

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141923.7 - phase: 0
(406 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
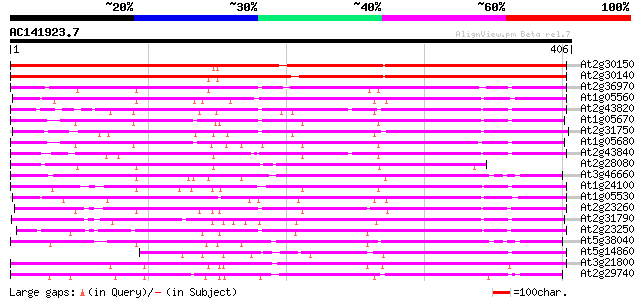
Score E
Sequences producing significant alignments: (bits) Value
At2g30150 putative glucosyltransferase 377 e-105
At2g30140 putative glucosyltransferase 371 e-103
At2g36970 putative glucosyltransferase 193 1e-49
At1g05560 UDP-Glucose Transferase (UGT75B2) 191 8e-49
At2g43820 putative glucosyltransferase 187 1e-47
At1g05670 UDP glycosyltransferase UGT74E1 185 3e-47
At2g31750 putative glucosyltransferase 185 4e-47
At1g05680 indole-3-acetate beta-glucosyltransferase like protein 182 3e-46
At2g43840 putative glucosyltransferase 175 4e-44
At2g28080 putative glucosyltransferase 167 1e-41
At3g46660 glucosyltransferase-like protein 163 2e-40
At1g24100 putative indole-3-acetate beta-glucosyltransferase 163 2e-40
At1g05530 hypothetical protein 161 7e-40
At2g23260 putative glucosyltransferase 160 2e-39
At2g31790 putative glucosyltransferase 159 2e-39
At2g23250 putative glucosyltransferase 154 8e-38
At5g38040 glucosyltransferase-like protein 145 3e-35
At5g14860 glucosyltransferase -like protein 142 3e-34
At3g21800 putative UDP-glucose glucosyltransferase 140 2e-33
At2g29740 putative flavonol 3-O-glucosyltransferase 136 2e-32
>At2g30150 putative glucosyltransferase
Length = 440
Score = 377 bits (968), Expect = e-105
Identities = 193/442 (43%), Positives = 273/442 (61%), Gaps = 45/442 (10%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
MP+P RG+INPM+NLCK LV + N+ V+FVVTEEWL FI S+PKP+ I F + N+IPS
Sbjct: 1 MPWPGRGHINPMLNLCKSLVRRDPNLTVTFVVTEEWLGFIGSDPKPNRIHFATLPNIIPS 60
Query: 61 ELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWAVVVANRRNIPAALF 120
EL+ D AF++ V+T++E PFE+LLD L+ PP+ I+ DT + WAV V +RNIP A F
Sbjct: 61 ELVRANDFIAFIDAVLTRLEEPFEQLLDRLNSPPTAIIADTYIIWAVRVGTKRNIPVASF 120
Query: 121 WPMPASIFSVFLHQHIFEQNGHYPVK------------YPG------------------- 149
W A+I S+F++ + +GH+P++ PG
Sbjct: 121 WTTSATILSLFINSDLLASHGHFPIEPSESKLDEIVDYIPGLSPTRLSDLQILHGYSHQV 180
Query: 150 -------FEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKP 202
F ++KA+YLLF S YELE +AID SK P+Y+ GP IP L
Sbjct: 181 FNIFKKSFGELYKAKYLLFPSAYELEPKAIDFFTSKFDFPVYSTGPLIPLEEL-----SV 235
Query: 203 SNTNHSY-YIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEAS 261
N N Y +WLD QP SVLYI+QGSF SVS AQ++EI + + V+F W+AR
Sbjct: 236 GNENRELDYFKWLDEQPESSVLYISQGSFLSVSEAQMEEIVVGVREAGVKFFWVARGGEL 295
Query: 262 RLKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQ 321
+LKE +G+++ WCDQLRVL H +IGGFW+HCG+NST E + +GVP LT P++ DQ
Sbjct: 296 KLKEALEGS-LGVVVSWCDQLRVLCHAAIGGFWTHCGYNSTLEGICSGVPLLTFPVFWDQ 354
Query: 322 PFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKI 381
N+KM+VE+W+VG ++ + + L+ D+I +LV FMD + E +++R R+ L +I
Sbjct: 355 FLNAKMIVEEWRVGMGIERKKQMELLIVSDEIKELVKRFMDGESEEGKEMRRRTCDLSEI 414
Query: 382 CLNSIANGGSAHTDFNAFISDV 403
C ++A GGS+ + +AFI D+
Sbjct: 415 CRGAVAKGGSSDANIDAFIKDI 436
>At2g30140 putative glucosyltransferase
Length = 455
Score = 371 bits (953), Expect = e-103
Identities = 195/441 (44%), Positives = 272/441 (61%), Gaps = 44/441 (9%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
MP+P RG+INPMMNLCK LV N+HV+FVVTEEWL FI +PKPD I F + N+IPS
Sbjct: 17 MPYPGRGHINPMMNLCKRLVRRYPNLHVTFVVTEEWLGFIGPDPKPDRIHFSTLPNLIPS 76
Query: 61 ELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPP-SIIVYDTLLYWAVVVANRRNIPAAL 119
EL+ +D F++ V T++E PFE+LLD L+ PP S+I DT + WAV V +RNIP
Sbjct: 77 ELVRAKDFIGFIDAVYTRLEEPFEKLLDSLNSPPPSVIFADTYVIWAVRVGRKRNIPVVS 136
Query: 120 FWPMPASIFSVFLHQHIFEQNGH------------------------YPVKYPG------ 149
W M A+I S FLH + +GH P + G
Sbjct: 137 LWTMSATILSFFLHSDLLISHGHALFEPSEEEVVDYVPGLSPTKLRDLPPIFDGYSDRVF 196
Query: 150 ------FEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSL-IKNDPKP 202
F+ + A+ LLF++ YELE +AID SKL +P+Y IGP IP L ++ND K
Sbjct: 197 KTAKLCFDELPGARSLLFTTAYELEHKAIDAFTSKLDIPVYAIGPLIPFEELSVQNDNKE 256
Query: 203 SNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASR 262
N YI+WL+ QP GSVLYI+QGSF SVS AQ++EI L S VRFLW+AR +
Sbjct: 257 PN-----YIQWLEEQPEGSVLYISQGSFLSVSEAQMEEIVKGLRESGVRFLWVARGGELK 311
Query: 263 LKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQP 322
LKE +G+++ WCDQLRVL H ++GGFW+HCG+NST E + +GVP L P++ DQ
Sbjct: 312 LKEALEGS-LGVVVSWCDQLRVLCHKAVGGFWTHCGFNSTLEGIYSGVPMLAFPLFWDQI 370
Query: 323 FNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKIC 382
