
Sbjct: 420 RGKEIRKNVEKLKVLAREAISEGGSSDKKIDEFV 453
>At2g43820 putative glucosyltransferase
Length = 449
Score = 332 bits (850), Expect = 3e-91
Identities = 179/456 (39%), Positives = 276/456 (60%), Gaps = 26/456 (5%)
Query: 9 KSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIETIS 68
K H L +P+P GH P +F K L + G+K TL T + +I + I+I TIS
Sbjct: 4 KRGHVLAVPYPTQGHITPFRQFCKRLHFK-GLKTTLALTTFVFNSINPDLSGPISIATIS 62
Query: 69 DGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAK 128
DG+D GG A YL F G +++A +I ++ + C++YD+F+PW LDVA+
Sbjct: 63 DGYDHGGFETADSIDDYLKDFKTSGSKTIADIIQKHQTSDNPITCIVYDAFLPWALDVAR 122
Query: 129 EFGIVGASFLTQNLVMNSIYY--HVHLGKLKPPFVEQEITLPALPQLQPRDMPSFYFTYE 186
EFG+V F TQ +N +YY +++ G L+ P E LP L+ +D+PSF+
Sbjct: 123 EFGLVATPFFTQPCAVNYVYYLSYINNGSLQLPIEE-------LPFLELQDLPSFFSVSG 175
Query: 187 QDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSN---FRTVGPCLPYTFL 243
P + ++ + QF N KAD++L NSF ELE + +WS T+GP +P +L
Sbjct: 176 SYPAYFEMVLQQFINFEKADFVLVNSFQELELHENE----LWSKACPVLTIGPTIPSIYL 231
Query: 244 DKRVKDDEDHSIAQLKSDES---IEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKD 300
D+R+K D + + +S + I WL+ +P+ S VYV+FGSMA L Q+EE+A + +
Sbjct: 232 DQRIKSDTGYDLNLFESKDDSFCINWLDTRPQGSVVYVAFGSMAQLTNVQMEELASAVSN 291
Query: 301 CGSYFLWVVKTSEETKLPKDFEK--KSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTL 358
FLWVV++SEE KLP F + E LV+ W PQL+VL+++AIGCF+THCGWNST+
Sbjct: 292 FS--FLWVVRSSEEEKLPSGFLETVNKEKSLVLKWSPQLQVLSNKAIGCFLTHCGWNSTM 349
Query: 359 EALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVD-EKQIVRKDPLKDCICEIMSMS 417
EAL+ GVP+VA+P ++DQ ++AK++ D+WK G+R + E I +++ ++ I E+M
Sbjct: 350 EALTFGVPMVAMPQWTDQPMNAKYIQDVWKAGVRVKTEKESGIAKREEIEFSIKEVME-G 408
Query: 418 EKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVN 453
E+ KE+ NV +W+ LA +++ + GS+ N+ FV+
Sbjct: 409 ERSKEMKKNVKKWRDLAVKSLNEGGSTDTNIDTFVS 444
>At2g43840 putative glucosyltransferase
Length = 449
Score = 328 bits (841), Expect = 4e-90
Identities = 172/452 (38%), Positives = 279/452 (61%), Gaps = 20/452 (4%)
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIETISDGF 71
H L +PFP+ GH P+ +F K L + G K T T + I P++ I+I TISDG+
Sbjct: 7 HVLAVPFPSQGHITPIRQFCKRLHSK-GFKTTHTLTTFIFNTIHLDPSSPISIATISDGY 65
Query: 72 DKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAKEFG 131
D+GG + A YL F G +++A +I + ++ + C++YDSFMPW LD+A +FG
Sbjct: 66 DQGGFSSAGSVPEYLQNFKTFGSKTVADIIRKHQSTDNPITCIVYDSFMPWALDLAMDFG 125
Query: 132 IVGASFLTQNLVMNSIYY--HVHLGKLKPPFVEQEITLPALPQLQPRDMPSFYFTYEQDP 189
+ A F TQ+ +N I Y +++ G L P + LP L+ +D+P+F
Sbjct: 126 LAAAPFFTQSCAVNYINYLSYINNGSLTLPIKD-------LPLLELQDLPTFVTPTGSHL 178
Query: 190 TFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLPYTFLDKRVKD 249
+ ++ + QF+N KAD++L NSF +L+ + K+ T+GP +P +LD+++K
Sbjct: 179 AYFEMVLQQFTNFDKADFVLVNSFHDLDLHEEELLSKVCPVL-TIGPTVPSMYLDQQIKS 237
Query: 250 DEDHSIAQLKSDESI---EWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCGSYFL 306
D D+ + E+ +WL+ +P+ S VY++FGSMA L+ EQ+EE+A + + +L
Sbjct: 238 DNDYDLNLFDLKEAALCTDWLDKRPEGSVVYIAFGSMAKLSSEQMEEIASAISNFS--YL 295
Query: 307 WVVKTSEETKLPKDFEKK--SENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIG 364
WVV+ SEE+KLP F + + LV+ W PQL+VL+++AIGCF+THCGWNST+E LS+G
Sbjct: 296 WVVRASEESKLPPGFLETVDKDKSLVLKWSPQLQVLSNKAIGCFMTHCGWNSTMEGLSLG 355
Query: 365 VPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVD-EKQIVRKDPLKDCICEIMSMSEKGKEI 423
VP+VA+P ++DQ ++AK++ D+WKVG+R + E I +++ ++ I E+M EK KE+
Sbjct: 356 VPMVAMPQWTDQPMNAKYIQDVWKVGVRVKAEKESGICKREEIEFSIKEVME-GEKSKEM 414
Query: 424 MNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
N +W+ LA +++ + GS+ N+ EFV+ +
Sbjct: 415 KENAGKWRDLAVKSLSEGGSTDININEFVSKI 446
>At2g31750 putative glucosyltransferase
Length = 456
Score = 324 bits (830), Expect = 7e-89
Identities = 184/452 (40%), Positives = 270/452 (59%), Gaps = 23/452 (5%)
Query: 14 LVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNS------ITIETI 67
LV FP GH NP+L+FSK L + V VT +TT S + +I + ++ I
Sbjct: 10 LVFSFPIQGHINPLLQFSKRLLSKN-VNVTFLTTSSTHNSILRRAITGGATALPLSFVPI 68
Query: 68 SDGF--DKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLD 125
DGF D + D Y KF + +SL+ LI++++ + + V +YDS +P+ LD
Sbjct: 69 DDGFEEDHPSTDTSPD---YFAKFQENVSRSLSELISSMDPKPNAV---VYDSCLPYVLD 122
Query: 126 VAKEF-GIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPRDMPSFYFT 184
V ++ G+ ASF TQ+ +N+ Y H G+ K + ++ LPA+P L+ D+P F +
Sbjct: 123 VCRKHPGVAAASFFTQSSTVNATYIHFLRGEFKE--FQNDVVLPAMPPLKGNDLPVFLYD 180
Query: 185 YEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLPYTFLD 244
++ +QF N+ D+ L NSF ELE EV W W + +GP +P +LD
Sbjct: 181 NNLCRPLFELISSQFVNVDDIDFFLVNSFDELEVEVLQWMKNQWP-VKNIGPMIPSMYLD 239
Query: 245 KRVKDDEDHSIAQLKS--DESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCG 302
KR+ D+D+ I + +E ++WL++KP S +YVSFGS+A L ++Q+ EVA LK G
Sbjct: 240 KRLAGDKDYGINLFNAQVNECLDWLDSKPPGSVIYVSFGSLAVLKDDQMIEVAAGLKQTG 299
Query: 303 SYFLWVVKTSEETKLPKDF-EKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEAL 361
FLWVV+ +E KLP ++ E + GL+V W PQL+VLAH++IGCF+THCGWNSTLEAL
Sbjct: 300 HNFLWVVRETETKKLPSNYIEDICDKGLIVNWSPQLQVLAHKSIGCFMTHCGWNSTLEAL 359
Query: 362 SIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCICEIM-SMSEKG 420
S+GV ++ +P YSDQ +AKF+ D+WKVG+R D+ V K+ + C+ E+M MSEKG
Sbjct: 360 SLGVALIGMPAYSDQPTNAKFIEDVWKVGVRVKADQNGFVPKEEIVRCVGEVMEDMSEKG 419
Query: 421 KEIMNNVMQWKTLATRAVGKDGSSHKNMIEFV 452
KEI N + A A+ G+S KN+ EFV
Sbjct: 420 KEIRKNARRLMEFAREALSDGGNSDKNIDEFV 451
>At1g24100 putative indole-3-acetate beta-glucosyltransferase
Length = 460
Score = 299 bits (765), Expect = 2e-81
Identities = 179/457 (39%), Positives = 258/457 (56%), Gaps = 22/457 (4%)
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSITIETISDGF 71
H ++LP+P GH NPM++F+K L + VKVT+ TT +I S+++E ISDGF
Sbjct: 11 HVVILPYPVQGHLNPMVQFAKRLVSKN-VKVTIATTTYTASSIT---TPSLSVEPISDGF 66
Query: 72 DKGGVA-EAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAKEF 130
D + Y F G ++L LI + + +DCLIYDSF+PW L+VA+
Sbjct: 67 DFIPIGIPGFSVDTYSESFKLNGSETLTLLIEKFKSTDSPIDCLIYDSFLPWGLEVARSM 126
Query: 131 GIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQE--ITLPALPQLQPRDMPSF----YFT 184
+ ASF T NL + S+ G P + LP L ++PSF + T
Sbjct: 127 ELSAASFFTNNLTVCSVLRKFSNGDFPLPADPNSAPFRIRGLPSLSYDELPSFVGRHWLT 186
Query: 185 YEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRT--VGPCLPYTF 242
+ + L + QF N ADW+ N F LE E D + +GP +P +
Sbjct: 187 HPEHGRVL---LNQFPNHENADWLFVNGFEGLE-ETQDCENGESDAMKATLIGPMIPSAY 242
Query: 243 LDKRVKDDEDHSIAQLK--SDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKD 300
LD R++DD+D+ + LK S E +EWL K +S +VSFGS L E+Q+ EVA L++
Sbjct: 243 LDDRMEDDKDYGASLLKPISKECMEWLETKQAQSVAFVSFGSFGILFEKQLAEVAIALQE 302
Query: 301 CGSYFLWVVKTSEETKLPKDF-EKKSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLE 359
FLWV+K + KLP+ F E + L+V+WC QLEVLAHE+IGCF+THCGWNSTLE
Sbjct: 303 SDLNFLWVIKEAHIAKLPEGFVESTKDRALLVSWCNQLEVLAHESIGCFLTHCGWNSTLE 362
Query: 360 ALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQ-IVRKDPLKDCICEIMSMSE 418
LS+GVP+V +P +SDQ DAKF+ ++WKVG R + + IV+ + L C+ +M E
Sbjct: 363 GLSLGVPMVGVPQWSDQMNDAKFVEEVWKVGYRAKEEAGEVIVKSEELVRCLKGVME-GE 421
Query: 419 KGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
+I + +WK LA +A+ + GSS +++ EF+ SL
Sbjct: 422 SSVKIRESSKKWKDLAVKAMSEGGSSDRSINEFIESL 458
>At4g15480 indole-3-acetate beta-glucosyltransferase like protein
Length = 490
Score = 271 bits (694), Expect = 4e-73
Identities = 166/473 (35%), Positives = 256/473 (54%), Gaps = 32/473 (6%)
Query: 7 STKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPK---------- 56
S +H +++ F GH NP+L KL+ + G+ VT VTT K + +
Sbjct: 14 SPNPIHVMLVSFQGQGHVNPLLRLGKLIASK-GLLVTFVTTELWGKKMRQANKIVDGELK 72
Query: 57 -LPNNSITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLI 115
+ + SI E + + + A DF LY+ VG + ++ L+ N+ V CLI
Sbjct: 73 PVGSGSIRFEFFDEEWAEDDDRRA-DFSLYIAHLESVGIREVSKLVRRYEEANEPVSCLI 131