N+KM+VEDW+VG R++ K + L+ +++I ++V FMD + E +++R R+ L +I
Sbjct: 371 LNAKMIVEDWRVGMRIERTKKNELLIGREEIKEVVKRFMDRESEEGKEMRRRACDLSEIS 430
Query: 383 LNSIANGGSAHTDFNAFISDV 403
++A GS++ + + F+ +
Sbjct: 431 RGAVAKSGSSNVNIDEFVRHI 451
>At2g36970 putative glucosyltransferase
Length = 490
Score = 193 bits (491), Expect = 1e-49
Identities = 123/471 (26%), Positives = 228/471 (48%), Gaps = 82/471 (17%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPD------------- 47
+P+P++G++ P ++L L S+ I +FV T+ IS+ + D
Sbjct: 14 IPYPLQGHVIPFVHLAIKLASHGFTI--TFVNTDSIHHHISTAHQDDAGDIFSAARSSGQ 71
Query: 48 -NISFRSGSNVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLL----DHPPSIIVYDTL 102
+I + + S+ P + +H F E ++ A ++L+ L D P + ++ DT
Sbjct: 72 HDIRYTTVSDGFPLDFDRSLNHDQFFEGILHVFSAHVDDLIAKLSRRDDPPVTCLIADTF 131
Query: 103 LYWAVVVANRRNIPAALFWPMPASIFSVFLHQHIFEQNGH-------------------- 142
W+ ++ ++ N+ FW PA + +++ H + NGH
Sbjct: 132 YVWSSMICDKHNLVNVSFWTEPALVLNLYYHMDLLISNGHFKSLDNRKDVIDYVPGVKAI 191
Query: 143 ----------------------YPVKYPGFEWIHKAQYLLFSSIYELESQAIDVLKSKLP 180
Y + + F+ + +A +++ +++ ELE ++ L++K P
Sbjct: 192 EPKDLMSYLQVSDKDVDTNTVVYRILFKAFKDVKRADFVVCNTVQELEPDSLSALQAKQP 251
Query: 181 LPIYTIGPTIPKFSLIKNDPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDE 240
+Y IGP S++ S S EWL +P GSVLY++ GS+ V +I E
Sbjct: 252 --VYAIGPVFSTDSVVPT----SLWAESDCTEWLKGRPTGSVLYVSFGSYAHVGKKEIVE 305
Query: 241 IAAALCASNVRFLWIARSE--ASRLKE------ICGAHHMGLIMEWCDQLRVLSHPSIGG 292
IA L S + F+W+ R + S + + + A GL+++WC Q+ V+S+P++GG
Sbjct: 306 IAHGLLLSGISFIWVLRPDIVGSNVPDFLPAGFVDQAQDRGLVVQWCCQMEVISNPAVGG 365
Query: 293 FWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDK 352
F++HCGWNS ES+ G+P L P+ DQ N K++V+DW +G + E + +D+
Sbjct: 366 FFTHCGWNSILESVWCGLPLLCYPLLTDQFTNRKLVVDDWCIGINLCE----KKTITRDQ 421
Query: 353 IVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
+ V M +GE + ++R +K+++ +++ GS+ T+FN F+S+V
Sbjct: 422 VSANVKRLM--NGETSSELRNNVEKVKRHLKDAVTTVGSSETNFNLFVSEV 470
>At1g05560 UDP-Glucose Transferase (UGT75B2)
Length = 469
Score = 191 bits (484), Expect = 8e-49
Identities = 127/450 (28%), Positives = 224/450 (49%), Gaps = 58/450 (12%)
Query: 3 FPVRGNINPMMNLCKLLVSNNSNIHVSFV--VTEEWLSFISSEPKPDNISFRSGSNVIPS 60
FP +G++NP + + L+ + V+FV V+ S I++ K +N+SF + S+
Sbjct: 11 FPAQGHVNPSLRFARRLIKR-TGARVTFVTCVSVFHNSMIANHNKVENLSFLTFSDGFDD 69
Query: 61 ELICG-RDHPAFMEDVMTKMEAPFEELLDLL---DHPPSIIVYDTLLYWAVVVANRRNIP 116
I D ++ + + ++ D P + ++Y LL WA VA R +P
Sbjct: 70 GGISTYEDRQKRSVNLKVNGDKALSDFIEATKNGDSPVTCLIYTILLNWAPKVARRFQLP 129
Query: 117 AALFWPMPASIFSVFL-----HQHIFE---------------------QNGHYPVKYPGF 150
+AL W PA +F+++ ++ +FE G Y
Sbjct: 130 SALLWIQPALVFNIYYTHFMGNKSVFELPNLSSLEIRDLPSFLTPSNTNKGAYDAFQEMM 189
Query: 151 EWIHKAQY--LLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSNTNHS 208
E++ K +L ++ LE +A+ + + + +GP +P + K S
Sbjct: 190 EFLIKETKPKILINTFDSLEPEALTAFPN---IDMVAVGPLLPTEIFSGSTNKSVKDQSS 246
Query: 209 YYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASR------ 262
Y WLDS+ SV+Y++ G+ +S QI+E+A AL FLW+ +++R
Sbjct: 247 SYTLWLDSKTESSVIYVSFGTMVELSKKQIEELARALIEGKRPFLWVITDKSNRETKTEG 306
Query: 263 -----LKEICGAHH----MGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFL 313
+++I G H +G+I+ WC Q+ VLSH ++G F +HCGW+ST ESLV GVP +
Sbjct: 307 EEETEIEKIAGFRHELEEVGMIVSWCSQIEVLSHRAVGCFVTHCGWSSTLESLVLGVPVV 366
Query: 314 TLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRE 373
P++ DQP N+K++ E WK G RV+E+ +D LV++ +I + + M+ E + ++RE
Sbjct: 367 AFPMWSDQPTNAKLLEESWKTGVRVREN--KDGLVERGEIRRCLEAVME---EKSVELRE 421
Query: 374 RSKKLQKICLNSIANGGSAHTDFNAFISDV 403
+KK +++ + + GGS+ + AF+ D+
Sbjct: 422 NAKKWKRLAMEAGREGGSSDKNMEAFVEDI 451
>At2g43820 putative glucosyltransferase
Length = 449
Score = 187 bits (474), Expect = 1e-47
Identities = 137/456 (30%), Positives = 225/456 (49%), Gaps = 73/456 (16%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+P+P +G+I P CK L + + + +T +F+ + PD SG I +
Sbjct: 11 VPYPTQGHITPFRQFCKRL--HFKGLKTTLALT----TFVFNSINPD----LSGPISIAT 60
Query: 61 ELICGRDHPAF---------MEDVMTKMEAPFEELLD---LLDHPPSIIVYDTLLYWAVV 108
+ G DH F ++D T +++ D+P + IVYD L WA+
Sbjct: 61 -ISDGYDHGGFETADSIDDYLKDFKTSGSKTIADIIQKHQTSDNPITCIVYDAFLPWALD 119
Query: 109 VANRRNIPAALFWPMPASIFSVFLHQHI-----------------------FEQNGHYPV 145
VA + A F+ P ++ V+ +I F +G YP
Sbjct: 120 VAREFGLVATPFFTQPCAVNYVYYLSYINNGSLQLPIEELPFLELQDLPSFFSVSGSYPA 179
Query: 146 KYP----GFEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSL---IKN 198
+ F KA ++L +S ELE ++ P + TIGPTIP L IK+