Query: 116 YDSFMPWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFV---EQEITLPALPQ 172
+ F+PW VA+EF I A Q+ S YYH G + P E ++ LP +P
Sbjct: 132 NNPFIPWVCHVAEEFNIPCAVLWVQSCACFSAYYHYQDGSVSFPTETEPELDVKLPCVPV 191
Query: 173 LQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFR 232
L+ ++PSF + F + QF N+ K+ +L +SF LE+EV D+ M +
Sbjct: 192 LKNDEIPSFLHPSSRFTGFRQAILGQFKNLSKSFCVLIDSFDSLEQEVIDY-MSSLCPVK 250
Query: 233 TVGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIE 292
TVGP + V D I + +D+ +EWL+++PK S VY+SFG++A L +EQIE
Sbjct: 251 TVGPLFKVA---RTVTSDVSGDICK-STDKCLEWLDSRPKSSVVYISFGTVAYLKQEQIE 306
Query: 293 EVAHCLKDCGSYFLWVVKTS------EETKLPKDFEKKSENG--LVVAWCPQLEVLAHEA 344
E+AH + G FLWV++ E LP++ ++ S G ++V WCPQ +VL+H +
Sbjct: 307 EIAHGVLKSGLSFLWVIRPPPHDLKVETHVLPQELKESSAKGKGMIVDWCPQEQVLSHPS 366
Query: 345 IGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIR--PLVDEKQIVR 402
+ CFVTHCGWNST+E+LS GVP+V P + DQ DA +L+D++K G+R E+++V
Sbjct: 367 VACFVTHCGWNSTMESLSSGVPVVCCPQWGDQVTDAVYLIDVFKTGVRLGRGATEERVVP 426
Query: 403 KDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
++ + + + E ++ EK +E+ N ++WK A AV GSS KN EFV L
Sbjct: 427 REEVAEKLLE-ATVGEKAEELRKNALKWKAEAEAAVAPGGSSDKNFREFVEKL 478
>At1g05560 UDP-Glucose Transferase (UGT75B2)
Length = 469
Score = 253 bits (646), Expect = 2e-67
Identities = 163/466 (34%), Positives = 242/466 (50%), Gaps = 41/466 (8%)
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNSI---TIETIS 68
H L++ FPA GH NP L F++ L ++ G +VT VT +S + N +N + + T S
Sbjct: 5 HFLLVTFPAQGHVNPSLRFARRLIKRTGARVTFVTCVSVFHNSMIANHNKVENLSFLTFS 64
Query: 69 DGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAK 128
DGFD GG++ +D + G ++L+ I + V CLIY + W VA+
Sbjct: 65 DGFDDGGISTYEDRQKRSVNLKVNGDKALSDFIEATKNGDSPVTCLIYTILLNWAPKVAR 124
Query: 129 EFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPRDMPSFYFTYEQD 188
F + A Q ++ +IYY +G + LP L L+ RD+PSF +
Sbjct: 125 RFQLPSALLWIQPALVFNIYYTHFMGN------KSVFELPNLSSLEIRDLPSFLTPSNTN 178
Query: 189 PTFLDI--GVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLPYTFL--- 243
D + +F IL N+F LE E I + VGP LP
Sbjct: 179 KGAYDAFQEMMEFLIKETKPKILINTFDSLEPEALTAFPNI--DMVAVGPLLPTEIFSGS 236
Query: 244 -DKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCG 302
+K VKD +S WL++K + S +YVSFG+M L+++QIEE+A L +
Sbjct: 237 TNKSVKD---------QSSSYTLWLDSKTESSVIYVSFGTMVELSKKQIEELARALIEGK 287
Query: 303 SYFLWVV--KTSEETKLPKDFEKKSEN-----------GLVVAWCPQLEVLAHEAIGCFV 349
FLWV+ K++ ETK + E + E G++V+WC Q+EVL+H A+GCFV
Sbjct: 288 RPFLWVITDKSNRETKTEGEEETEIEKIAGFRHELEEVGMIVSWCSQIEVLSHRAVGCFV 347
Query: 350 THCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDC 409
THCGW+STLE+L +GVP+VA P++SDQ +AK L + WK G+R ++ +V + ++ C
Sbjct: 348 THCGWSSTLESLVLGVPVVAFPMWSDQPTNAKLLEESWKTGVRVRENKDGLVERGEIRRC 407
Query: 410 ICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
+ + M EK E+ N +WK LA A + GSS KNM FV +
Sbjct: 408 LEAV--MEEKSVELRENAKKWKRLAMEAGREGGSSDKNMEAFVEDI 451
>At1g05530 hypothetical protein
Length = 455
Score = 249 bits (635), Expect = 3e-66
Identities = 163/466 (34%), Positives = 235/466 (49%), Gaps = 36/466 (7%)
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTIS--NYKNIPKLPN-NSITIETIS 68
H L++ FPA GH NP L F++ L + G +VT T +S + IP N +++ T S
Sbjct: 5 HFLLVTFPAQGHVNPSLRFARRLIKTTGARVTFATCLSVIHRSMIPNHNNVENLSFLTFS 64
Query: 69 DGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLDVAK 128
DGFD G ++ D + L F + G ++L+ I + V CLIY W VA+
Sbjct: 65 DGFDDGVISNTDDVQNRLVHFERNGDKALSDFIEANQNGDSPVSCLIYTILPNWVPKVAR 124
Query: 129 EFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPRDMPSFYFTYEQD 188
F + Q IYY+ G P LP L+ RD+PSF +
Sbjct: 125 RFHLPSVHLWIQPAFAFDIYYNYSTGN------NSVFEFPNLPSLEIRDLPSFLSPSNTN 178
Query: 189 PTFLDI--GVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCLP---YTFL 243
+ + F IL N+F LE E I VGP LP +T
Sbjct: 179 KAAQAVYQELMDFLKEESNPKILVNTFDSLEPEFLTAIPNI--EMVAVGPLLPAEIFTGS 236
Query: 244 DKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLKDCGS 303
+ DH +S WL++K + S +YVSFG+M L+++QIEE+A L + G
Sbjct: 237 ESGKDLSRDH-----QSSSYTLWLDSKTESSVIYVSFGTMVELSKKQIEELARALIEGGR 291
Query: 304 YFLWVV--KTSEETKLPKDFEKKSEN-----------GLVVAWCPQLEVLAHEAIGCFVT 350
FLWV+ K + E K+ + E + E G++V+WC Q+EVL H AIGCF+T
Sbjct: 292 PFLWVITDKLNREAKIEGEEETEIEKIAGFRHELEEVGMIVSWCSQIEVLRHRAIGCFLT 351
Query: 351 HCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCI 410
HCGW+S+LE+L +GVP+VA P++SDQ +AK L +IWK G+R + + +V + + C+
Sbjct: 352 HCGWSSSLESLVLGVPVVAFPMWSDQPANAKLLEEIWKTGVRVRENSEGLVERGEIMRCL 411
Query: 411 CEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSLF 456
+ M K E+ N +WK LAT A + GSS KN+ FV SLF
Sbjct: 412 EAV--MEAKSVELRENAEKWKRLATEAGREGGSSDKNVEAFVKSLF 455
>At4g15490 indole-3-acetate beta-glucosyltransferase like protein
Length = 479
Score = 248 bits (634), Expect = 4e-66
Identities = 162/473 (34%), Positives = 247/473 (51%), Gaps = 35/473 (7%)
Query: 8 TKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNS------ 61
++ H +++ FP GH NP+L KL+ + G+ VT VTT + + N
Sbjct: 4 SRHTHVMLVSFPGQGHVNPLLRLGKLIASK-GLLVTFVTTEKPWGKKMRQANKIQDGVLK 62
Query: 62 ------ITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLI 115
I E SDGF + DF + VG Q + +L+ N + V CLI
Sbjct: 63 PVGLGFIRFEFFSDGFADDD-EKRFDFDAFRPHLEAVGKQEIKNLVKRYN--KEPVTCLI 119
Query: 116 YDSFMPWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLK-PPFVEQEIT--LPALPQ 172
++F+PW DVA+E I A Q+ + YY+ H +K P E +I+ +P LP
Sbjct: 120 NNAFVPWVCDVAEELHIPSAVLWVQSCACLTAYYYYHHRLVKFPTKTEPDISVEIPCLPL 179
Query: 173 LQPRDMPSFYFTYEQDPTFLDIGVAQFSNI--HKADWILCNSFFELEKEVADWTMKIWSN 230
L+ ++PSF F DI + Q HK+ ++ ++F ELEK++ D ++
Sbjct: 180 LKHDEIPSFLHPSSPYTAFGDIILDQLKRFENHKSFYLFIDTFRELEKDIMDHMSQLCPQ 239
Query: 231 FRTVGPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQ 290
+ P P + + + D I++ SD +EWL+++ S VY+SFG++A+L +EQ
Sbjct: 240 -AIISPVGPLFKMAQTLSSDVKGDISEPASD-CMEWLDSREPSSVVYISFGTIANLKQEQ 297
Query: 291 IEEVAHCLKDCGSYFLWVVKTSEETK------LPKDFEKKSENGLVVAWCPQLEVLAHEA 344
+EE+AH + G LWVV+ E LP++ E+K G +V WCPQ VLAH A
Sbjct: 298 MEEIAHGVLSSGLSVLWVVRPPMEGTFVEPHVLPRELEEK---GKIVEWCPQERVLAHPA 354
Query: 345 IGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIR--PLVDEKQIVR 402
I CF++HCGWNST+EAL+ GVP+V P + DQ DA +L D++K G+R E+ IV
Sbjct: 355 IACFLSHCGWNSTMEALTAGVPVVCFPQWGDQVTDAVYLADVFKTGVRLGRGAAEEMIVS 414
Query: 403 KDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
++ + + + E ++ EK E+ N +WK A AV GSS N EFV+ L
Sbjct: 415 REVVAEKLLE-ATVGEKAVELRENARRWKAEAEAAVADGGSSDMNFKEFVDKL 466
>At4g15550 UDP-glucose:indole-3-acetate beta-D-glucosyltransferase
(iaglu)
Length = 474
Score = 231 bits (590), Expect = 5e-61
Identities = 164/493 (33%), Positives = 245/493 (49%), Gaps = 67/493 (13%)
Query: 3 NKTISTKSVHCLVLPFPAHGHTNPMLEFSKLLQQQ-EGVKVTLVTTISNY-------KNI 54
N + S H L + FPA GH NP LE +K L G +VT +IS Y +N+
Sbjct: 4 NNSNSPTGPHFLFVTFPAQGHINPSLELAKRLAGTISGARVTFAASISAYNRRMFSTENV 63
Query: 55 PKLPNNSITIETISDGFDKGGVAEAKDFKL-------YLNKFWQVGPQSLAHLINNLNAR 107
P+ ++ T SDG D G + A K ++++ + G ++L LI + +
Sbjct: 64 PE----TLIFATYSDGHDDGFKSSAYSDKSRQDATGNFMSEMRRRGKETLTELIEDNRKQ 119
Query: 108 NDHVDCLIYDSFMPWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHLG-------KLKPPF 160
N C++Y + W ++A+EF + A Q + + SI+YH G P
Sbjct: 120 NRPFTCVVYTILLTWVAELAREFHLPSALLWVQPVTVFSIFYHYFNGYEDAISEMANTP- 178
Query: 161 VEQEITLPALPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKA--DWILCNSFFELEK 218
I LP+LP L RD+PSF + L Q ++ + IL N+F ELE
Sbjct: 179 -SSSIKLPSLPLLTVRDIPSFIVSSNVYAFLLPAFREQIDSLKEEINPKILINTFQELEP 237
Query: 219 EVADWTMKIWSNFRTV--GPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAV 276
E + NF+ V GP L D S E IEWL+ K S +
Sbjct: 238 EAMS---SVPDNFKIVPVGPLLTLR---------TDFS----SRGEYIEWLDTKADSSVL 281
Query: 277 YVSFGSMASLNEEQIEEVAHCLKDCGSYFLWVVKTSEETKLPKDFEKKSEN--------- 327
YVSFG++A L+++Q+ E+ L FLWV+ T + + +D ++K E+