Sbjct: 180 YFEMVLQQFINFEKADFVLVNSFQELELHENELWSKACP--VLTIGPTIPSIYLDQRIKS 237
Query: 199 DPKPS-----NTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFL 253
D + + S+ I WLD++P GSV+Y+A GS +++ Q++E+A+A+ SN FL
Sbjct: 238 DTGYDLNLFESKDDSFCINWLDTRPQGSVVYVAFGSMAQLTNVQMEELASAV--SNFSFL 295
Query: 254 WIARSEASR------LKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLV 307
W+ RS L+ + L+++W QL+VLS+ +IG F +HCGWNST E+L
Sbjct: 296 WVVRSSEEEKLPSGFLETV--NKEKSLVLKWSPQLQVLSNKAIGCFLTHCGWNSTMEALT 353
Query: 308 AGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGEL 367
GVP + +P + DQP N+K + + WK G RVK + K + K+++I + E M +GE
Sbjct: 354 FGVPMVAMPQWTDQPMNAKYIQDVWKAGVRVKTE-KESGIAKREEIEFSIKEVM--EGER 410
Query: 368 TRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
++++++ KK + + + S+ GGS T+ + F+S V
Sbjct: 411 SKEMKKNVKKWRDLAVKSLNEGGSTDTNIDTFVSRV 446
>At1g05670 UDP glycosyltransferase UGT74E1
Length = 453
Score = 185 bits (470), Expect = 3e-47
Identities = 138/458 (30%), Positives = 219/458 (47%), Gaps = 74/458 (16%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKP------DNISFRSG 54
+PFP +G+I PM CK L S + I L +S +P P D I+
Sbjct: 10 LPFPAQGHITPMSQFCKRLASKSLKIT---------LVLVSDKPSPPYKTEHDTITVVPI 60
Query: 55 SNVIPSELICGRDHPAFMEDVMTKMEAPFEELLD---LLDHPPSIIVYDTLLYWAVVVAN 111
SN D +ME V + ++ +L++ L +PP +VYD+ + W + VA+
Sbjct: 61 SNGFQEGQERSEDLDEYMERVESSIKNRLPKLIEDMKLSGNPPRALVYDSTMPWLLDVAH 120
Query: 112 RRNIPAALFWPMPASIFSVFLHQHIFEQNGHYP-VKY-------------------PGF- 150
+ A+F+ P + +++ H +F+ + P KY P F
Sbjct: 121 SYGLSGAVFFTQPWLVSAIYYH--VFKGSFSVPSTKYGHSTLASFPSLPILNANDLPSFL 178
Query: 151 -----------------EWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKF 193
I + +L ++ +LE + + +KS P + IGPT+P
Sbjct: 179 CESSSYPYILRTVIDQLSNIDRVDIVLCNTFDKLEEKLLKWIKSVWP--VLNIGPTVPSM 236
Query: 194 SLIKNDPKPSNTNHSYY-------IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALC 246
L K + N S + +EWL+S+ SV+Y++ GS + Q+ E+AA L
Sbjct: 237 YLDKRLAEDKNYGFSLFGAKIAECMEWLNSKQPSSVVYVSFGSLVVLKKDQLIELAAGLK 296
Query: 247 ASNVRFLWIAR-SEASRLKE--ICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTK 303
S FLW+ R +E +L E I GL + W QL VL+H SIG F +HCGWNST
Sbjct: 297 QSGHFFLWVVRETERRKLPENYIEEIGEKGLTVSWSPQLEVLTHKSIGCFVTHCGWNSTL 356
Query: 304 ESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDL 363
E L GVP + +P + DQP N+K M + WKVG RVK D D V++++ V+ V E M
Sbjct: 357 EGLSLGVPMIGMPHWADQPTNAKFMEDVWKVGVRVKAD--SDGFVRREEFVRRVEEVM-- 412
Query: 364 DGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFIS 401
+ E ++IR+ ++K + + +++ GGS+ + N F+S
Sbjct: 413 EAEQGKEIRKNAEKWKVLAQEAVSEGGSSDKNINEFVS 450
>At2g31750 putative glucosyltransferase
Length = 456
Score = 185 bits (469), Expect = 4e-47
Identities = 130/457 (28%), Positives = 224/457 (48%), Gaps = 70/457 (15%)
Query: 3 FPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPSEL 62
FP++G+INP++ K L+S N N V+F+ T + I + G+ +P
Sbjct: 14 FPIQGHINPLLQFSKRLLSKNVN--VTFLTTSSTHNSILRR------AITGGATALPLSF 65
Query: 63 I-----CGRDHPA------FMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWAVVVAN 111
+ DHP+ + + EL+ +D P+ +VYD+ L + + V
Sbjct: 66 VPIDDGFEEDHPSTDTSPDYFAKFQENVSRSLSELISSMDPKPNAVVYDSCLPYVLDVCR 125
Query: 112 RR-NIPAALFWPMPASIFSVFLH---------QHIFEQNGHYPVK--------------Y 147
+ + AA F+ +++ + ++H Q+ P+K
Sbjct: 126 KHPGVAAASFFTQSSTVNATYIHFLRGEFKEFQNDVVLPAMPPLKGNDLPVFLYDNNLCR 185
Query: 148 PGFEWIHKA-------QYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDP 200
P FE I + L +S ELE + + +K++ P + IGP IP L K
Sbjct: 186 PLFELISSQFVNVDDIDFFLVNSFDELEVEVLQWMKNQWP--VKNIGPMIPSMYLDKRLA 243
Query: 201 KPS-------NTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFL 253
N + ++WLDS+P GSV+Y++ GS + Q+ E+AA L + FL
Sbjct: 244 GDKDYGINLFNAQVNECLDWLDSKPPGSVIYVSFGSLAVLKDDQMIEVAAGLKQTGHNFL 303
Query: 254 WIARSEASR------LKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLV 307
W+ R ++ +++IC GLI+ W QL+VL+H SIG F +HCGWNST E+L
Sbjct: 304 WVVRETETKKLPSNYIEDICDK---GLIVNWSPQLQVLAHKSIGCFMTHCGWNSTLEALS 360
Query: 308 AGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGEL 367
GV + +P Y DQP N+K + + WKVG RVK D ++ V K++IV+ V E M+ E
Sbjct: 361 LGVALIGMPAYSDQPTNAKFIEDVWKVGVRVKAD--QNGFVPKEEIVRCVGEVMEDMSEK 418
Query: 368 TRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVM 404
++IR+ +++L + ++++GG++ + + F++ ++
Sbjct: 419 GKEIRKNARRLMEFAREALSDGGNSDKNIDEFVAKIV 455
>At1g05680 indole-3-acetate beta-glucosyltransferase like protein
Length = 453