Sbjct: 282 YVSFGTLAVLSKKQLVELCKALIQSRRPFLWVI-TDKSYRNKEDEQEKEEDCISSFREEL 340
Query: 328 ---GLVVAWCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLV 384
G+VV+WC Q VL H +IGCFVTHCGWNSTLE+L GVP+VA P ++DQ ++AK L
Sbjct: 341 DEIGMVVSWCDQFRVLNHRSIGCFVTHCGWNSTLESLVSGVPVVAFPQWNDQMMNAKLLE 400
Query: 385 DIWKVGIRPLVDEKQ----IVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGK 440
D WK G+R + +++ +V + ++ CI E+ M +K +E N +WK LA AV +
Sbjct: 401 DCWKTGVRVMEKKEEEGVVVVDSEEIRRCIEEV--MEDKAEEFRGNATRWKDLAAEAVRE 458
Query: 441 DGSSHKNMIEFVN 453
GSS ++ FV+
Sbjct: 459 GGSSFNHLKAFVD 471
>At3g21560 UDP-glucose:indole-3-acetate beta-D-glucosyltransferase,
putative
Length = 496
Score = 228 bits (580), Expect = 7e-60
Identities = 144/469 (30%), Positives = 240/469 (50%), Gaps = 33/469 (7%)
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTT--------ISNY---KNIPKLPNN 60
H +++ FP GH NP+L KLL + G+ +T VTT ISN + + +
Sbjct: 12 HVMLVSFPGQGHVNPLLRLGKLLASK-GLLITFVTTESWGKKMRISNKIQDRVLKPVGKG 70
Query: 61 SITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLN-ARNDHVDCLIYDSF 119
+ + DG + A + + VG + + +L+ V CLI + F
Sbjct: 71 YLRYDFFDDGLPEDDEASRTNLTILRPHLELVGKREIKNLVKRYKEVTKQPVTCLINNPF 130
Query: 120 MPWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFV---EQEITLPALPQLQPR 176
+ W DVA++ I A Q+ + YY+ H + P E ++ + +P L+
Sbjct: 131 VSWVCDVAEDLQIPCAVLWVQSCACLAAYYYYHHNLVDFPTKTEPEIDVQISGMPLLKHD 190
Query: 177 DMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADW--TMKIWSNFRTV 234
++PSF ++ + Q +HK I ++F LEK++ D T+ + R +
Sbjct: 191 EIPSFIHPSSPHSALREVIIDQIKRLHKTFSIFIDTFNSLEKDIIDHMSTLSLPGVIRPL 250
Query: 235 GPCLPYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEV 294
GP + K V D +D +EWL+++P S VY+SFG++A L +EQI+E+
Sbjct: 251 GPLYK---MAKTVAYDVVKVNISEPTDPCMEWLDSQPVSSVVYISFGTVAYLKQEQIDEI 307
Query: 295 AHCLKDCGSYFLWVVK------TSEETKLPKDFEKKSENGLVVAWCPQLEVLAHEAIGCF 348
A+ + + FLWV++ E+ LP++ + K G +V WC Q +VL+H ++ CF
Sbjct: 308 AYGVLNADVTFLWVIRQQELGFNKEKHVLPEEVKGK---GKIVEWCSQEKVLSHPSVACF 364
Query: 349 VTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPL--VDEKQIVRKDPL 406
VTHCGWNST+EA+S GVP V P + DQ DA +++D+WK G+R E+++V ++ +
Sbjct: 365 VTHCGWNSTMEAVSSGVPTVCFPQWGDQVTDAVYMIDVWKTGVRLSRGEAEERLVPREEV 424
Query: 407 KDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
+ + E+ + EK E+ N ++WK A AV + GSS +N+ +FV L
Sbjct: 425 AERLREV-TKGEKAIELKKNALKWKEEAEAAVARGGSSDRNLEKFVEKL 472
>At4g15500 indole-3-acetate beta-glucosyltransferase like protein
Length = 475
Score = 227 bits (578), Expect = 1e-59
Identities = 149/468 (31%), Positives = 243/468 (51%), Gaps = 32/468 (6%)
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNN-------SITI 64
H +++ FP GH +P+L K++ + G+ VT VTT + NN + +
Sbjct: 9 HVMLVSFPGQGHISPLLRLGKIIASK-GLIVTFVTTEEPLGKKMRQANNIQDGVLKPVGL 67
Query: 65 ETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCL 124
+ F + G +DF L G + + +L+ + V CLI ++F+PW
Sbjct: 68 GFLRFEFFEDGFVYKEDFDLLQKSLEVSGKREIKNLVKKYEKQP--VRCLINNAFVPWVC 125
Query: 125 DVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLK-PPFVEQEITL--PALP-QLQPRDMPS 180
D+A+E I A Q+ + YY+ H +K P E EIT+ P P L+ ++PS
Sbjct: 126 DIAEELQIPSAVLWVQSCACLAAYYYYHHQLVKFPTETEPEITVDVPFKPLTLKHDEIPS 185
Query: 181 FYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWS--NFRTVGPCL 238
F + + Q +HK +L +F ELEK+ D ++ NF +GP
Sbjct: 186 FLHPSSPLSSIGGTILEQIKRLHKPFSVLIETFQELEKDTIDHMSQLCPQVNFNPIGPLF 245
Query: 239 PYTFLDKRVKDDEDHSIAQLKSDESIEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCL 298
+ K ++ D I++ SD IEWL+++ S VY+SFG++A L + QI+E+AH +
Sbjct: 246 T---MAKTIRSDIKGDISKPDSD-CIEWLDSREPSSVVYISFGTLAFLKQNQIDEIAHGI 301
Query: 299 KDCGSYFLWVVKTS------EETKLPKDFEKKSENGLVVAWCPQLEVLAHEAIGCFVTHC 352
+ G LWV++ E LP + E+K G +V WC Q +VLAH A+ CF++HC
Sbjct: 302 LNSGLSCLWVLRPPLEGLAIEPHVLPLELEEK---GKIVEWCQQEKVLAHPAVACFLSHC 358
Query: 353 GWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPL--VDEKQIVRKDPLKDCI 410
GWNST+EAL+ GVP++ P + DQ +A +++D++K G+R +++IV ++ + + +
Sbjct: 359 GWNSTMEALTSGVPVICFPQWGDQVTNAVYMIDVFKTGLRLSRGASDERIVPREEVAERL 418
Query: 411 CEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSLFQV 458
E ++ EK E+ N +WK A AV G+S +N EFV+ L V
Sbjct: 419 LE-ATVGEKAVELRENARRWKEEAESAVAYGGTSERNFQEFVDKLVDV 465
>At4g14090 glucosyltransferase like protein
Length = 456
Score = 225 bits (573), Expect = 4e-59
Identities = 153/462 (33%), Positives = 238/462 (51%), Gaps = 31/462 (6%)
Query: 7 STKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNNS-ITIE 65
S + H L++ FPA GH NP L+ + L G VT T +S ++ + + P+ ++
Sbjct: 8 SHRRPHYLLVTFPAQGHINPALQLANRLIHH-GATVTYSTAVSAHRRMGEPPSTKGLSFA 66
Query: 66 TISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLIN-NLNA--RNDHVDCLIYDSFMPW 122
+DGFD G + +D K+Y+++ + G +L +I NL+A + + +IY +PW
Sbjct: 67 WFTDGFDDG-LKSFEDQKIYMSELKRCGSNALRDIIKANLDATTETEPITGVIYSVLVPW 125
Query: 123 CLDVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEITLPALPQLQPRDMPSFY 182
VA+EF + + + IYY+ K F + I LP LP + D+PSF
Sbjct: 126 VSTVAREFHLPTTLLWIEPATVLDIYYYYFNTSYKHLFDVEPIKLPKLPLITTGDLPSFL 185
Query: 183 FTYEQDPTFLDIGVAQFSNIHKADW-----ILCNSFFELEKEVADWTMKIWSNFRTVGPC 237
+ P+ L V +I + IL N+F LE + K+ +GP
Sbjct: 186 QPSKALPSAL---VTLREHIEALETESNPKILVNTFSALEHDALTSVEKL--KMIPIGPL 240
Query: 238 LPYTFLDKRVKDDEDHSIAQLKSDESI-EWLNNKPKRSAVYVSFGSMAS-LNEEQIEEVA 295
V E + SDE +WL++K +RS +Y+S G+ A L E+ +E +
Sbjct: 241 ---------VSSSEGKTDLFKSSDEDYTKWLDSKLERSVIYISLGTHADDLPEKHMEALT 291
Query: 296 HCLKDCGSYFLWVV--KTSEETKLPKDFE--KKSENGLVVAWCPQLEVLAHEAIGCFVTH 351
H + FLW+V K EE K + E + S+ GLVV WC Q VLAH A+GCFVTH
Sbjct: 292 HGVLATNRPFLWIVREKNPEEKKKNRFLELIRGSDRGLVVGWCSQTAVLAHCAVGCFVTH 351
Query: 352 CGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIVRKDPLKDCIC 411
CGWNSTLE+L GVP+VA P ++DQ AK + D W++G++ V E+ V + ++ C+
Sbjct: 352 CGWNSTLESLESGVPVVAFPQFADQCTTAKLVEDTWRIGVKVKVGEEGDVDGEEIRRCLE 411
Query: 412 EIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVN 453
++MS E+ +E+ N +WK +A A + G S N+ FV+
Sbjct: 412 KVMSGGEEAEEMRENAEKWKAMAVDAAAEGGPSDLNLKGFVD 453
>At2g23260 putative glucosyltransferase
Length = 456
Score = 218 bits (555), Expect = 5e-57
Identities = 148/459 (32%), Positives = 238/459 (51%), Gaps = 24/459 (5%)
Query: 9 KSVHCLVLPFPAHGHTNPMLEFSKLLQ-QQEGVKVTLVTTIS--NYKNIPKLPNNSITIE 65
+ H L++ P GH NPML+ +K L + + + L T S + + + P + +
Sbjct: 7 QETHVLMVTLPFQGHINPMLKLAKHLSLSSKNLHINLATIESARDLLSTVEKPRYPVDLV 66
Query: 66 TISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDSFMPWCLD 125
SDG K + LNK VG +L+ +I C+I F PW
Sbjct: 67 FFSDGLPKEDPKAPETLLKSLNK---VGAMNLSKIIEE-----KRYSCIISSPFTPWVPA 118
Query: 126 VAKEFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVE---QEITLPALPQLQPRDMPSFY 182
VA I A Q S+YY ++ P +E Q + LPALP L+ RD+PSF
Sbjct: 119 VAASHNISCAILWIQACGAYSVYYRYYMKTNSFPDLEDLNQTVELPALPLLEVRDLPSFM 178
Query: 183 FTYEQDPTFLDIGVAQFSN-IHKADWILCNSFFELEKEVADWTMKIWSNFRTVGPCL-PY 240
F ++ +A+F++ + W+L NSF+ELE E+ + +M +GP + P+
Sbjct: 179 LP-SGGAHFYNL-MAEFADCLRYVKWVLVNSFYELESEIIE-SMADLKPVIPIGPLVSPF 235
Query: 241 TFLDKRVKDDEDHSIAQLKSDES-IEWLNNKPKRSAVYVSFGSMASLNEEQIEEVAHCLK 299
D + + ++ KSD+ +EWL+ + + S VY+SFGSM E Q+E +A LK
Sbjct: 236 LLGDGEEETLDGKNLDFCKSDDCCMEWLDKQARSSVVYISFGSMLETLENQVETIAKALK 295
Query: 300 DCGSYFLWVVKTSEETKLPKDFEK--KSENGLVVAWCPQLEVLAHEAIGCFVTHCGWNST 357
+ G FLWV++ E+ + ++ K G+V+ W PQ ++L+HEAI CFVTHCGWNST
Sbjct: 296 NRGLPFLWVIRPKEKAQNVAVLQEMVKEGQGVVLEWSPQEKILSHEAISCFVTHCGWNST 355
Query: 358 LEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQ-IVRKDPLKDCICEIMSM 416
+E + GVP+VA P ++DQ IDA+ LVD++ +G+R D ++ + ++ CI E ++
Sbjct: 356 METVVAGVPVVAYPSWTDQPIDARLLVDVFGIGVRMRNDSVDGELKVEEVERCI-EAVTE 414
Query: 417 SEKGKEIMNNVMQWKTLATRAVGKDGSSHKNMIEFVNSL 455
+I + K +A A+ GSS +N+ F++ +
Sbjct: 415 GPAAVDIRRRAAELKRVARLALAPGGSSTRNLDLFISDI 453
>At1g22360 unknown protein
Length = 481
Score = 216 bits (550), Expect = 2e-56
Identities = 151/478 (31%), Positives = 241/478 (49%), Gaps = 44/478 (9%)