Score = 182 bits (462), Expect = 3e-46
Identities = 138/456 (30%), Positives = 218/456 (47%), Gaps = 70/456 (15%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKP------DNISFRSG 54
+PFP +G+I PM CK L S + L +S +P P D+I+
Sbjct: 10 LPFPGQGHITPMSQFCKRLASKGLKLT---------LVLVSDKPSPPYKTEHDSITVFPI 60
Query: 55 SNVIPSELICGRDHPAFMEDVMTKMEAPFEELLD---LLDHPPSIIVYDTLLYWAVVVAN 111
SN +D +ME V T ++ +L++ L +PP IVYD+ + W + VA+
Sbjct: 61 SNGFQEGEEPLQDLDDYMERVETSIKNTLPKLVEDMKLSGNPPRAIVYDSTMPWLLDVAH 120
Query: 112 RRNIPAALFWPMPASIFSVFLHQHIFEQNGHYP---------VKYPGFEWIHK---AQYL 159
+ A+F+ P + +++ H +F+ + P +P F + +L
Sbjct: 121 SYGLSGAVFFTQPWLVTAIYYH--VFKGSFSVPSTKYGHSTLASFPSFPMLTANDLPSFL 178
Query: 160 LFSSIYE------------------LESQAIDVLKSKLPL------PIYTIGPTIPKFSL 195
SS Y + D L+ KL P+ IGPT+P L
Sbjct: 179 CESSSYPNILRIVVDQLSNIDRVDIVLCNTFDKLEEKLLKWVQSLWPVLNIGPTVPSMYL 238
Query: 196 IKNDPKPSNTNHSYY-------IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCAS 248
K + N S + +EWL+S+ SV+Y++ GS + Q+ E+AA L S
Sbjct: 239 DKRLSEDKNYGFSLFNAKVAECMEWLNSKEPNSVVYLSFGSLVILKEDQMLELAAGLKQS 298
Query: 249 NVRFLWIAR-SEASRLKE--ICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKES 305
FLW+ R +E +L + GLI+ W QL VL+H SIG F +HCGWNST E
Sbjct: 299 GRFFLWVVRETETHKLPRNYVEEIGEKGLIVSWSPQLDVLAHKSIGCFLTHCGWNSTLEG 358
Query: 306 LVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDG 365
L GVP + +P + DQP N+K M + WKVG RVK + D V++++I++ V E M +G
Sbjct: 359 LSLGVPMIGMPHWTDQPTNAKFMQDVWKVGVRVK--AEGDGFVRREEIMRSVEEVM--EG 414
Query: 366 ELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFIS 401
E ++IR+ ++K + + +++ GGS+ N F+S
Sbjct: 415 EKGKEIRKNAEKWKVLAQEAVSEGGSSDKSINEFVS 450
>At2g43840 putative glucosyltransferase
Length = 449
Score = 175 bits (444), Expect = 4e-44
Identities = 133/454 (29%), Positives = 222/454 (48%), Gaps = 69/454 (15%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+PFP +G+I P+ CK L S F T +FI + I S + +
Sbjct: 11 VPFPSQGHITPIRQFCKRLHSKG------FKTTHTLTTFIFN-----TIHLDPSSPISIA 59
Query: 61 ELICGRDH---------PAFMEDVMT---KMEAPFEELLDLLDHPPSIIVYDTLLYWAVV 108
+ G D P ++++ T K A D+P + IVYD+ + WA+
Sbjct: 60 TISDGYDQGGFSSAGSVPEYLQNFKTFGSKTVADIIRKHQSTDNPITCIVYDSFMPWALD 119
Query: 109 VANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHYPVK---------------------- 146
+A + AA F+ ++ + +I + P+K
Sbjct: 120 LAMDFGLAAAPFFTQSCAVNYINYLSYINNGSLTLPIKDLPLLELQDLPTFVTPTGSHLA 179
Query: 147 -----YPGFEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPK 201
F KA ++L +S ++L+ ++L SK+ P+ TIGPT+P L +
Sbjct: 180 YFEMVLQQFTNFDKADFVLVNSFHDLDLHEEELL-SKV-CPVLTIGPTVPSMYLDQQIKS 237
Query: 202 PSNTNHSYY--------IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFL 253
++ + + + +WLD +P GSV+YIA GS +SS Q++EIA+A+ SN +L
Sbjct: 238 DNDYDLNLFDLKEAALCTDWLDKRPEGSVVYIAFGSMAKLSSEQMEEIASAI--SNFSYL 295
Query: 254 WIAR-SEASRLKE---ICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAG 309
W+ R SE S+L L+++W QL+VLS+ +IG F +HCGWNST E L G
Sbjct: 296 WVVRASEESKLPPGFLETVDKDKSLVLKWSPQLQVLSNKAIGCFMTHCGWNSTMEGLSLG 355
Query: 310 VPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTR 369
VP + +P + DQP N+K + + WKVG RVK + K + K+++I + E M +GE ++
Sbjct: 356 VPMVAMPQWTDQPMNAKYIQDVWKVGVRVKAE-KESGICKREEIEFSIKEVM--EGEKSK 412
Query: 370 DIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
+++E + K + + + S++ GGS + N F+S +
Sbjct: 413 EMKENAGKWRDLAVKSLSEGGSTDININEFVSKI 446
>At2g28080 putative glucosyltransferase
Length = 482
Score = 167 bits (423), Expect = 1e-41
Identities = 115/408 (28%), Positives = 186/408 (45%), Gaps = 68/408 (16%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPD-----------NI 49
+P+P +G++NP ++L L S I V+FV T I++ D +I
Sbjct: 22 IPYPFQGHVNPFVHLAIKLASQG--ITVTFVNTHYIHHQITNGSDGDIFAGVRSESGLDI 79
Query: 50 SFRSGSNVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLL---DHPPSIIVYDTLLYWA 106
+ + S+ +P +H + ++ A EEL+ L D ++++ DT W
Sbjct: 80 RYATVSDGLPVGFDRSLNHDTYQSSLLHVFYAHVEELVASLVGGDGGVNVMIADTFFVWP 139
Query: 107 VVVANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHYPVK-------------------- 146
VVA + + FW A +FS++ H + +GH+ +
Sbjct: 140 SVVARKFGLVCVSFWTEAALVFSLYYHMDLLRIHGHFGAQETRSDLIDYIPGVAAINPKD 199
Query: 147 ------------------YPGFEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGP 188
+ FE + K ++L ++I + E + I L +K+P Y IGP
Sbjct: 200 TASYLQETDTSSVVHQIIFKAFEDVKKVDFVLCNTIQQFEDKTIKALNTKIPF--YAIGP 257
Query: 189 TIPKFSLIKNDPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCAS 248
IP F+ S + S +WL+++P SVLYI+ GS+ V+ + EIA + S
Sbjct: 258 IIP-FNNQTGSVTTSLWSESDCTQWLNTKPKSSVLYISFGSYAHVTKKDLVEIAHGILLS 316
Query: 249 NVRFLWIARSEASRLKEI--------CGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWN 300
V F+W+ R + E A G+++ WC Q+ VLSH S+GGF +HCGWN
Sbjct: 317 KVNFVWVVRPDIVSSDETNPLPEGFETEAGDRGIVIPWCCQMTVLSHESVGGFLTHCGWN 376
Query: 301 STKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVG---CRVKEDVKRD 345
S E++ VP L P+ DQ N K++V+DW++G C K D RD
Sbjct: 377 SILETIWCEVPVLCFPLLTDQVTNRKLVVDDWEIGINLCEDKSDFGRD 424
>At3g46660 glucosyltransferase-like protein
Length = 458
Score = 163 bits (412), Expect = 2e-40
Identities = 126/450 (28%), Positives = 207/450 (46%), Gaps = 65/450 (14%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+PFP +G+I+PMM L K L +I V V ++ F S+ + F + +P
Sbjct: 18 VPFPAQGHISPMMQLAKTLHLKGFSITV---VQTKFNYFSPSDDFTHDFQFVTIPESLPE 74
Query: 61 ELICGRDHPAFMEDVMTKMEAPFEELLDLL----DHPPSIIVYDTLLYWAVVVANRRNIP 116
F+ + + + F++ L L + S ++YD +Y+A A +P
Sbjct: 75 SDFKNLGPIQFLFKLNKECKVSFKDCLGQLVLQQSNEISCVIYDEFMYFAEAAAKECKLP 134
Query: 117 AALFWPMPASIF---SVF-------LHQHIFEQNGH--------YPVKYPGFEWIHKAQY 158
+F A+ F SVF + + E G YP++Y F A
Sbjct: 135 NIIFSTTSATAFACRSVFDKLYANNVQAPLKETKGQQEELVPEFYPLRYKDFPVSRFASL 194
Query: 159 LLFSSIYE------------------LESQAIDVLKSK-LPLPIYTIGPTIPKFSLIKND 199
+Y LES ++ L+ + L +P+Y IGP ++ +
Sbjct: 195 ESIMEVYRNTVDKRTASSVIINTASCLESSSLSFLQQQQLQIPVYPIGP----LHMVASA 250
Query: 200 PKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSE 259
P + IEWL+ Q + SV+YI+ GS + +I E+A+ L ASN FLW+ R
Sbjct: 251 PTSLLEENKSCIEWLNKQKVNSVIYISMGSIALMEINEIMEVASGLAASNQHFLWVIRPG 310
Query: 260 ASRLKEICGAH---------HMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGV 310
+ E + G I++W Q VLSHP++GGFWSHCGWNST ES+ GV
Sbjct: 311 SIPGSEWIESMPEEFSKMVLDRGYIVKWAPQKEVLSHPAVGGFWSHCGWNSTLESIGQGV 370
Query: 311 PFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRD 370
P + P DQ N++ + WK+G +V+ ++ R + ++ VK + +D +GE +
Sbjct: 371 PMICRPFSGDQKVNARYLECVWKIGIQVEGELDRGVV---ERAVKRL--MVDEEGE---E 422
Query: 371 IRERSKKLQKICLNSIANGGSAHTDFNAFI 400
+R+R+ L++ S+ +GGS+H F+
Sbjct: 423 MRKRAFSLKEQLRASVKSGGSSHNSLEEFV 452
>At1g24100 putative indole-3-acetate beta-glucosyltransferase
Length = 460
Score = 163 bits (412), Expect = 2e-40
Identities = 132/465 (28%), Positives = 213/465 (45%), Gaps = 83/465 (17%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVS---FVVTEEWLSFISSEPKPDNISFRSGSNV 57
+P+PV+G++NPM+ K LVS N + ++ + + +S EP D F
Sbjct: 15 LPYPVQGHLNPMVQFAKRLVSKNVKVTIATTTYTASSITTPSLSVEPISDGFDF------ 68
Query: 58 IPSELICGRDHPAFMEDVMTK-MEAPFEELLDLL-------DHPPSIIVYDTLLYWAVVV 109
IP + P F D ++ + E L LL D P ++YD+ L W + V
Sbjct: 69 IPIGI------PGFSVDTYSESFKLNGSETLTLLIEKFKSTDSPIDCLIYDSFLPWGLEV 122
Query: 110 ANRRNIPAALFW------------------PMPASIFSV-----------------FLHQ 134
A + AA F+ P+PA S F+ +
Sbjct: 123 ARSMELSAASFFTNNLTVCSVLRKFSNGDFPLPADPNSAPFRIRGLPSLSYDELPSFVGR 182
Query: 135 HIFEQNGHYPV---KYPGFE---WIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGP 188
H H V ++P E W+ + + E+ D +K+ L IGP
Sbjct: 183 HWLTHPEHGRVLLNQFPNHENADWLFVNGFEGLEETQDCENGESDAMKATL------IGP 236
Query: 189 TIPKFSLIKNDPKPSNTNHSYY-------IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEI 241
IP L + S +EWL+++ SV +++ GSF + Q+ E+
Sbjct: 237 MIPSAYLDDRMEDDKDYGASLLKPISKECMEWLETKQAQSVAFVSFGSFGILFEKQLAEV 296
Query: 242 AAALCASNVRFLWIAR-SEASRLKE--ICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCG 298
A AL S++ FLW+ + + ++L E + L++ WC+QL VL+H SIG F +HCG
Sbjct: 297 AIALQESDLNFLWVIKEAHIAKLPEGFVESTKDRALLVSWCNQLEVLAHESIGCFLTHCG 356
Query: 299 WNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVH 358
WNST E L GVP + +P + DQ ++K + E WKVG R KE+ + +VK +++V+ +
Sbjct: 357 WNSTLEGLSLGVPMVGVPQWSDQMNDAKFVEEVWKVGYRAKEEA-GEVIVKSEELVRCLK 415
Query: 359 EFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
M +GE + IRE SKK + + + +++ GGS+ N FI +
Sbjct: 416 GVM--EGESSVKIRESSKKWKDLAVKAMSEGGSSDRSINEFIESL 458
>At1g05530 hypothetical protein
Length = 455
Score = 161 bits (407), Expect = 7e-40
Identities = 122/451 (27%), Positives = 210/451 (46%), Gaps = 57/451 (12%)
Query: 3 FPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWL--SFISSEPKPDNISFRSGSNVIPS 60
FP +G++NP + + L+ + V+F + S I + +N+SF + S+
Sbjct: 11 FPAQGHVNPSLRFARRLIKT-TGARVTFATCLSVIHRSMIPNHNNVENLSFLTFSDGFDD 69
Query: 61 ELICGRDHP----AFMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWAVVVANRRNIP 116
+I D E K + F E D P S ++Y L W VA R ++P