Query: 12 HCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKL--PNN-----SITI 64
H + +P+PA GH NPM++ +KLL + G +T V T+ N+ + + PN S
Sbjct: 10 HVVCVPYPAQGHINPMMKVAKLLYAK-GFHITFVNTVYNHNRLLRSRGPNAVDGLPSFRF 68
Query: 65 ETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARND--HVDCLIYDSFMPW 122
E+I DG + V +D + L+ +NAR+D V C++ D M +
Sbjct: 69 ESIPDGLPETDVDVTQDIPTLCESTMKHCLAPFKELLRQINARDDVPPVSCIVSDGCMSF 128
Query: 123 CLDVAKEFGIVGASFLTQNLV--MNSIYYHVHLGK-LKPPFVEQEIT----------LPA 169
LD A+E G+ F T + + +YY+ + K L P E +T +P+
Sbjct: 129 TLDAAEELGVPEVLFWTTSACGFLAYLYYYRFIEKGLSPIKDESYLTKEHLDTKIDWIPS 188
Query: 170 LPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWS 229
+ L+ +D+PSF T D L+ + + +A I+ N+F +LE +V I
Sbjct: 189 MKNLRLKDIPSFIRTTNPDDIMLNFIIREADRAKRASAIILNTFDDLEHDVIQSMKSIVP 248
Query: 230 NFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSD------ESIEWLNNKPKRSAVYVSFGSM 283
++GP L ++ + E I + S+ E ++WLN K + S VYV+FGS+
Sbjct: 249 PVYSIGPL----HLLEKQESGEYSEIGRTGSNLWREETECLDWLNTKARNSVVYVNFGSI 304
Query: 284 ASLNEEQIEEVAHCLKDCGSYFLWVVK----TSEETKLPKDF-EKKSENGLVVAWCPQLE 338
L+ +Q+ E A L G FLWV++ +E +P +F ++ ++ +WCPQ +
Sbjct: 305 TVLSAKQLVEFAWGLAATGKEFLWVIRPDLVAGDEAMVPPEFLTATADRRMLASWCPQEK 364
Query: 339 VLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEK 398
VL+H AIG F+THCGWNSTLE+L GVP+V P +++Q + KF D W+VGI D
Sbjct: 365 VLSHPAIGGFLTHCGWNSTLESLCGGVPMVCWPFFAEQQTNCKFSRDEWEVGIEIGGD-- 422
Query: 399 QIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAV-GKDGSSHKNMIEFVNSL 455
V+++ ++ + E+M EKGK + +W+ LA A K GSS N VN +
Sbjct: 423 --VKREEVEAVVRELMD-EEKGKNMREKAEEWRRLANEATEHKHGSSKLNFEMLVNKV 477
>At1g22400 Putative UDP-glucose glucosyltransferase
Length = 489
Score = 214 bits (545), Expect = 8e-56
Identities = 142/476 (29%), Positives = 240/476 (49%), Gaps = 45/476 (9%)
Query: 7 STKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNIPKLPNN------ 60
+++ H + +P+PA GH NPM+ +KLL + G VT V T+ N+ + +
Sbjct: 8 NSQKPHVVCVPYPAQGHINPMMRVAKLLHAR-GFYVTFVNTVYNHNRFLRSRGSNALDGL 66
Query: 61 -SITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARND--HVDCLIYD 117
S E+I+DG + + +D + L+ +NA ++ V C++ D
Sbjct: 67 PSFRFESIADGLPETDMDATQDITALCESTMKNCLAPFRELLQRINAGDNVPPVSCIVSD 126
Query: 118 SFMPWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHL------------GKLKPPFVEQEI 165
M + LDVA+E G+ F T + Y H +L L ++E +
Sbjct: 127 GCMSFTLDVAEELGVPEVLFWTTSGCAFLAYLHFYLFIEKGLCPLKDESYLTKEYLEDTV 186
Query: 166 T--LPALPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADW 223
+P + ++ +D+PSF T D + + + +A I+ N+F +LE +V
Sbjct: 187 IDFIPTMKNVKLKDIPSFIRTTNPDDVMISFALRETERAKRASAIILNTFDDLEHDVVHA 246
Query: 224 TMKIWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQLKSD------ESIEWLNNKPKRSAVY 277
I +VGP + ++ + +E I + S+ E ++WL+ K + S +Y
Sbjct: 247 MQSILPPVYSVGPL--HLLANREI--EEGSEIGMMSSNLWKEEMECLDWLDTKTQNSVIY 302
Query: 278 VSFGSMASLNEEQIEEVAHCLKDCGSYFLWVVK----TSEETKLPKDFEKKSEN-GLVVA 332
++FGS+ L+ +Q+ E A L G FLWV++ EE +P DF ++++ ++ +
Sbjct: 303 INFGSITVLSVKQLVEFAWGLAGSGKEFLWVIRPDLVAGEEAMVPPDFLMETKDRSMLAS 362
Query: 333 WCPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIR 392
WCPQ +VL+H AIG F+THCGWNS LE+LS GVP+V P ++DQ ++ KF D W VGI
Sbjct: 363 WCPQEKVLSHPAIGGFLTHCGWNSILESLSCGVPMVCWPFFADQQMNCKFCCDEWDVGIE 422
Query: 393 PLVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAV-GKDGSSHKN 447
D V+++ ++ + E+M EKGK++ ++W+ LA +A K GSS N
Sbjct: 423 IGGD----VKREEVEAVVRELMD-GEKGKKMREKAVEWQRLAEKATEHKLGSSVMN 473
>At1g78270 similar to glucoronosyl transferase-like protein; similar
to ESTs gb|AI996767.1, emb|Z46520.1, gb|T44500.1, and
gb|AI099822.1
Length = 489
Score = 206 bits (524), Expect = 2e-53
Identities = 150/483 (31%), Positives = 238/483 (49%), Gaps = 44/483 (9%)
Query: 7 STKSVHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTTISNYKNI-----PKLPNN- 60
S++ H + +P+PA GH NPML+ +KLL + G VT V T N++ I P N