Sbjct: 70 GVISNTDDVQNRLVHFERNGDKALSDFIEANQNGDSPVSCLIYTILPNWVPKVARRFHLP 129
Query: 117 AALFWPMPASIFSVFLHQHIFEQNGHYPVKYPGFEWIHKAQYLLFSSIYELESQAI---- 172
+ W PA F ++ + + P E I L S +QA+
Sbjct: 130 SVHLWIQPAFAFDIYYNYSTGNNSVFEFPNLPSLE-IRDLPSFLSPSNTNKAAQAVYQEL 188
Query: 173 -DVLKSK--------------------LP-LPIYTIGPTIPKFSLIKNDP-KPSNTNH-- 207
D LK + +P + + +GP +P ++ K + +H
Sbjct: 189 MDFLKEESNPKILVNTFDSLEPEFLTAIPNIEMVAVGPLLPAEIFTGSESGKDLSRDHQS 248
Query: 208 SYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASR----- 262
S Y WLDS+ SV+Y++ G+ +S QI+E+A AL FLW+ + +R
Sbjct: 249 SSYTLWLDSKTESSVIYVSFGTMVELSKKQIEELARALIEGGRPFLWVITDKLNREAKIE 308
Query: 263 ------LKEICGAHH----MGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPF 312
+++I G H +G+I+ WC Q+ VL H +IG F +HCGW+S+ ESLV GVP
Sbjct: 309 GEEETEIEKIAGFRHELEEVGMIVSWCSQIEVLRHRAIGCFLTHCGWSSSLESLVLGVPV 368
Query: 313 LTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIR 372
+ P++ DQP N+K++ E WK G RV+E+ + LV++ +I++ + M+ + ++R
Sbjct: 369 VAFPMWSDQPANAKLLEEIWKTGVRVREN--SEGLVERGEIMRCLEAVMEAK---SVELR 423
Query: 373 ERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
E ++K +++ + GGS+ + AF+ +
Sbjct: 424 ENAEKWKRLATEAGREGGSSDKNVEAFVKSL 454
>At2g23260 putative glucosyltransferase
Length = 456
Score = 160 bits (404), Expect = 2e-39
Identities = 119/452 (26%), Positives = 208/452 (45%), Gaps = 67/452 (14%)
Query: 4 PVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKP----DNISFRSGSNVIP 59
P +G+INPM+ L K L ++ N+H++ E +S+ KP D + F G +P
Sbjct: 17 PFQGHINPMLKLAKHLSLSSKNLHINLATIESARDLLSTVEKPRYPVDLVFFSDG---LP 73
Query: 60 SELICGRDHPAFMEDVMTKMEAPFEELLDLL--DHPPSIIVYDTLLYWAVVVANRRNIPA 117
E P E ++ + L + + S I+ W VA NI
Sbjct: 74 KE------DPKAPETLLKSLNKVGAMNLSKIIEEKRYSCIISSPFTPWVPAVAASHNISC 127
Query: 118 ALFWPMPASIFSVFLHQHI----------FEQNGHYPV-------KYPGF---------- 150
A+ W +SV+ ++ Q P P F
Sbjct: 128 AILWIQACGAYSVYYRYYMKTNSFPDLEDLNQTVELPALPLLEVRDLPSFMLPSGGAHFY 187
Query: 151 -------EWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPS 203
+ + +++L +S YELES+ I+ + P + IGP + F L + +
Sbjct: 188 NLMAEFADCLRYVKWVLVNSFYELESEIIESMADLKP--VIPIGPLVSPFLLGDGEEETL 245
Query: 204 NTNHSYY-------IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIA 256
+ + + +EWLD Q SV+YI+ GS Q++ IA AL + FLW+
Sbjct: 246 DGKNLDFCKSDDCCMEWLDKQARSSVVYISFGSMLETLENQVETIAKALKNRGLPFLWVI 305
Query: 257 RSEASRLKEICGAHHM-----GLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVP 311
R + + + + M G+++EW Q ++LSH +I F +HCGWNST E++VAGVP
Sbjct: 306 RPK-EKAQNVAVLQEMVKEGQGVVLEWSPQEKILSHEAISCFVTHCGWNSTMETVVAGVP 364
Query: 312 FLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDI 371
+ P + DQP +++++V+ + +G R++ D D +K +++ + + +G DI
Sbjct: 365 VVAYPSWTDQPIDARLLVDVFGIGVRMRND-SVDGELKVEEVERCIEAV--TEGPAAVDI 421
Query: 372 RERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
R R+ +L+++ ++A GGS+ + + FISD+
Sbjct: 422 RRRAAELKRVARLALAPGGSSTRNLDLFISDI 453
>At2g31790 putative glucosyltransferase
Length = 457
Score = 159 bits (403), Expect = 2e-39
Identities = 125/454 (27%), Positives = 209/454 (45%), Gaps = 66/454 (14%)
Query: 2 PFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPSE 61
P+P++G+INPM+ L K L + + + S + + G P E
Sbjct: 13 PYPLQGHINPMIQLAKRLSKKGITSTLIIASKDHREPYTSDDYSITVHTIHDG--FFPHE 70
Query: 62 LICGRDHPAFME-----DVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWAVVVANRRNIP 116
H F++ + ++ F L D+PP ++YD + +A+ +A ++
Sbjct: 71 ----HPHAKFVDLDRFHNSTSRSLTDFISSAKLSDNPPKALIYDPFMPFALDIAKDLDLY 126
Query: 117 AALFWPMPASIFSVFLHQHIFEQNGHYPV---------KYPGFEWI----------HKAQ 157
++ P + ++ HI E PV +PGF + K
Sbjct: 127 VVAYFTQPW--LASLVYYHINEGTYDVPVDRHENPTLASFPGFPLLSQDDLPSFACEKGS 184
Query: 158 YLL--------FSSIYELES---QAIDVLKSKL------PLPIYTIGPTIPKFSLIKNDP 200
Y L FS++ + + D L+ K+ P+ IGP +P L P
Sbjct: 185 YPLLHEFVVRQFSNLLQADCILCNTFDQLEPKVVKWMNDQWPVKNIGPVVPSKFLDNRLP 244
Query: 201 KPSNTN--------HSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRF 252
+ + ++WL ++P SV+Y+A G+ ++S Q+ EIA A+ + F
Sbjct: 245 EDKDYELENSKTEPDESVLKWLGNRPAKSVVYVAFGTLVALSEKQMKEIAMAISQTGYHF 304
Query: 253 LWIAR-SEASRLK----EICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLV 307
LW R SE S+L E GL+ +W QL VL+H SIG F SHCGWNST E+L
Sbjct: 305 LWSVRESERSKLPSGFIEEAEEKDSGLVAKWVPQLEVLAHESIGCFVSHCGWNSTLEALC 364
Query: 308 AGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGEL 367
GVP + +P + DQP N+K + + WK+G RV+ D + L K++I + + E M +GE
Sbjct: 365 LGVPMVGVPQWTDQPTNAKFIEDVWKIGVRVRTD--GEGLSSKEEIARCIVEVM--EGER 420
Query: 368 TRDIRERSKKLQKICLNSIANGGSAHTDFNAFIS 401
++IR+ +KL+ + +I+ GGS+ + F++
Sbjct: 421 GKEIRKNVEKLKVLAREAISEGGSSDKKIDEFVA 454
>At2g23250 putative glucosyltransferase
Length = 438
Score = 154 bits (389), Expect = 8e-38
Identities = 118/442 (26%), Positives = 209/442 (46%), Gaps = 57/442 (12%)
Query: 6 RGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISS---EP-KPDNISFRSGSNVIPSE 61
+G++NPM+ K L +N+H + TE+ +SS EP +P +++F S +P +
Sbjct: 7 QGHLNPMLKFAKHLA--RTNLHFTLATTEQARDLLSSTADEPHRPVDLAFFSDG--LPKD 62
Query: 62 LICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWAVVVANRRNIPAALFW 121
RD + + ++++ + I+ W VA NIP A+ W
Sbjct: 63 --DPRDPDTLAKSLKKDGAKNLSKIIE--EKRFDCIISVPFTPWVPAVAAAHNIPCAILW 118
Query: 122 PMPASIFSVFLHQHIFE------QNGHYPVKYPGF------------------------- 150
FSV+ ++ ++ + V+ P
Sbjct: 119 IQACGAFSVYYRYYMKTNPFPDLEDLNQTVELPALPLLEVRDLPSLMLPSQGANVNTLMA 178
Query: 151 ---EWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSNT-- 205
+ + +++L +S YELES+ I+ + P I IGP + F L ++ K +
Sbjct: 179 EFADCLKDVKWVLVNSFYELESEIIESMSDLKP--IIPIGPLVSPFLLGNDEEKTLDMWK 236
Query: 206 NHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIAR----SEAS 261
Y +EWLD Q SV+YI+ GS Q++ IA AL V FLW+ R E
Sbjct: 237 VDDYCMEWLDKQARSSVVYISFGSILKSLENQVETIATALKNRGVPFLWVIRPKEKGENV 296
Query: 262 RLKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQ 321
++ + G++ EW Q ++LSH +I F +HCGWNST E++V GVP + P +IDQ
Sbjct: 297 QVLQEMVKEGKGVVTEWGQQEKILSHMAISCFITHCGWNSTIETVVTGVPVVAYPTWIDQ 356
Query: 322 PFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKI 381
P +++++V+ + +G R+K D D +K ++ + + +G D+R R+ +L+
Sbjct: 357 PLDARLLVDVFGIGVRMKNDA-IDGELKVAEVERCIEAV--TEGPAAADMRRRATELKHA 413
Query: 382 CLNSIANGGSAHTDFNAFISDV 403
++++ GGS+ + ++FISD+
Sbjct: 414 ARSAMSPGGSSAQNLDSFISDI 435
>At5g38040 glucosyltransferase-like protein
Length = 449
Score = 145 bits (367), Expect = 3e-35
Identities = 118/454 (25%), Positives = 208/454 (44%), Gaps = 75/454 (16%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHV-----SFVVTEEWLSFISSEPKPDNISFRSGS 55
+P P +G+I PM+ L K L S +I V +++ LS P+N+
Sbjct: 14 VPVPAQGHITPMIQLAKALHSKGFSITVVQTKFNYLNPSNDLSDFQFVTIPENLPVSDLK 73
Query: 56 NVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLL----DHPPSIIVYDTLLYWAVVVAN 111
N+ P F+ + + F++LL L + + ++YD +Y+ V
Sbjct: 74 NLGPGR---------FLIKLANECYVSFKDLLGQLLVNEEEEIACVIYDEFMYFVEVAVK 124
Query: 112 RRNIPAALFWPMPASIFSV-FLHQHIFEQNG-----------------HYPVKYPG---- 149
+ + A+ F F+ ++ ++G YP++Y
Sbjct: 125 EFKLRNVILSTTSATAFVCRFVMCELYAKDGLAQLKEGGEREVELVPELYPIRYKDLPSS 184
Query: 150 -FEWIHKAQYLLFSSIYE-------------LESQAIDVLKSKLPLPIYTIGPTIPKFSL 195
F + + L ++ Y+ LE +++ L+ +L +P+Y+IGP +
Sbjct: 185 VFASVESSVELFKNTCYKGTASSVIINTVRCLEMSSLEWLQQELEIPVYSIGPL---HMV 241
Query: 196 IKNDPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWI 255
+ P + IEWL+ Q SV+YI+ GSF + + ++ E+A +SN FLW+
Sbjct: 242 VSAPPTSLLEENESCIEWLNKQKPSSVIYISLGSFTLMETKEMLEMAYGFVSSNQHFLWV 301
Query: 256 AR---------SEASRLKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESL 306
R SE LK++ G I++W Q +VL+H ++G FWSHCGWNST ESL
Sbjct: 302 IRPGSICGSEISEEELLKKMV-ITDRGYIVKWAPQKQVLAHSAVGAFWSHCGWNSTLESL 360
Query: 307 VAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGE 366
GVP + P DQ N++ + WKVG +V+ +++R + ++ VK + +D +GE
Sbjct: 361 GEGVPLICRPFTTDQKGNARYLECVWKVGIQVEGELERGAI---ERAVKRL--MVDEEGE 415
Query: 367 LTRDIRERSKKLQKICLNSIANGGSAHTDFNAFI 400
+++ R+ L++ S+ GS+H + FI
Sbjct: 416 ---EMKRRALSLKEKLKASVLAQGSSHKSLDDFI 446
>At5g14860 glucosyltransferase -like protein
Length = 492
Score = 142 bits (359), Expect = 3e-34
Identities = 105/362 (29%), Positives = 176/362 (48%), Gaps = 65/362 (17%)
Query: 95 SIIVYDTLLYWAVVVANRRNIPAALFWPM----PASIFSVFLHQHIFE----QNGHYPVK 146
S +V D L+W A + IP F+ M A ++ +H+ + ++ PV
Sbjct: 126 SFMVSDGFLWWTSESAAKFEIPRLAFYGMNSYASAMCSAISVHELFTKPESVKSDTEPVT 185
Query: 147 YPGFEWI-------------------------------HKAQYLLFSSIYELESQAID-- 173
P F WI K++ ++ +S YELES +D
Sbjct: 186 VPDFPWICVKKCEFDPVLTEPDQSDPAFELLIDHLMSTKKSRGVIVNSFYELESTFVDYR 245
Query: 174 VLKSKLPLPIYTIGPTIPKFSLIKNDPKPSNTNHSYYIEWLDS--QPIGSVLYIAQGSFF 231
+ + P P + +GP + N PKP + + I WLD + V+Y+A G+
Sbjct: 246 LRDNDEPKP-WCVGPLC-----LVNPPKPESDKPDW-IHWLDRKLEERCPVMYVAFGTQA 298
Query: 232 SVSSAQIDEIAAALCASNVRFLWIARSEASRLKEICGA--------HHMGLIMEWCDQLR 283
+S+ Q+ EIA L S V FLW+ R + L+E+ G H ++ +W DQ
Sbjct: 299 EISNEQLKEIALGLEDSKVNFLWVTRKD---LEEVTGGLGFEKRVKEHGMIVRDWVDQWE 355