Sbjct: 8 SSQKPHAMCIPYPAQGHINPMLKLAKLLHAR-GFHVTFVNTDYNHRRILQSRGPHALNGL 66
Query: 61 -SITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARND--HVDCLIYD 117
S ETI DG V +D ++ LI LN+ +D V C+I D
Sbjct: 67 PSFRFETIPDGLPWTDVDAKQDMLKLIDSTINNCLAPFKDLILRLNSGSDIPPVSCIISD 126
Query: 118 SFMPWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHL------------GKLKPPFVEQEI 165
+ M + +D A+E I T + +Y H LK +
Sbjct: 127 ASMSFTIDAAEELKIPVVLLWTNSATALILYLHYQKLIEKEIIPLKDSSDLKKHLETEID 186
Query: 166 TLPALPQLQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTM 225
+P++ +++ +D P F T + + I +A I N+F +LE V
Sbjct: 187 WIPSMKKIKLKDFPDFVTTTNPQDPMISFILHVTGRIKRASAIFINTFEKLEHNVLLSLR 246
Query: 226 KIWSNFRTVGPCLPYTFLDKRVKDDEDHSIAQL------KSDESIEWLNNKPKRSAVYVS 279
+ +VGP + L+ R + D++ I +L + ES++WL+ K +++ +YV+
Sbjct: 247 SLLPQIYSVGP---FQILENR-EIDKNSEIRKLGLNLWEEETESLDWLDTKAEKAVIYVN 302
Query: 280 FGSMASLNEEQIEEVAHCLKDCGSYFLWVVKTS----EETKLPKDF--EKKSENGLVVAW 333
FGS+ L EQI E A L G FLWVV++ +++ LP +F E K+ L+ W
Sbjct: 303 FGSLTVLTSEQILEFAWGLARSGKEFLWVVRSGMVDGDDSILPAEFLSETKNRGMLIKGW 362
Query: 334 CPQLEVLAHEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRP 393
C Q +VL+H AIG F+THCGWNSTLE+L GVP++ P ++DQ + KF + W +G+
Sbjct: 363 CSQEKVLSHPAIGGFLTHCGWNSTLESLYAGVPMICWPFFADQLTNRKFCCEDWGIGME- 421
Query: 394 LVDEKQIVRKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKD-GSSHKNMIEFV 452
+ E+ V+++ ++ + E+M EKGK + V++W+ LA A GSS+ N V
Sbjct: 422 -IGEE--VKRERVETVVKELMD-GEKGKRLREKVVEWRRLAEEASAPPLGSSYVNFETVV 477
Query: 453 NSL 455
N +
Sbjct: 478 NKV 480
>At2g28080 putative glucosyltransferase
Length = 482
Score = 203 bits (517), Expect = 1e-52
Identities = 145/475 (30%), Positives = 240/475 (50%), Gaps = 50/475 (10%)
Query: 11 VHCLVLPFPAHGHTNPMLEFSKLLQQQEGVKVTLVTT------ISN------YKNIPKLP 58
+H L++P+P GH NP + + L Q G+ VT V T I+N + +
Sbjct: 17 LHALLIPYPFQGHVNPFVHLAIKLASQ-GITVTFVNTHYIHHQITNGSDGDIFAGVRSES 75
Query: 59 NNSITIETISDGFDKGGVAEAKDFKLYLNKFWQVGPQSLAHLINNLNARNDHVDCLIYDS 118
I T+SDG G + + Y + V + L+ +L + V+ +I D+
Sbjct: 76 GLDIRYATVSDGLPVG-FDRSLNHDTYQSSLLHVFYAHVEELVASLVGGDGGVNVMIADT 134
Query: 119 FMPWCLDVAKEFGIVGASFLTQNLVMNSIYYHVHLGKLKPPFVEQEIT------LPALPQ 172
F W VA++FG+V SF T+ ++ S+YYH+ L ++ F QE +P +
Sbjct: 135 FFVWPSVVARKFGLVCVSFWTEAALVFSLYYHMDLLRIHGHFGAQETRSDLIDYIPGVAA 194
Query: 173 LQPRDMPSFYFTYEQDPTFLDIGVAQFSNIHKADWILCNSFFELEKEVADWTMKIWSN-- 230
+ P+D S+ + I F ++ K D++LCN+ + E D T+K +
Sbjct: 195 INPKDTASYLQETDTSSVVHQIIFKAFEDVKKVDFVLCNTIQQFE----DKTIKALNTKI 250
Query: 231 -FRTVGPCLPYTFLDKRVKDDEDHSIAQLKSDES--IEWLNNKPKRSAVYVSFGSMASLN 287
F +GP +P+ +++ S+ ES +WLN KPK S +Y+SFGS A +
Sbjct: 251 PFYAIGPIIPF--------NNQTGSVTTSLWSESDCTQWLNTKPKSSVLYISFGSYAHVT 302
Query: 288 EEQIEEVAHCLKDCGSYFLWVVK----TSEETK-LPKDFEKKS-ENGLVVAWCPQLEVLA 341
++ + E+AH + F+WVV+ +S+ET LP+ FE ++ + G+V+ WC Q+ VL+
Sbjct: 303 KKDLVEIAHGILLSKVNFVWVVRPDIVSSDETNPLPEGFETEAGDRGIVIPWCCQMTVLS 362
Query: 342 HEAIGCFVTHCGWNSTLEALSIGVPIVAIPLYSDQGIDAKFLVDIWKVGIRPLVDEKQIV 401
HE++G F+THCGWNS LE + VP++ PL +DQ + K +VD W++GI L ++K
Sbjct: 363 HESVGGFLTHCGWNSILETIWCEVPVLCFPLLTDQVTNRKLVVDDWEIGIN-LCEDKSDF 421
Query: 402 RKDPLKDCICEIMSMSEKGKEIMNNVMQWKTLATRAVGKDGSSHK-NMIEFVNSL 455
+D + I +M K K + + K AV GSS + N+ F++ L
Sbjct: 422 GRDEVGRNINRLMCGVSKEK-----IGRVKMSLEGAVRNSGSSSEMNLGLFIDGL 471
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,881,081
Number of Sequences: 26719
Number of extensions: 482012
Number of successful extensions: 1713
Number of sequences better than 10.0: 126
Number of HSP's better than 10.0 without gapping: 118
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1191
Number of HSP's gapped (non-prelim): 169
length of query: 458
length of database: 11,318,596
effective HSP length: 103
effective length of query: 355
effective length of database: 8,566,539
effective search space: 3041121345
effective search space used: 3041121345
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC142095.2