Query: 284 VLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVK-EDV 342
+LSH S+ GF SHCGWNS +ES+ AGVP L P+ +QP N+K++VE+ K+G R++ EDV
Sbjct: 356 ILSHKSVKGFLSHCGWNSAQESICAGVPLLAWPMMAEQPLNAKLVVEELKIGVRIETEDV 415
Query: 343 KRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANG-GSAHTDFNAFIS 401
V ++++ + V + M +GE+ + + K+ K+ ++A G GS+ ++ +
Sbjct: 416 SVKGFVTREELSRKVKQLM--EGEMGKTTMKNVKEYAKMAKKAMAQGTGSSWKSLDSLLE 473
Query: 402 DV 403
++
Sbjct: 474 EL 475
>At3g21800 putative UDP-glucose glucosyltransferase
Length = 480
Score = 140 bits (352), Expect = 2e-33
Identities = 121/470 (25%), Positives = 204/470 (42%), Gaps = 70/470 (14%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+PFP+ G++ + KLLV + + +S ++ S + + ++ +
Sbjct: 9 VPFPILGHLKSTAEMAKLLVEQETRLSISIIILPLLSGDDVSASAYISALSAASNDRLHY 68
Query: 61 ELICGRDHPAF---MEDVMTKMEAPFEELLDLLDHPPSI-----IVYDTLLYWAVVVANR 112
E+I D P +++ + ++ +L+D P +V D + VAN
Sbjct: 69 EVISDGDQPTVGLHVDNHIPMVKRTVAKLVDDYSRRPDSPRLAGLVVDMFCISVIDVANE 128
Query: 113 RNIPAALFWPMPASIFSVFLH-QHIFEQNGH----------------------YPVKYPG 149
++P LF+ I ++ LH Q +F++ + YPVK
Sbjct: 129 VSVPCYLFYTSNVGILALGLHIQMLFDKKEYSVSETDFEDSEVVLDVPSLTCPYPVKCLP 188
Query: 150 F-----EWI----------HKAQYLLFSSIYELESQAIDVLKSKLPLP-IYTIGPTIPKF 193
+ EW+ + + +L ++ ELE A++ L S P Y +GP +
Sbjct: 189 YGLATKEWLPMYLNQGRRFREMKGILVNTFAELEPYALESLHSSGDTPRAYPVGPLL--- 245
Query: 194 SLIKNDPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFL 253
L + + S + WLD QP SV+++ GS + Q E+A AL S RFL
Sbjct: 246 HLENHVDGSKDEKGSDILRWLDEQPPKSVVFLCFGSIGGFNEEQAREMAIALERSGHRFL 305
Query: 254 WIAR-----------SEASRLKEICG------AHHMGLIMEWCDQLRVLSHPSIGGFWSH 296
W R E L+EI G ++ W Q+ VL+ P+IGGF +H
Sbjct: 306 WSLRRASRDIDKELPGEFKNLEEILPEGFFDRTKDKGKVIGWAPQVAVLAKPAIGGFVTH 365
Query: 297 CGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKL 356
CGWNS ESL GVP P+Y +Q FN+ +MVE+ + ++++ + D LV ++
Sbjct: 366 CGWNSILESLWFGVPIAPWPLYAEQKFNAFVMVEELGLAVKIRKYWRGDQLVGTATVIVT 425
Query: 357 VHEF---MDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
E + E D+R R K++ K C ++ +GGS+ + FI DV
Sbjct: 426 AEEIERGIRCLMEQDSDVRNRVKEMSKKCHMALKDGGSSQSALKLFIQDV 475
>At2g29740 putative flavonol 3-O-glucosyltransferase
Length = 474
Score = 136 bits (343), Expect = 2e-32
Identities = 128/475 (26%), Positives = 206/475 (42%), Gaps = 90/475 (18%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNN-SNIH--------VSFVVTEEWLSFISS----EPKPD 47
+PFP+ G+I + L K L+S+ S IH + F+ + ++F+ S E +
Sbjct: 12 IPFPIPGHILATIELAKRLISHQPSRIHTITILHWSLPFLPQSDTIAFLKSLIETESRIR 71
Query: 48 NISFRSGSNVIPSELICGRDHPAFMEDV---MTKMEAPFEELLDLLDHPPSI----IVYD 100
I+ N P EL +E V + + LL D S+ +V D
Sbjct: 72 LITLPDVQNPPPMELFVKASESYILEYVKKMVPLVRNALSTLLSSRDESDSVHVAGLVLD 131
Query: 101 TLLYWAVVVANRRNIPAALFWPMPASIFSVFLHQHIFEQN-----------GHYPVKYPG 149
+ V N N+P+ +F AS + +++ E+N + PG
Sbjct: 132 FFCVPLIDVGNEFNLPSYIFLTCSASFLGMM--KYLLERNRETKPELNRSSDEETISVPG 189
Query: 150 F--------------------EWIH------KAQYLLFSSIYELESQAIDVLKSKLP--L 181
F W+ +A+ +L +S LE A D +
Sbjct: 190 FVNSVPVKVLPPGLFTTESYEAWVEMAERFPEAKGILVNSFESLERNAFDYFDRRPDNYP 249
Query: 182 PIYTIGPTIPKFSLIKND-PKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDE 240
P+Y IGP L ND P + ++WLD QP SV+++ GS S++++QI E
Sbjct: 250 PVYPIGPI-----LCSNDRPNLDLSERDRILKWLDDQPESSVVFLCFGSLKSLAASQIKE 304
Query: 241 IAAALCASNVRFLWIARSEASR------------LKEICGAHHMGLIMEWCDQLRVLSHP 288
IA AL +RFLW R++ + + G +GL+ W Q+ +L+H
Sbjct: 305 IAQALELVGIRFLWSIRTDPKEYASPNEILPDGFMNRVMG---LGLVCGWAPQVEILAHK 361
Query: 289 SIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRD--T 346
+IGGF SHCGWNS ESL GVP T P+Y +Q N+ +V++ + ++ D +
Sbjct: 362 AIGGFVSHCGWNSILESLRFGVPIATWPMYAEQQLNAFTIVKELGLALEMRLDYVSEYGE 421
Query: 347 LVKKDKIVKLVHEFMDLDGELTRDI-RERSKKLQKICLNSIANGGSAHTDFNAFI 400
+VK D+I V M DGE D+ R + K++ + ++ +GGS+ FI
Sbjct: 422 IVKADEIAGAVRSLM--DGE---DVPRRKLKEIAEAGKEAVMDGGSSFVAVKRFI 471
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,680,880
Number of Sequences: 26719
Number of extensions: 427666
Number of successful extensions: 1414
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 118
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 974
Number of HSP's gapped (non-prelim): 199
length of query: 406
length of database: 11,318,596
effective HSP length: 102
effective length of query: 304
effective length of database: 8,593,258
effective search space: 2612350432
effective search space used: 2612350432
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC141923.7


